The Conserved YPX3L Motif in the BK Polyomavirus VP1 Protein Is Important for Viral Particle Assembly but Not for Its Secretion into Extracellular Vesicles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Cultures, Antibodies and Plasmids
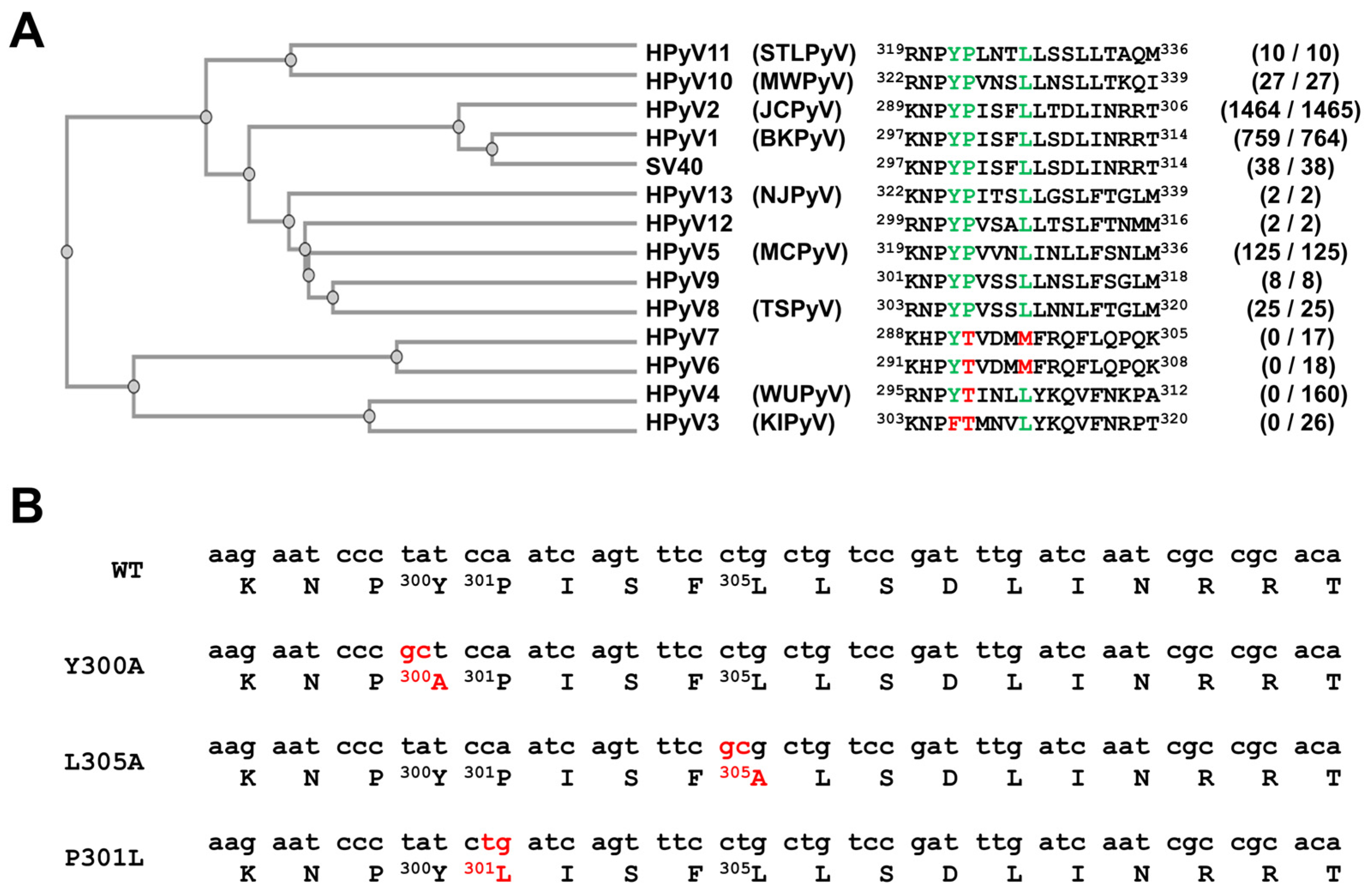
2.2. Site-Directed Mutagenesis
2.3. Pseudovirion Production and Infection
2.4. Reporter Plasmid Quantification by Real-Time PCR
2.5. Buoyant Density Iodixanol Gradient Ultracentrifugation
2.6. Neuraminidase Treatment
2.7. Disruption of Pseudovirions into Pentamers
2.8. Western Blotting
2.9. Electron Microscopy
2.10. Statistical Analyses
3. Results
3.1. A YPX3L Motif Is Conserved in the VP1 Protein Sequence of Not Only the BK Polyomavirus but Also Most Other Human Polyomaviruses
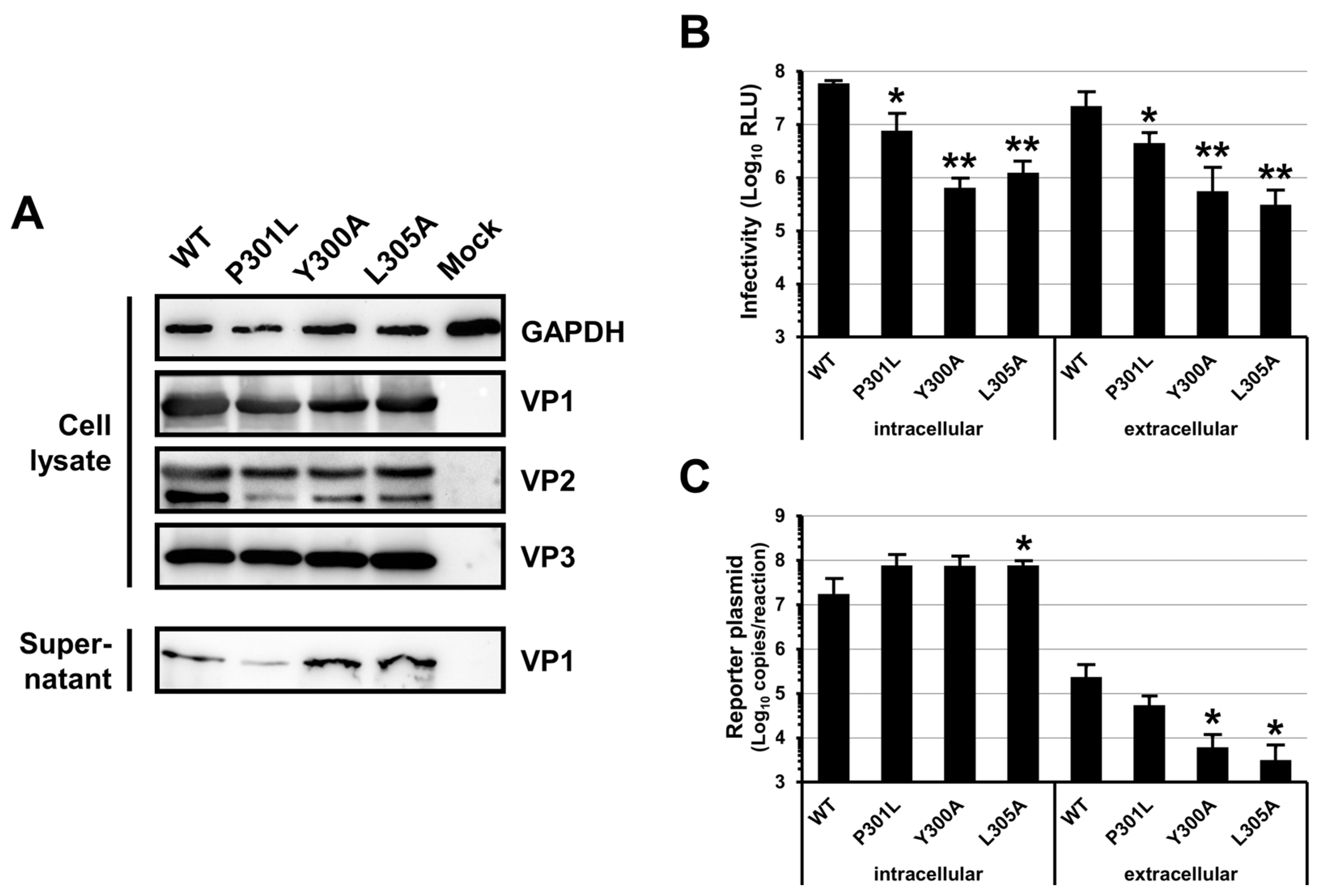
3.2. Mutations of the BKPyV VP1 YPX3L Motif Impair the Assembly of Viral Particles
3.3. Mutations of the BKPyV VP1 YPX3L Motif Do Not Impair the Association of Infectious Particles with EVs
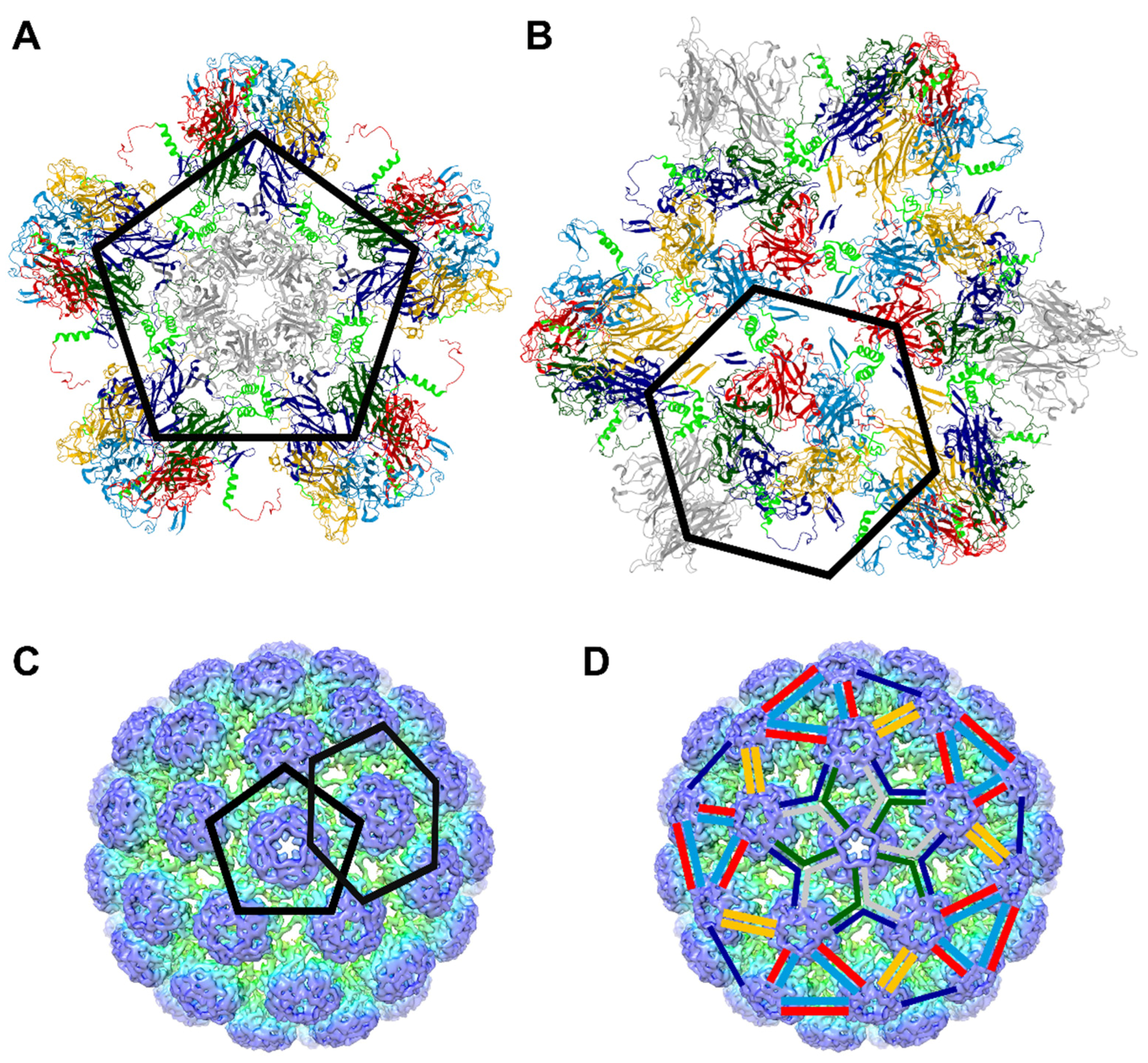
3.4. The P301L, Y300A and L305A Mutations in the BKPyV VP1 Protein May Impair the Assembly of VP1 Pentamers into Capsid
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rinaldo, C.H.; Tylden, G.D.; Sharma, B.N. The human polyomavirus BK (BKPyV): Virological background and clinical implications. Apmis 2013, 121, 728–745. [Google Scholar] [CrossRef] [PubMed]
- Helle, F.; Brochot, E.; Handala, L.; Martin, E.; Castelain, S.; Francois, C.; Duverlie, G. Biology of the BKPyV: An Update. Viruses 2017, 9, 327. [Google Scholar] [CrossRef] [PubMed]
- Kean, J.M.; Rao, S.; Wang, M.; Garcea, R.L. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009, 5, e1000363. [Google Scholar] [CrossRef] [PubMed]
- Chong, S.; Antoni, M.; Macdonald, A.; Reeves, M.; Harber, M.; Magee, C.N. BK virus: Current understanding of pathogenicity and clinical disease in transplantation. Rev. Med. Virol. 2019, 29, e2044. [Google Scholar] [CrossRef] [PubMed]
- Demey, B.; Tinez, C.; Francois, C.; Helle, F.; Choukroun, G.; Duverlie, G.; Castelain, S.; Brochot, E. Risk factors for BK virus viremia and nephropathy after kidney transplantation: A systematic review. J. Clin. Virol. 2018, 109, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Hurdiss, D.L.; Frank, M.; Snowden, J.S.; Macdonald, A.; Ranson, N.A. The Structure of an Infectious Human Polyomavirus and Its Interactions with Cellular Receptors. Structure 2018, 26, 839–847.e3. [Google Scholar] [CrossRef] [PubMed]
- Hurdiss, D.L.; Morgan, E.L.; Thompson, R.F.; Prescott, E.L.; Panou, M.M.; Macdonald, A.; Ranson, N.A. New Structural Insights into the Genome and Minor Capsid Proteins of BK Polyomavirus using Cryo-Electron Microscopy. Structure 2016, 24, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.J.; Di Mayorca, G. Virion polypeptide composition of the human papovavirus BK: Comparison with simian virus 40 and polyoma virus. J. Virol. 1975, 15, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Handala, L.; Blanchard, E.; Raynal, P.I.; Roingeard, P.; Morel, V.; Descamps, V.; Castelain, S.; Francois, C.; Duverlie, G.; Brochot, E.; et al. BK polyomavirus hijacks extracellular vesicles for en bloc transmission. J. Virol. 2020, 94, e01834-19. [Google Scholar] [CrossRef]
- Morris-Love, J.; Gee, G.V.; O’Hara, B.A.; Assetta, B.; Atkinson, A.L.; Dugan, A.S.; Haley, S.A.; Atwood, W.J. JC polyomavirus uses extracellular vesicles to infect target cells. mBio 2019, 10, e00379-19. [Google Scholar] [CrossRef]
- O’Hara, B.A.; Morris-Love, J.; Gee, G.V.; Haley, S.A.; Atwood, W.J. JC Virus infected choroid plexus epithelial cells produce extracellular vesicles that infect glial cells independently of the virus attachment receptor. PLoS Pathog. 2020, 16, e1008371. [Google Scholar] [CrossRef] [PubMed]
- Couch, Y.; Buzas, E.I.; Di Vizio, D.; Gho, Y.S.; Harrison, P.; Hill, A.F.; Lotvall, J.; Raposo, G.; Stahl, P.D.; Thery, C.; et al. A brief history of nearly EV-erything—The rise and rise of extracellular vesicles. J. Extracell. Vesicles 2021, 10, e12144. [Google Scholar] [CrossRef]
- Tkach, M.; Thery, C. Communication by extracellular vesicles: Where we are and where we need to go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.H. ESCRTs are everywhere. EMBO J. 2015, 34, 2398–2407. [Google Scholar] [CrossRef] [PubMed]
- Schoneberg, J.; Lee, I.H.; Iwasa, J.H.; Hurley, J.H. Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 2017, 18, 5–17. [Google Scholar] [CrossRef]
- Vietri, M.; Radulovic, M.; Stenmark, H. The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 2020, 21, 25–42. [Google Scholar] [CrossRef]
- Bird, S.W.; Maynard, N.D.; Covert, M.W.; Kirkegaard, K. Nonlytic viral spread enhanced by autophagy components. Proc. Natl. Acad. Sci. USA 2014, 111, 13081–13086. [Google Scholar] [CrossRef]
- Chen, Y.H.; Du, W.; Hagemeijer, M.C.; Takvorian, P.M.; Pau, C.; Cali, A.; Brantner, C.A.; Stempinski, E.S.; Connelly, P.S.; Ma, H.C.; et al. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 2015, 160, 619–630. [Google Scholar] [CrossRef]
- Robinson, S.M.; Tsueng, G.; Sin, J.; Mangale, V.; Rahawi, S.; McIntyre, L.L.; Williams, W.; Kha, N.; Cruz, C.; Hancock, B.M.; et al. Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS Pathog. 2014, 10, e1004045. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Wu, J.; Fang, D.; Qiu, Y.; Zou, X.; Jia, X.; Yin, Y.; Shen, L.; Mao, L. Exosomes cloak the virion to transmit Enterovirus 71 non-lytically. Virulence 2020, 11, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Wu, J.; Shen, L.; Yang, J.; Chen, J.; Xu, H. Enterovirus 71 transmission by exosomes establishes a productive infection in human neuroblastoma cells. Virus Genes 2016, 52, 189–194. [Google Scholar] [CrossRef] [PubMed]
- van der Grein, S.G.; Defourny, K.A.Y.; Rabouw, H.H.; Galiveti, C.R.; Langereis, M.A.; Wauben, M.H.M.; Arkesteijn, G.J.A.; van Kuppeveld, F.J.M.; Nolte-’t Hoen, E.N.M. Picornavirus infection induces temporal release of multiple extracellular vesicle subsets that differ in molecular composition and infectious potential. PLoS Pathog. 2019, 15, e1007594. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Hensley, L.; McKnight, K.L.; Hu, F.; Madden, V.; Ping, L.; Jeong, S.H.; Walker, C.; Lanford, R.E.; Lemon, S.M. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 2013, 496, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, S.; Jirintai, S.; Takahashi, M.; Kobayashi, T.; Tanggis; Nishizawa, T.; Kouki, T.; Yashiro, T.; Okamoto, H. Hepatitis E virus egress depends on the exosomal pathway, with secretory exosomes derived from multivesicular bodies. J. Gen. Virol. 2014, 95, 2166–2175. [Google Scholar] [CrossRef] [PubMed]
- Santiana, M.; Ghosh, S.; Ho, B.A.; Rajasekaran, V.; Du, W.L.; Mutsafi, Y.; De Jesus-Diaz, D.A.; Sosnovtsev, S.V.; Levenson, E.A.; Parra, G.I.; et al. Vesicle-Cloaked Virus Clusters Are Optimal Units for Inter-organismal Viral Transmission. Cell Host Microbe 2018, 24, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Helle, F.; Handala, L.; Bentz, M.; Duverlie, G.; Brochot, E. Intercellular transmission of naked viruses through extracellular vesicles: Focus on polyomaviruses. Viruses 2020, 12, 1086. [Google Scholar] [CrossRef]
- Welker, L.; Paillart, J.C.; Bernacchi, S. Importance of Viral Late Domains in Budding and Release of Enveloped RNA Viruses. Viruses 2021, 13, 1559. [Google Scholar] [CrossRef]
- Votteler, J.; Sundquist, W.I. Virus budding and the ESCRT pathway. Cell Host Microbe 2013, 14, 232–241. [Google Scholar] [CrossRef]
- Ren, X.; Hurley, J.H. Proline-rich regions and motifs in trafficking: From ESCRT interaction to viral exploitation. Traffic 2011, 12, 1282–1290. [Google Scholar] [CrossRef]
- Gonzalez-Lopez, O.; Rivera-Serrano, E.E.; Hu, F.; Hensley, L.; McKnight, K.L.; Ren, J.; Stuart, D.I.; Fry, E.E.; Lemon, S.M. Redundant late domain functions of tandem VP2 YPX3L motifs in nonlytic cellular egress of quasi-enveloped hepatitis A virus. J. Virol. 2018, 92, e01308-18. [Google Scholar] [CrossRef]
- Jiang, W.; Ma, P.; Deng, L.; Liu, Z.; Wang, X.; Liu, X.; Long, G. Hepatitis A virus structural protein pX interacts with ALIX and promotes the secretion of virions and foreign proteins through exosome-like vesicles. J. Extracell. Vesicles 2020, 9, 1716513. [Google Scholar] [CrossRef]
- Nagashima, S.; Takahashi, M.; Jirintai; Tanaka, T.; Yamada, K.; Nishizawa, T.; Okamoto, H. A PSAP motif in the ORF3 protein of hepatitis E virus is necessary for virion release from infected cells. J. Gen. Virol. 2011, 92, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, S.; Takahashi, M.; Jirintai, S.; Tanaka, T.; Nishizawa, T.; Yasuda, J.; Okamoto, H. Tumour susceptibility gene 101 and the vacuolar protein sorting pathway are required for the release of hepatitis E virions. J. Gen. Virol. 2011, 92, 2838–2848. [Google Scholar] [CrossRef] [PubMed]
- Shirasaki, T.; Feng, H.; Duyvesteyn, H.M.E.; Fusco, W.G.; McKnight, K.L.; Xie, L.; Boyce, M.; Kumar, S.; Barouch-Bentov, R.; Gonzalez-Lopez, O.; et al. Nonlytic cellular release of hepatitis A virus requires dual capsid recruitment of the ESCRT-associated Bro1 domain proteins HD-PTP and ALIX. PLoS Pathog. 2022, 18, e1010543. [Google Scholar] [CrossRef]
- Brady, J.N.; Winston, V.D.; Consigli, R.A. Dissociation of polyoma virus by the chelation of calcium ions found associated with purified virions. J. Virol. 1977, 23, 717–724. [Google Scholar] [CrossRef]
- Brady, J.N.; Winston, V.D.; Consigli, R.A. Characterization of a DNA-protein complex and capsomere subunits derived from polyoma virus by treatment with ethyleneglycol-bis-N,N’-tetraacetic acid and dithiothreitol. J. Virol. 1978, 27, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.S.; Stehle, T.; Harrison, S.C. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J. 1998, 17, 3233–3240. [Google Scholar] [CrossRef]
- Stehle, T.; Gamblin, S.J.; Yan, Y.; Harrison, S.C. The structure of simian virus 40 refined at 3.1 A resolution. Structure 1996, 4, 165–182. [Google Scholar] [CrossRef]
- Ben-nun-Shaul, O.; Bronfeld, H.; Reshef, D.; Schueler-Furman, O.; Oppenheim, A. The SV40 capsid is stabilized by a conserved pentapeptide hinge of the major capsid protein VP1. J. Mol. Biol. 2009, 386, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodova, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koca, J.; Rose, A.S. Mol* Viewer: Modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 2021, 49, W431–W437. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M.; et al. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021, 49, D437–D451. [Google Scholar] [CrossRef] [PubMed]
- Bekker, G.J.; Yokochi, M.; Suzuki, H.; Ikegawa, Y.; Iwata, T.; Kudou, T.; Yura, K.; Fujiwara, T.; Kawabata, T.; Kurisu, G. Protein Data Bank Japan: Celebrating our 20th anniversary during a global pandemic as the Asian hub of three dimensional macromolecular structural data. Protein Sci. 2022, 31, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Dores, M.R.; Grimsey, N.J.; Mendez, F.; Trejo, J. ALIX Regulates the Ubiquitin-Independent Lysosomal Sorting of the P2Y1 Purinergic Receptor via a YPX3L Motif. PLoS ONE 2016, 11, e0157587. [Google Scholar] [CrossRef]
- Dores, M.R.; Chen, B.; Lin, H.; Soh, U.J.; Paing, M.M.; Montagne, W.A.; Meerloo, T.; Trejo, J. ALIX binds a YPX(3)L motif of the GPCR PAR1 and mediates ubiquitin-independent ESCRT-III/MVB sorting. J. Cell Biol. 2012, 197, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ren, J.; Gao, Q.; Hu, Z.; Sun, Y.; Li, X.; Rowlands, D.J.; Yin, W.; Wang, J.; Stuart, D.I.; et al. Hepatitis A virus and the origins of picornaviruses. Nature 2015, 517, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.C.; Wu, R. BK virus DNA: Complete nucleotide sequence of a human tumor virus. Science 1979, 206, 456–462. [Google Scholar] [CrossRef]
- Morris-Love, J.; O’Hara, B.A.; Gee, G.V.; Dugan, A.S.; O’Rourke, R.S.; Armstead, B.E.; Assetta, B.; Haley, S.A.; Atwood, W.J. Biogenesis of JC polyomavirus associated extracellular vesicles. J. Extracell. Biol. 2022, 1, e43. [Google Scholar] [CrossRef]
- Liddington, R.C.; Yan, Y.; Moulai, J.; Sahli, R.; Benjamin, T.L.; Harrison, S.C. Structure of simian virus 40 at 3.8-A resolution. Nature 1991, 354, 278–284. [Google Scholar] [CrossRef]
- Yokoyama, N.; Kawano, M.A.; Tsukamoto, H.; Enomoto, T.; Inoue, T.; Takahashi, R.U.; Nakanishi, A.; Imai, T.; Wada, T.; Handa, H. Mutational analysis of the carboxyl-terminal region of the SV40 major capsid protein VP1. J. Biochem. 2007, 141, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Osipov, E.M.; Munawar, A.H.; Beelen, S.; Fearon, D.; Douangamath, A.; Wild, C.; Weeks, S.D.; Van Aerschot, A.; von Delft, F.; Strelkov, S.V. Discovery of novel druggable pockets on polyomavirus VP1 through crystallographic fragment-based screening to develop capsid assembly inhibitors. RSC Chem. Biol. 2022, 3, 1013–1027. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bentz, M.; Collet, L.; Morel, V.; Descamps, V.; Blanchard, E.; Lambert, C.; Demey, B.; Brochot, E.; Helle, F. The Conserved YPX3L Motif in the BK Polyomavirus VP1 Protein Is Important for Viral Particle Assembly but Not for Its Secretion into Extracellular Vesicles. Viruses 2024, 16, 1124. https://doi.org/10.3390/v16071124
Bentz M, Collet L, Morel V, Descamps V, Blanchard E, Lambert C, Demey B, Brochot E, Helle F. The Conserved YPX3L Motif in the BK Polyomavirus VP1 Protein Is Important for Viral Particle Assembly but Not for Its Secretion into Extracellular Vesicles. Viruses. 2024; 16(7):1124. https://doi.org/10.3390/v16071124
Chicago/Turabian StyleBentz, Marine, Louison Collet, Virginie Morel, Véronique Descamps, Emmanuelle Blanchard, Caroline Lambert, Baptiste Demey, Etienne Brochot, and Francois Helle. 2024. "The Conserved YPX3L Motif in the BK Polyomavirus VP1 Protein Is Important for Viral Particle Assembly but Not for Its Secretion into Extracellular Vesicles" Viruses 16, no. 7: 1124. https://doi.org/10.3390/v16071124






