Chimeric Viruses Enable Study of Antibody Responses to Human Rotaviruses in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Virus Quantification by Fluorescent Focus Assay (FFA)
2.3. Rotavirus Infection of Mice
2.4. Quantification of Virus Shedding by RT-qPCR
2.5. ELISAs
2.6. Western Blot
2.7. Extracellular Neutralisation Assay
2.8. Intracellular Neutralisation Assay
2.9. Isolation of Single Cells from Lymph Nodes and Spleens
2.10. Flow Cytometry
2.11. ELISpot Assay
2.12. Statistics
3. Results
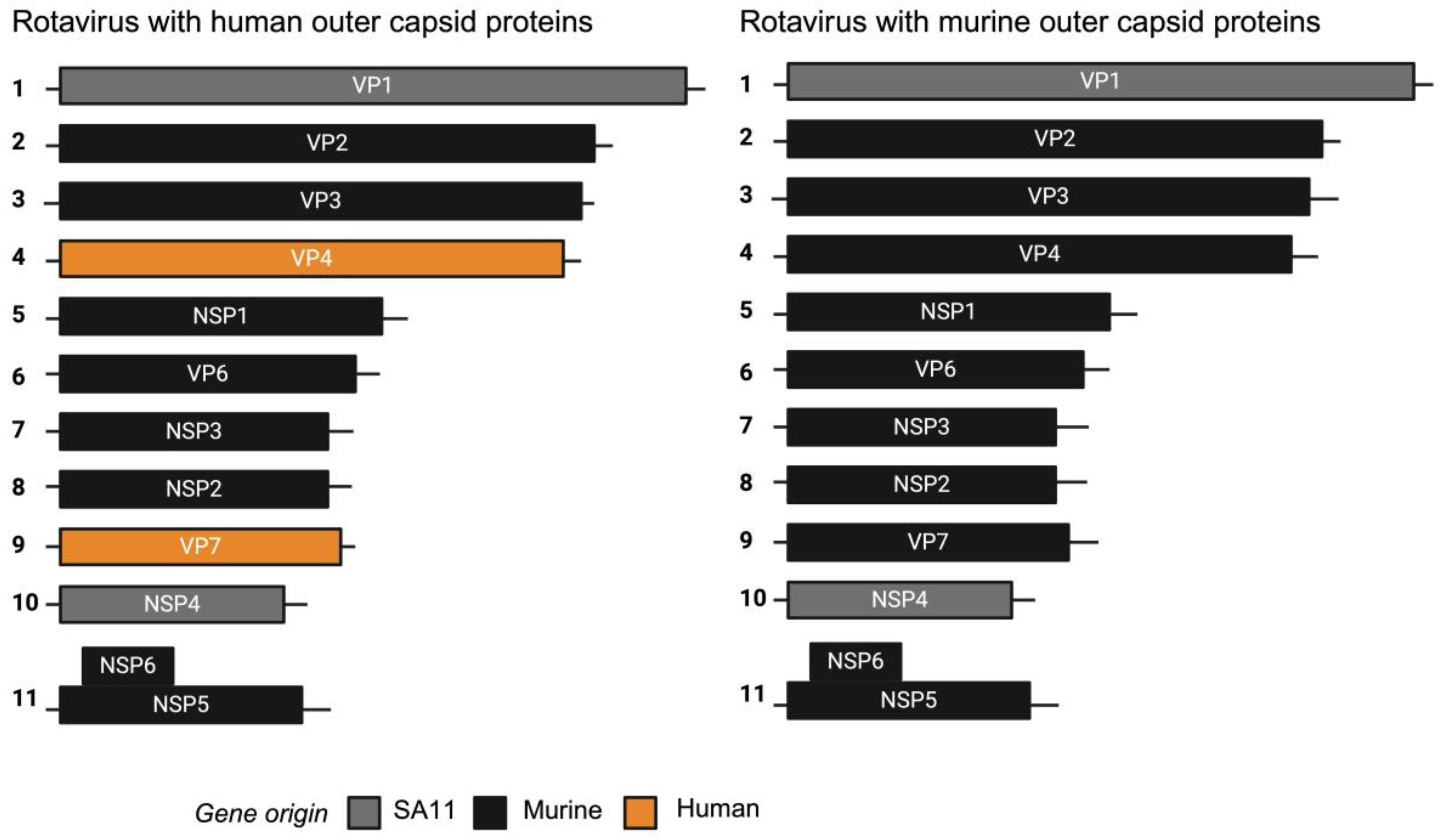
3.1. Construction and Characterisation of Chimeric Rotaviruses Expressing Murine or Human Strain Outer Capsid Proteins
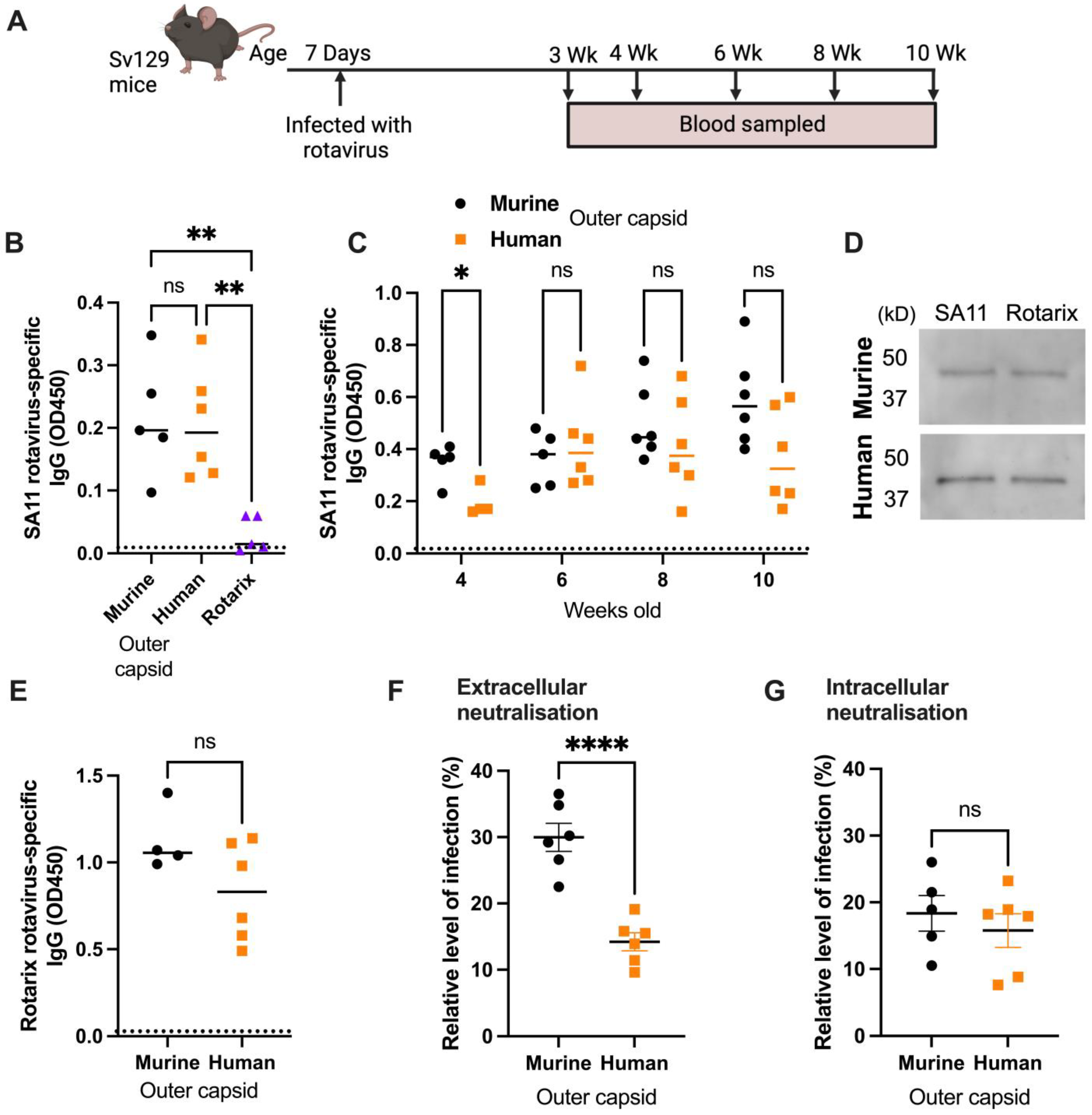
3.2. Chimeric Rotaviruses with Human Outer Capsid Proteins Replicate in Mice
3.3. Human Rotavirus-Specific Antibody Responses Are Generated in Mice Infected with Chimeric Rotaviruses
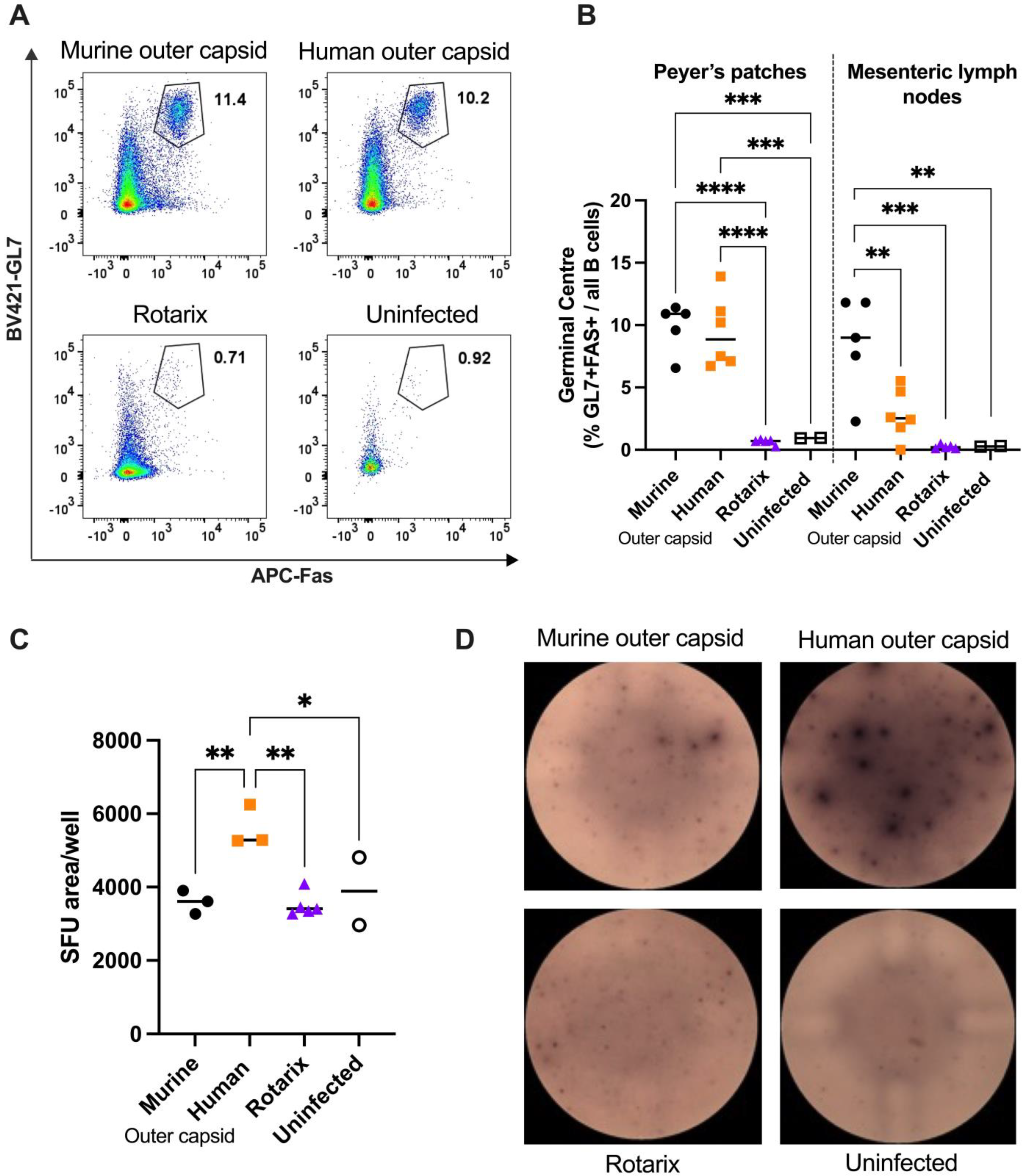
3.4. Chimeric Viruses Induced Human-Rotavirus-Specific B Cell Responses in Mice
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruiz-Palacios, G.M.; Pérez-Schael, I.; Velázquez, F.R.; Abate, H.; Breuer, T.; Clemens, S.C.; Cheuvart, B.; Espinoza, F.; Gillard, P.; Innis, B.L.; et al. Safety and Efficacy of an Attenuated Vaccine against Severe Rotavirus Gastroenteritis. N. Engl. J. Med. 2006, 354, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Vesikari, T.; Matson, D.O.; Dennehy, P.; Van Damme, P.; Santosham, M.; Rodriguez, Z.; Dallas, M.J.; Heyse, J.F.; Goveia, M.G.; Black, S.B.; et al. Safety and Efficacy of a Pentavalent Human–Bovine (WC3) Reassortant Rotavirus Vaccine. N. Engl. J. Med. 2006, 354, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Cates, J.E.; Tate, J.E.; Parashar, U. Rotavirus vaccines: Progress and new developments. Expert Opin. Biol. Ther. 2022, 22, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Desselberger, U. Differences of Rotavirus Vaccine Effectiveness by Country: Likely Causes and Contributing Factors. Pathogens 2017, 6, 65. [Google Scholar] [CrossRef] [PubMed]
- Caddy, S.; Papa, G.; Borodavka, A.; Desselberger, U. Rotavirus research: 2014–2020. Virus Res. 2021, 304, 198499. [Google Scholar] [CrossRef] [PubMed]
- Feng, N.; Yasukawa, L.L.; Sen, A.; Greenberg, H.B. Permissive Replication of Homologous Murine Rotavirus in the Mouse Intestine Is Primarily Regulated by VP4 and NSP1. J. Virol. 2013, 87, 8307–8316. [Google Scholar] [CrossRef]
- Cook, N. The zoonotic potential of rotavirus. J. Infect. 2004, 48, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Komoto, S.; Kawagishi, T.; Nouda, R.; Nagasawa, N.; Onishi, M.; Matsuura, Y.; Taniguchi, K.; Kobayashi, T. Entirely plasmid-based reverse genetics system for rotaviruses. Proc. Natl. Acad. Sci. USA 2017, 114, 2349–2354. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Kobayashi, T. Rotavirus reverse genetics systems: Development and application. Virus Res. 2021, 295, 198296. [Google Scholar] [CrossRef]
- Desselberger, U. Rotaviruses. Virus Res. 2014, 190, 75–96. [Google Scholar] [CrossRef]
- Sánchez-Tacuba, L.; Kawagishi, T.; Feng, N.; Jiang, B.; Ding, S.; Greenberg, H.B. The Role of the VP4 Attachment Protein in Rotavirus Host Range Restriction in an In Vivo Suckling Mouse Model. J. Virol. 2022, 96, e00550-22. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Tacuba, L.; Feng, N.; Meade, N.J.; Mellits, K.H.; Jaïs, P.H.; Yasukawa, L.L.; Resch, T.K.; Jiang, B.; López, S.; Ding, S.; et al. An Optimized Reverse Genetics System Suitable for Efficient Recovery of Simian, Human, and Murine-Like Rotaviruses. J. Virol. 2020, 94, e01294-20. [Google Scholar] [CrossRef] [PubMed]
- Caddy, S.L.; Vaysburd, M.; Wing, M.; Foss, S.; Andersen, J.T.; O’Connell, K.; Mayes, K.; Higginson, K.; Iturriza-Gómara, M.; Desselberger, U.; et al. Intracellular neutralisation of rotavirus by VP6-specific IgG. PLoS Pathog. 2020, 16, e1008732. [Google Scholar] [CrossRef] [PubMed]
- Ishida, S.; Feng, N.; Gilbert, J.M.; Tang, B.; Greenberg, H.B. Immune Responses to Individual Rotavirus Proteins following Heterologous and Homologous Rotavirus Infection in Mice. J. Infect. Dis. 1997, 175, 1317–1323. [Google Scholar] [CrossRef] [PubMed]
- Winkler, C.W.; Peterson, K.E. Using immunocompromised mice to identify mechanisms of Zika virus transmission and pathogenesis. Immunology 2018, 153, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Kutle, I.; Dittrich, A.; Wirth, D. Mouse Models for Human Herpesviruses. Pathogens 2023, 12, 953. [Google Scholar] [CrossRef] [PubMed]
- Vancott, J.L.; McNeal, M.M.; Choi, A.H.C.; Ward, R.L. The Role of Interferons in Rotavirus Infections and Protection. J. Interferon Cytokine Res. 2003, 23, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Otero, C.E.; Langel, S.N.; Blasi, M.; Permar, S.R. Maternal antibody interference contributes to reduced rotavirus vaccine efficacy in developing countries. PLoS Pathog. 2020, 16, e1009010. [Google Scholar] [CrossRef]
- Mwila, K.; Chilengi, R.; Simuyandi, M.; Permar, S.R.; Becker-Dreps, S. Contribution of Maternal Immunity to Decreased Rotavirus Vaccine Performance in Low- and Middle-Income Countries. Clin. Vaccine Immunol. 2017, 24. [Google Scholar] [CrossRef]
- Roier, S.; Mangala Prasad, V.; McNeal, M.M.; Lee, K.K.; Petsch, B.; Rauch, S. mRNA-based VP8* nanoparticle vaccines against rotavirus are highly immunogenic in rodents. NPJ Vaccines 2023, 8, 190. [Google Scholar] [CrossRef]
- Langel, S.; Steppe, J.; Chang, J.; Travieso, T.; Webster, H.; Otero, C.; Williamson, L.; Crowe, J.; Greenberg, H.; Wu, H.; et al. Protective Transfer: Maternal Passive Immunization with a Rotavirus-Neutralizing Dimeric IgA Protects against Rotavirus Disease in Suckling Neonates. bioRxiv 2021. [Google Scholar] [CrossRef]
- Zha, M.; Yang, J.; Zhou, L.; Wang, H.; Pan, X.; Deng, Z.; Yang, Y.; Li, W.; Wang, B.; Li, M. Preparation of mouse anti-human rotavirus VP7 monoclonal antibody and its protective effect on rotavirus infection. Exp. Ther. Med. 2019, 18, 1384–1390. [Google Scholar] [CrossRef] [PubMed]
- Crawford, S.E.; Ramani, S.; Tate, J.E.; Parashar, U.D.; Svensson, L.; Hagbom, M.; Franco, M.A.; Greenberg, H.B.; O’Ryan, M.; Kang, G.; et al. Rotavirus infection. Nat. Rev. Dis. Primers 2017, 3, 17083. [Google Scholar] [CrossRef]
- Svensson, L.; Sheshberadaran, H.; Vene, S.; Norrby, E.; Grandien, M.; Wadell, G. Serum Antibody Responses to Individual Viral Polypeptides in Human Rotavirus Infections. J. Gen. Virol. 1987, 68, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Johansen, K.; Granqvist, L.; Karlén, K.; Stintzing, G.; Uhnoo, I.; Svensson, L. Serum IgA immune response to individual rotavirus polypeptides in young children with rotavirus infection. Arch. Virol. 1994, 138, 247–259. [Google Scholar] [CrossRef]
- Burns, J.W.; Siadat-Pajouh, M.; Krishnaney, A.A.; Greenberg, H.B. Protective Effect of Rotavirus VP6-Specific IgA Monoclonal Antibodies That Lack Neutralizing Activity. Science 1996, 272, 104–107. [Google Scholar] [CrossRef]

| Reagent | Source | Identifier |
|---|---|---|
| Antibodies and Dyes | ||
| Alexa Fluor 488 Anti-Sheep IgG (Donkey) | Invitrogen, Carlsbad, CA, USA | CAT#A-11015 |
| Anti-Mouse IgG Biotin (Goat) | Mabtech, Cincinnati, OH, USA | CAT#3825-6 |
| Anti-Mouse IgG HRP (Goat) | Sigma Aldrich, St. Louis, MO, USA | CAT#A0168 |
| CD45 (BUV395 Rat Anti-Mouse) | BD Biosciences, San Jose, CA, USA | CAT#564279; Clone: 30-F11 |
| CD45R (PerCP/Cyanine 5.5 Anti-Mouse/Human CD45R/B220) | BioLegend, San Diego, CA, USA | CAT#103236; Clone: RA3-6B2 |
| CD95 (APC Anti-Mouse (Fas)) | BioLegend, San Diego, CA, USA | CAT#152603; Clone: SA367H8 |
| Clarity Western ECL Substrate | Bio-Rad, Hercules, CA, USA | CAT#170-5060S |
| Fc Block (TruStain FcX Anti-Mouse CD16/32) | BioLegend, San Diego, CA, USA | CAT#101320; Clone: 93 |
| Fixable Viability Dye eFluor 780 | Invitrogen, Carlsbad, CA, USA | CAT#65-0865 |
| GL7 Antigen (Pacific Blue Anti-Mouse/Human) | BioLegend, San Diego, CA, USA | CAT#144614; Clone: GL7 |
| Hoechst 33342 | Invitrogen, Carlsbad, CA, USA | CAT#H3570 |
| Rotavirus Polyclonal Antibody (Sheep) | Invitrogen, Carlsbad, CA, USA | CAT#PA1-85845 |
| Streptavidin–ALP | Mabtech, Cincinnati, OH, USA | CAT#3310-10 |
| Chemicals | ||
| BCIP/NBT-plus for ALP | Mabtech, Cincinnati, OH, USA | CAT#3650-10 |
| Dulbecco’s Modified Eagle’s Medium (DMEM) | Corning, Corning, NY, USA | CAT#10-013 |
| Fetal Bovine Serum (FBS) | Corning, Corning, NY, USA | CAT#28622001 |
| Laemmli Sample Buffer (4×) | Bio-Rad, Hercules, CA, USA | CAT#1610747 |
| Pierce Protease Inhibitor Mini Tablets | Thermo Fisher Scientific, Waltham, MA, USA | CAT#A32953 |
| Red Blood Cell Lysis Buffer (10×) | BioLegend, San Diego, CA, USA | CAT#420301 |
| RIPA Lysis and Extraction Buffer | Thermo Fisher Scientific, Waltham, MA, USA | CAT#89900 |
| RPMI 1640 | Corning, Corning, NY, USA | CAT#10-040-CV |
| TPCK-Treated Trypsin | Worthington Biochemical, Lakewood, NJ, USA | CAT#LS003740 |
| 3,3’,5,5’-Tetramethylbenzidine (TMB) Membrane Peroxidase Substrate Plus | Avantor, Radnor, PA, USA | CAT#K830 |
| Commercial Assays | ||
| BCA Protein Assay Kit | Thermo Fisher Scientific, Waltham, MA, USA | CAT#23227 |
| Luna Universal One-Step RT-qPCR Kit | New England Biolabs, Ipswich, MA, USA | CAT#E3006 |
| Monarch Total RNA Miniprep Kit | New England Biolabs, Ipswich, MA, USA | CAT#T2010S |
| Neon Transfection System 100 μL Kit | Invitrogen, Carlsbad, CA, USA | CAT#MPK10025 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woodyear, S.; Chandler, T.L.; Kawagishi, T.; Lonergan, T.M.; Patel, V.A.; Williams, C.A.; Permar, S.R.; Ding, S.; Caddy, S.L. Chimeric Viruses Enable Study of Antibody Responses to Human Rotaviruses in Mice. Viruses 2024, 16, 1145. https://doi.org/10.3390/v16071145
Woodyear S, Chandler TL, Kawagishi T, Lonergan TM, Patel VA, Williams CA, Permar SR, Ding S, Caddy SL. Chimeric Viruses Enable Study of Antibody Responses to Human Rotaviruses in Mice. Viruses. 2024; 16(7):1145. https://doi.org/10.3390/v16071145
Chicago/Turabian StyleWoodyear, Sarah, Tawny L. Chandler, Takahiro Kawagishi, Tom M. Lonergan, Vanshika A. Patel, Caitlin A. Williams, Sallie R. Permar, Siyuan Ding, and Sarah L. Caddy. 2024. "Chimeric Viruses Enable Study of Antibody Responses to Human Rotaviruses in Mice" Viruses 16, no. 7: 1145. https://doi.org/10.3390/v16071145
APA StyleWoodyear, S., Chandler, T. L., Kawagishi, T., Lonergan, T. M., Patel, V. A., Williams, C. A., Permar, S. R., Ding, S., & Caddy, S. L. (2024). Chimeric Viruses Enable Study of Antibody Responses to Human Rotaviruses in Mice. Viruses, 16(7), 1145. https://doi.org/10.3390/v16071145





