Discovery of Novel Viruses in Culicoides Biting Midges in Chihuahua, Mexico
Abstract
1. Introduction
2. Materials and Methods
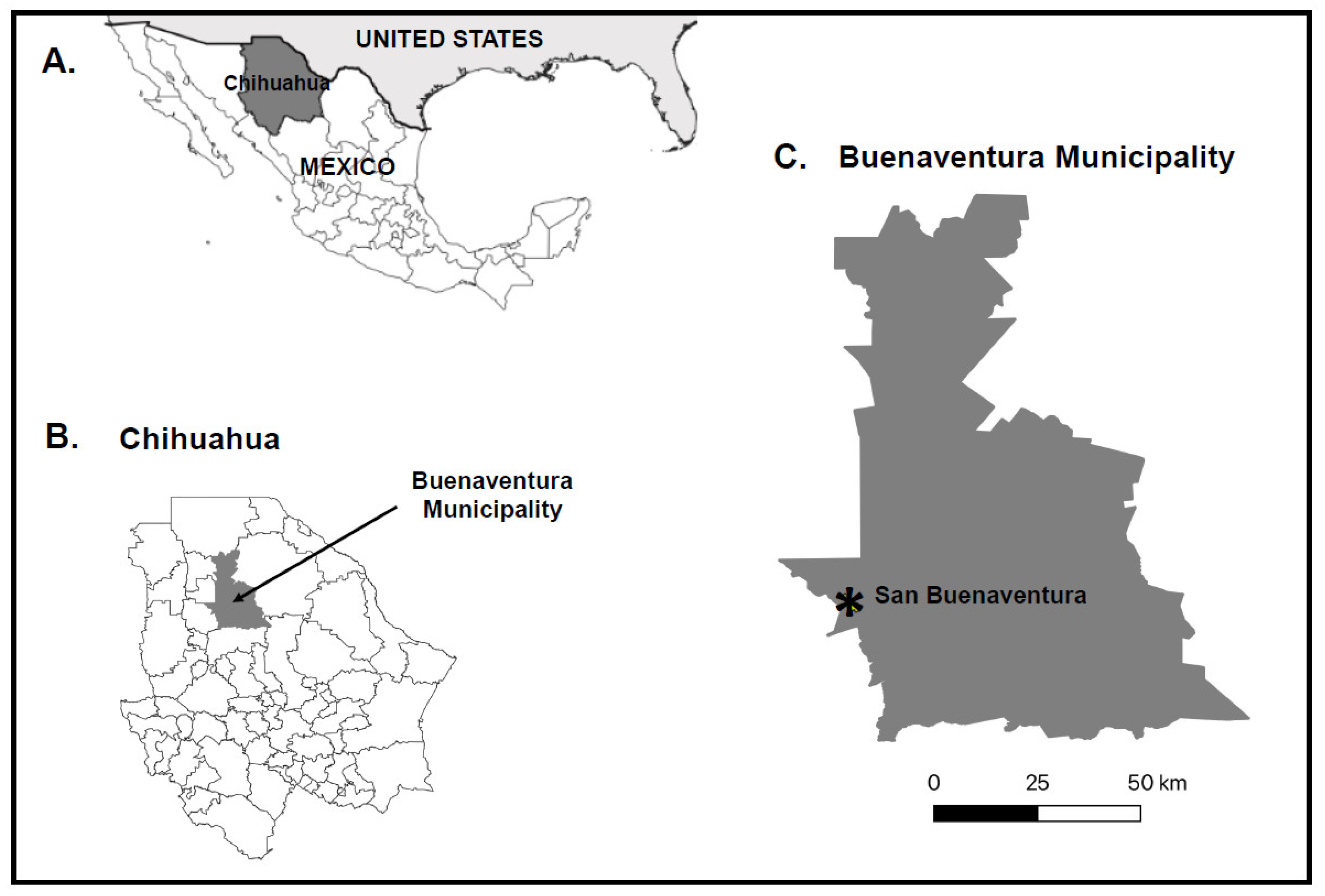
2.1. Study Sites and Midge Collections
2.2. Homogenizations
2.3. Polyethylene Glycol Precipitation
2.4. Unbiased High-Throughput Sequencing
2.5. Bioinformatics
2.6. RT-PCR and Sanger Sequencing
2.7. Virus Isolation in Cell Culture
2.8. Phylogenetic Analysis
3. Results
3.1. Midge Collections and Virus Identification
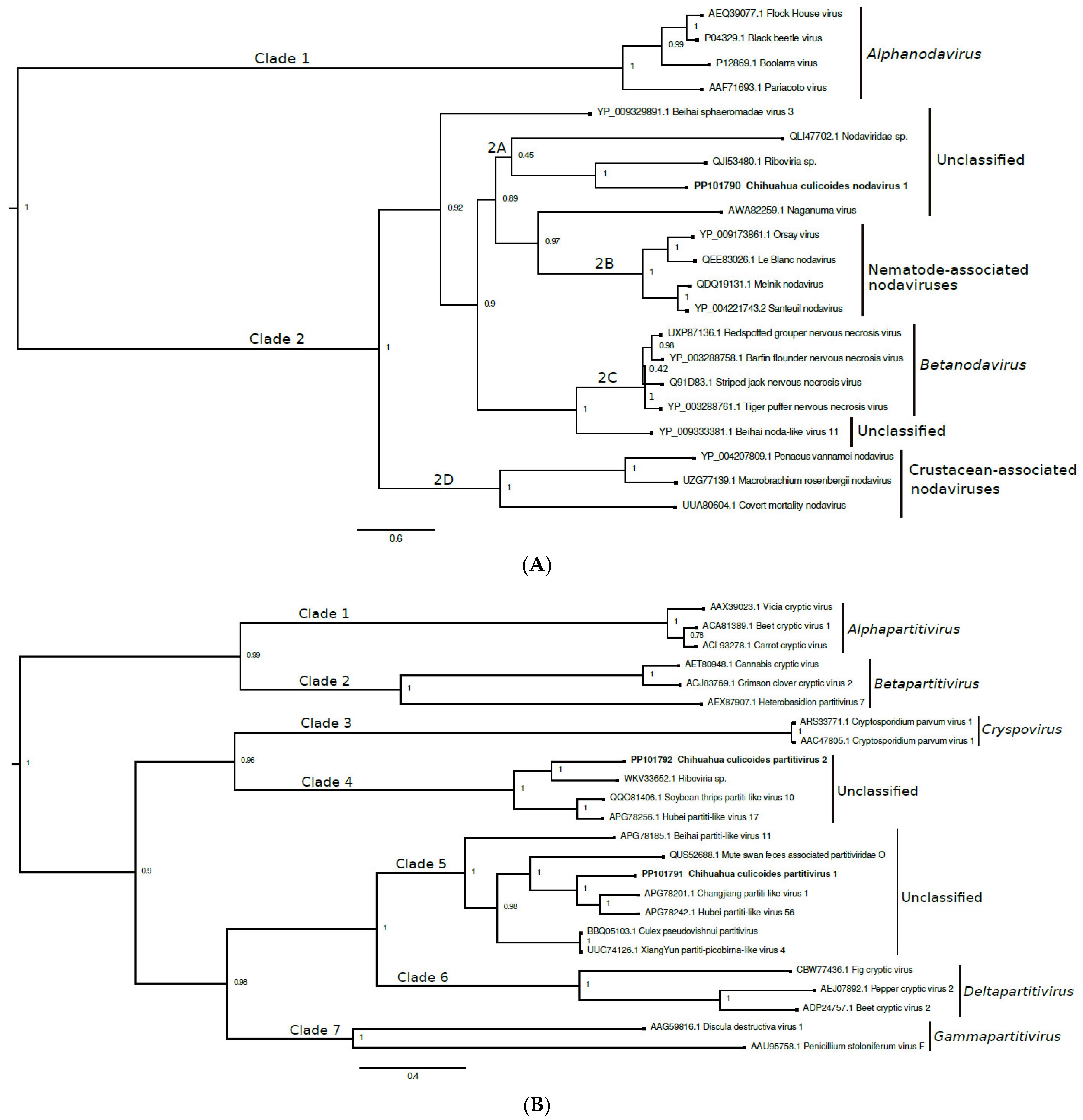
3.2. Nodaviridae
3.3. Partitiviridae
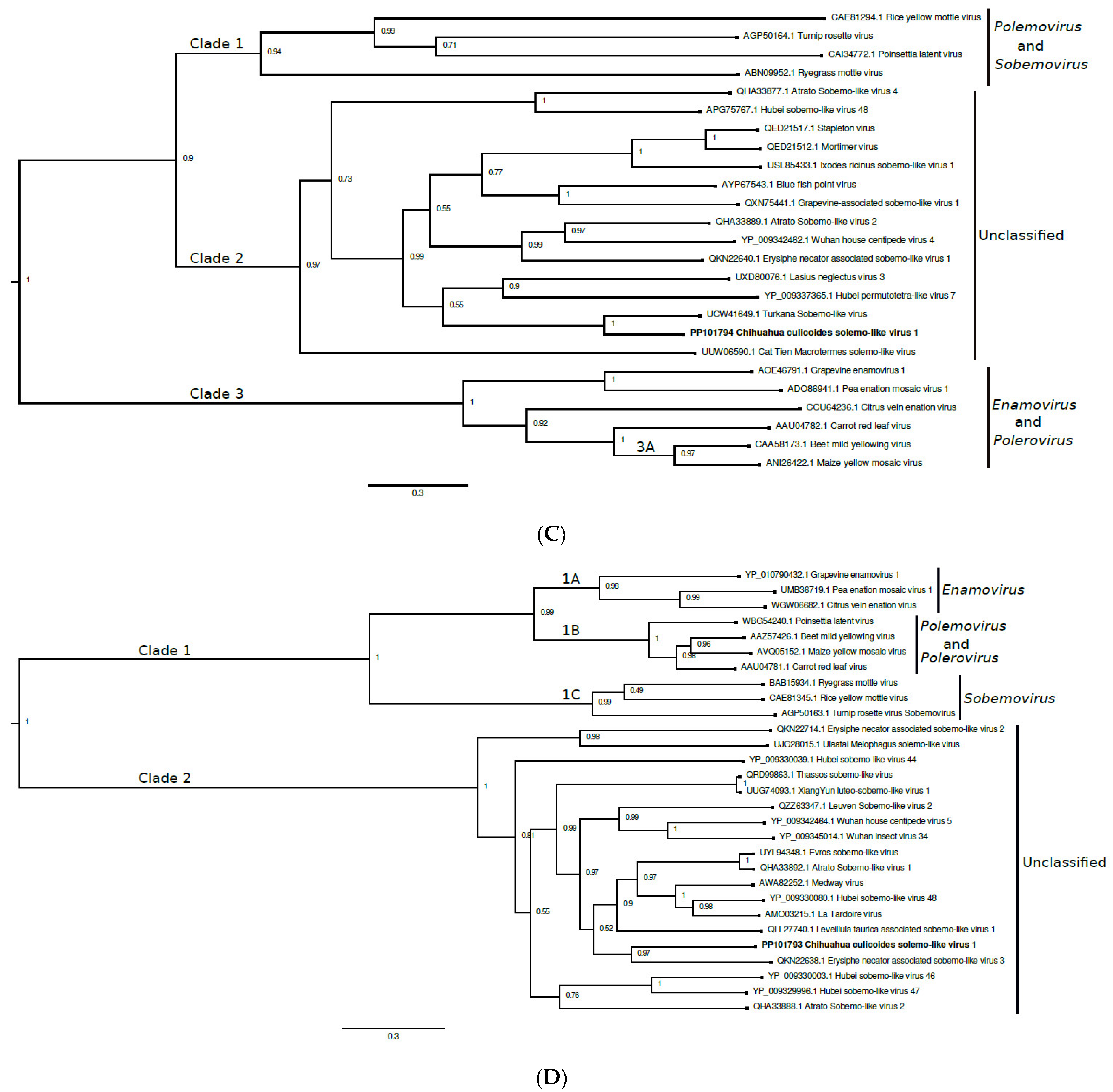
3.4. Solemoviridae
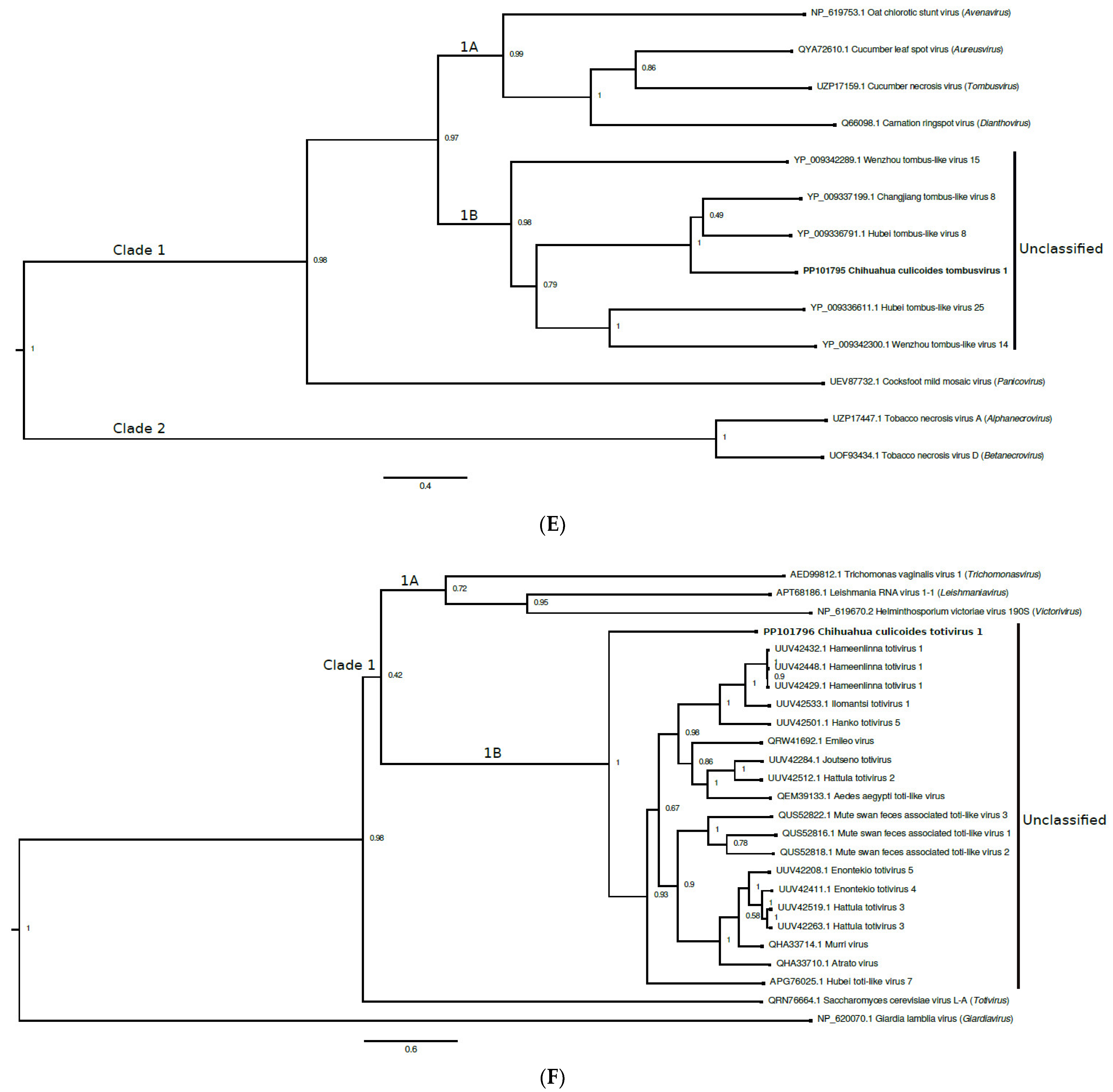
3.5. Tombusviridae
3.6. Totiviridae
3.7. Unclassified Virus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sick, F.; Beer, M.; Kampen, H.; Wernike, K. Culicoides Biting Midges-Underestimated Vectors for Arboviruses of Public Health and Veterinary Importance. Viruses 2019, 11, 376. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Elbers, A.R.; Koenraadt, C.J.; Meiswinkel, R. Mosquitoes and Culicoides biting midges: Vector range and the influence of climate change. Rev. Sci. Tech. 2015, 34, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Purse, B.V.; Carpenter, S.; Venter, G.J.; Bellis, G.; Mullens, B.A. Bionomics of temperate and tropical Culicoides midges: Knowledge gaps and consequences for transmission of Culicoides-borne viruses. Annu. Rev. Entomol. 2015, 60, 373–392. [Google Scholar] [CrossRef] [PubMed]
- Sakkas, H.; Bozidis, P.; Franks, A.; Papadopoulou, C. Oropouche Fever: A Review. Viruses 2018, 10, 175. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Da Rosa, J.F.T.; De Souza, W.M.; de Paula Pinheiro, F.; Figueiredo, M.L.; Cardoso, J.F.; Acrani, G.O.; Nunes, M.R.T. Oropouche Virus: Clinical, Epidemiological, and Molecular Aspects of a Neglected Orthobunyavirus. Am. J. Trop. Med. Hyg. 2017, 96, 1019–1030. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Carpenter, S.; Mellor, P.S.; Fall, A.G.; Garros, C.; Venter, G.J. African Horse Sickness Virus: History, Transmission, and Current Status. Annu. Rev. Entomol. 2017, 62, 343–358. [Google Scholar] [CrossRef] [PubMed]
- Lee, F. Bovine Ephemeral Fever in Asia: Recent Status and Research Gaps. Viruses 2019, 11, 412. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- De Regge, N. Akabane, Aino and Schmallenberg virus-where do we stand and what do we know about the role of domestic ruminant hosts and Culicoides vectors in virus transmission and overwintering? Curr. Opin. Virol. 2017, 27, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Endalew, A.D.; Faburay, B.; Wilson, W.C.; Richt, J.A. Schmallenberg Disease-A Newly Emerged Culicoides-borne Viral Disease of Ruminants. Viruses 2019, 11, 1065. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Maclachlan, N.J.; Zientara, S.; Wilson, W.C.; Richt, J.A.; Savini, G. Bluetongue and epizootic hemorrhagic disease viruses: Recent developments with these globally re-emerging arboviral infections of ruminants. Curr. Opin. Virol. 2019, 34, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Cabello, L.; Utrilla-Trigo, S.; Lorenzo, G.; Ortego, J.; Calvo-Pinilla, E. Epizootic Hemorrhagic Disease Virus: Current Knowledge and Emerging Perspectives. Microorganisms 2023, 11, 1339. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lipkin, W.I.; Firth, C. Viral surveillance and discovery. Curr. Opin. Virol. 2013, 3, 199–204. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bassi, C.; Guerriero, P.; Pierantoni, M.; Callegari, E.; Sabbioni, S. Novel Virus Identification through Metagenomics: A Systematic Review. Life 2022, 12, 2048. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Greninger, A.L. A decade of RNA virus metagenomics is (not) enough. Virus Res. 2018, 244, 218–229. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Santiago-Rodriguez, T.M.; Hollister, E.B. Unraveling the viral dark matter through viral metagenomics. Front. Immunol. 2022, 13, 1005107. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sadeghi, M.; Altan, E.; Deng, X.; Barker, C.M.; Fang, Y.; Coffey, L.L.; Delwart, E. Virome of > 12 thousand Culex mosquitoes from throughout California. Virology 2018, 523, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Tangudu, C.S.; Hargett, A.M.; Laredo-Tiscareno, S.V.; Smith, R.C.; Blitvich, B.J. Isolation of a novel rhabdovirus and detection of multiple novel viral sequences in Culex species mosquitoes in the United States. Arch. Virol. 2022, 167, 2577–2590. [Google Scholar] [CrossRef] [PubMed]
- Abílio, A.P.; Silva, M.; Kampango, A.; Narciso, I.; Gudo, E.S.; Das Neves, L.C.B.; Sidat, M.; Fafetine, J.M.; De Almeida, A.P.G.; Parreira, R. A survey of RNA viruses in mosquitoes from Mozambique reveals novel genetic lineages of flaviviruses and phenuiviruses, as well as frequent flavivirus-like viral DNA forms in Mansonia. BMC Microbiol. 2020, 20, 225. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chandra, S.; Harvey, E.; Emery, D.; Holmes, E.C.; Slapeta, J. Unbiased Characterization of the Microbiome and Virome of Questing Ticks. Front. Microbiol. 2021, 12, 627327. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pettersson, J.H.O.; Shi, M.; Bohlin, J.; Eldholm, V.; Brynildsrud, O.B.; Paulsen, K.M.; Andreassen, Å.; Holmes, E.C. Characterizing the virome of Ixodes ricinus ticks from northern Europe. Sci. Rep. 2017, 7, 10870. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yang, Z.; Wang, H.; Yang, S.; Wang, X.; Shen, Q.; Ji, L.; Zeng, J.; Zhang, W.; Gong, H.; Shan, T. Virome diversity of ticks feeding on domestic mammals in China. Virol. Sin. 2023, 38, 208–221. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Konstantinidis, K.; Bampali, M.; Williams, M.d.C.; Dovrolis, N.; Gatzidou, E.; Papazilakis, P.; Nearchou, A.; Veletza, S.; Karakasiliotis, I. Dissecting the Species-Specific Virome in Culicoides of Thrace. Front. Microbiol. 2022, 13, 802577. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Langat, S.K.; Eyase, F.; Bulimo, W.; Lutomiah, J.; Oyola, S.O.; Imbuga, M.; Sang, R. Profiling of RNA Viruses in Biting Midges (Ceratopogonidae) and Related Diptera from Kenya Using Metagenomics and Metabarcoding Analysis. mSphere 2021, 6, e0055121. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, L.; Shen, Q.; Li, N.; He, Y.; Han, N.; Wang, X.; Meng, J.; Peng, Y.; Pan, M.; Jin, Y.; et al. Comparative viromes of Culicoides and mosquitoes reveal their consistency and diversity in viral profiles. Brief. Bioinform. 2021, 22, bbaa323. [Google Scholar] [CrossRef] [PubMed]
- Modha, S.; Hughes, J.; Bianco, G.; Ferguson, H.M.; Helm, B.; Tong, L.; Wilkie, G.S.; Kohl, A.; Schnettler, E. Metaviromics Reveals Unknown Viral Diversity in the Biting Midge Culicoides impunctatus. Viruses 2019, 11, 865. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Temmam, S.; Monteil-Bouchard, S.; Robert, C.; Baudoin, J.-P.; Sambou, M.; Aubadie-Ladrix, M.; Labas, N.; Raoult, D.; Mediannikov, O.; Desnues, C. Characterization of Viral Communities of Biting Midges and Identification of Novel Thogotovirus Species and Rhabdovirus Genus. Viruses 2016, 8, 77. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Temmam, S.; Monteil-Bouchard, S.; Sambou, M.; Aubadie-Ladrix, M.; Azza, S.; Decloquement, P.; Khalil, J.Y.B.; Baudoin, J.-P.; Jardot, P.; Robert, C.; et al. Faustovirus-Like Asfarvirus in Hematophagous Biting Midges and Their Vertebrate Hosts. Front. Microbiol. 2015, 6, 1406. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kobayashi, D.; Murota, K.; Faizah, A.N.; Amoa-Bosompem, M.; Higa, Y.; Hayashi, T.; Tsuda, Y.; Sawabe, K.; Isawa, H. RNA virome analysis of hematophagous Chironomoidea flies (Diptera: Ceratopogonidae and Simuliidae) collected in Tokyo, Japan. Med. Entomol. Zool. 2020, 71, 225–243. [Google Scholar] [CrossRef]
- Borkent, A.; Dominiak, P. Catalog of the Biting Midges of the World (Diptera: Ceratopogonidae). Zootaxa 2020, 4787, 1–377. [Google Scholar] [CrossRef] [PubMed]
- Borkent, A.; Grogan, W.L. Catalog of the New World Biting Midges North of Mexico (Diptera: Ceratopogonidae). Zootaxa 2009, 2273, 1–48. [Google Scholar] [CrossRef]
- Grogan, W.L.; Spinelli, G.R.; Phillips, R.A.; Woodward, D.L. The male of Culicoides reevesi Wirth, with a redescription of the female and new seasonal activity, distribution, and biting records (Diptera: Ceratopogonidae). West. N. Am. Nat. 2004, 64, 433–438. [Google Scholar]
- Laredo-Tiscareño, S.A.; Garza-Hernández, J.A.; Tangudu, C.S.; Rodríguez-Alarcón, C.A.; Gonzalez-Peña, R.; Adame-Gallegos, J.R.; Beristain-Ruiz, D.M.; Barajas-López, I.N.; Hargett, A.M.; Munderloh, U.G.; et al. Detection of multiple novel viruses in argasid and ixodid ticks in Mexico Ticks and tick-borne viruses. Ticks Tick-Borne Dis. 2024; under review. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Morales-Hojas, R.; Hinsley, M.; Armean, I.M.; Silk, R.; Harrup, L.E.; Gonzalez-Uriarte, A.; Veronesi, E.; Campbell, L.; Nayduch, D.; Saski, C.; et al. The genome of the biting midge Culicoides sonorensis and gene expression analyses of vector competence for bluetongue virus. BMC Genom. 2018, 19, 624. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kopylova, E.; Noe, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- McHolland, L.E.; Mecham, J.O. Characterization of cell lines developed from field populations of Culicoides sonorensis (Diptera: Ceratopogonidae). J. Med. Entomol. 2003, 40, 348–351. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sahul Hameed, A.S.; Ninawe, A.S.; Nakai, T.; Chi, S.C.; Johnson, K.L.; Ictv Report, C. ICTV Virus Taxonomy Profile: Nodaviridae. J. Gen. Virol. 2019, 100, 3–4. [Google Scholar] [CrossRef] [PubMed]
- Félix, M.-A.; Ashe, A.; Piffaretti, J.; Wu, G.; Nuez, I.; Bélicard, T.; Jiang, Y.; Zhao, G.; Franz, C.J.; Goldstein, L.D.; et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011, 9, e1000586. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ho, K.L.; Gabrielsen, M.; Beh, P.L.; Kueh, C.L.; Thong, Q.X.; Streetley, J.; Tan, W.S.; Bhella, D. Structure of the Macrobrachium rosenbergii nodavirus: A new genus within the Nodaviridae? PLoS Biol. 2018, 16, e3000038. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- NaveenKumar, S.; Shekar, M.; Karunasagar, I.; Karunasagar, I. Genetic analysis of RNA1 and RNA2 of Macrobrachium rosenbergii nodavirus (MrNV) isolated from India. Virus Res. 2013, 173, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.F.; Pantoja, C.R.; Redman, R.M.; Lightner, D.V. Development of in situ hybridization and RT-PCR assay for the detection of a nodavirus (PvNV) that causes muscle necrosis in Penaeus vannamei. Dis. Aquat. Organ. 2007, 75, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lin, X.-D.; Tian, J.-H.; Chen, L.-J.; Chen, X.; Li, C.-X.; Qin, X.-C.; Li, J.; Cao, J.-P.; Eden, J.-S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Vainio, E.J.; Chiba, S.; Ghabrial, S.A.; Maiss, E.; Roossinck, M.; Sabanadzovic, S.; Suzuki, N.; Xie, J.; Nibert, M.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Partitiviridae. J. Gen. Virol. 2018, 99, 17–18. [Google Scholar] [CrossRef] [PubMed]
- Ternovoi, V.A.; Shvalov, A.N.; Kartashov, M.Y.; Ponomareva, E.P.; Tupota, N.L.; Khoroshavin, Y.A.; Bayandin, R.B.; Gladysheva, A.V.; Mikryukova, T.P.; Tregubchak, T.V.; et al. The Viromes of Mosquitoes from the Natural Landscapes of Western Siberia. Viruses 2023, 15, 1896. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Qin, T.; Shi, M.; Zhang, M.; Liu, Z.; Feng, H.; Sun, Y. Diversity of RNA viruses of three dominant tick species in North China. Front. Vet. Sci. 2022, 9, 1057977. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Faizah, A.N.; Kobayashi, D.; Isawa, H.; Amoa-Bosompem, M.; Murota, K.; Higa, Y.; Futami, K.; Shimada, S.; Kim, K.S.; Itokawa, K.; et al. Deciphering the Virome of Culex vishnui Subgroup Mosquitoes, the Major Vectors of Japanese Encephalitis, in Japan. Viruses 2020, 12, 264. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sõmera, M.; Fargette, D.; Hébrard, E.; Sarmiento, C.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Solemoviridae 2021. J. Gen. Virol. 2021, 102, 001707. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ribeiro, G.d.O.; Morais, V.S.; Monteiro, F.J.C.; Ribeiro, E.S.D.; Rego, M.O.d.S.; Souto, R.N.P.; Villanova, F.; Tahmasebi, R.; Hefford, P.M.; Deng, X.; et al. Aedes aegypti from Amazon Basin Harbor High Diversity of Novel Viral Species. Viruses 2020, 12, 866. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Aus dem Siepen, M.; Pohl, J.O.; Koo, B.J.; Wege, C.; Jeske, H. Poinsettia latent virus is not a cryptic virus, but a natural polerovirus-sobemovirus hybrid. Virology 2005, 336, 240–250. [Google Scholar] [CrossRef] [PubMed]
- ICTV Report Consortium. Current ICTV Taxonomy Release: Taxonomy Browser. Available online: https://ictv.global/taxonomy (accessed on 8 December 2023).
- Medd, N.C.; Fellous, S.; Waldron, F.M.; Xuéreb, A.; Nakai, M.; Cross, J.V.; Obbard, D.J. The virome of Drosophila suzukii, an invasive pest of soft fruit. Virus Evol. 2018, 4, vey009. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Boonham, N.; Henry, C.M.; Wood, K.R. The nucleotide sequence and proposed genome organization of oat chlorotic stunt virus, a new soil-borne virus of cereals. J. Gen. Virol. 1995, 76 Pt 8, 2025–2034. [Google Scholar] [CrossRef] [PubMed]
- Hillman, B.I.; Cohen, A.B. Totiviruses (Totiviridae). In Encyclopedia of Virology, 4th ed.; Zuckerman, M., Bamford, D.H., Eds.; Academic Press: Oxford, UK, 2021; pp. 648–657. [Google Scholar]
- Zhai, Y.; Attoui, H.; Jaafar, F.M.; Wang, H.-Q.; Cao, Y.-X.; Fan, S.-P.; Sun, Y.-X.; Liu, L.-D.; Mertens, P.P.C.; Meng, W.-S.; et al. Isolation and full-length sequence analysis of Armigeres subalbatus totivirus, the first totivirus isolate from mosquitoes representing a proposed novel genus (Artivirus) of the family Totiviridae. J. Gen. Virol. 2010, 91 Pt 11, 2836–2845. [Google Scholar] [CrossRef] [PubMed]
- Isawa, H.; Kuwata, R.; Hoshino, K.; Tsuda, Y.; Sakai, K.; Watanabe, S.; Nishimura, M.; Satho, T.; Kataoka, M.; Nagata, N.; et al. Identification and molecular characterization of a new nonsegmented double-stranded RNA virus isolated from Culex mosquitoes in Japan. Virus Res. 2011, 155, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Naim, S.; Brown, J.K.; Nibert, M.L. Genetic diversification of penaeid shrimp infectious myonecrosis virus between Indonesia and Brazil. Virus Res. 2014, 189, 97–105. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tighe, A.J.; Ruane, N.M.; Carlsson, J. Potential origins of fish toti-like viruses in invertebrates. J. Gen. Virol. 2022, 103, 001775. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Xu, L.; Bowers, H.; Schott, E.J. Characterization of Two Novel Toti-Like Viruses Co-infecting the Atlantic Blue Crab, Callinectes sapidus, in Its Northern Range of the United States. Front. Microbiol. 2022, 13, 855750. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cholleti, H.; de Jong, J.; Blomstrom, A.L.; Berg, M. Characterization of Pipistrellus pygmaeus Bat Virome from Sweden. Viruses 2022, 14, 1654. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Remnant, E.J.; Baty, J.W.; Bulgarella, M.; Dobelmann, J.; Quinn, O.; Gruber, M.A.M.; Lester, P.J. A Diverse Viral Community from Predatory Wasps in Their Native and Invaded Range, with a New Virus Infectious to Honey Bees. Viruses 2021, 13, 1431. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bell-Sakyi, L.; Jaafar, F.M.; Monsion, B.; Luu, L.; Denison, E.; Carpenter, S.; Attoui, H.; Mertens, P.P.C. Continuous Cell Lines from the European Biting Midge Culicoides nubeculosus (Meigen, 1830). Microorganisms 2020, 8, 825. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wechsler, S.J.; McHolland, L.E.; Tabachnick, W.J. Cell lines from Culicoides variipennis (Diptera: Ceratopogonidae) support replication of bluetongue virus. J. Invertebr. Pathol. 1989, 54, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Nasar, F.; Erasmus, J.H.; Haddow, A.D.; Tesh, R.B.; Weaver, S.C. Eilat virus induces both homologous and heterologous interference. Virology 2015, 484, 51–58. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Olmo, R.P.; Todjro, Y.M.H.; Aguiar, E.R.G.R.; de Almeida, J.P.P.; Ferreira, F.V.; Armache, J.N.; de Faria, I.J.S.; Ferreira, A.G.A.; Amadou, S.C.G.; Silva, A.T.S.; et al. Mosquito vector competence for dengue is modulated by insect-specific viruses. Nat. Microbiol. 2023, 8, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Hobson-Peters, J.; Yam, A.W.Y.; Lu, J.W.F.; Setoh, Y.X.; May, F.J.; Kurucz, N.; Walsh, S.; Prow, N.A.; Davis, S.S.; Weir, R.; et al. A new insect-specific flavivirus from northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells. PLoS ONE 2013, 8, e56534. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Romo, H.; Kenney, J.L.; Blitvich, B.J.; Brault, A.C. Restriction of Zika virus infection and transmission in Aedes aegypti mediated by an insect-specific flavivirus. Emerg. Microbes Infect. 2018, 7, 181. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bolling, B.G.; Olea-Popelka, F.J.; Eisen, L.; Moore, C.G.; Blair, C.D. Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colony and effects of co-infection on vector competence for West Nile virus. Virology 2012, 427, 90–97. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kenney, J.L.; Solberg, O.D.; Langevin, S.A.; Brault, A.C. Characterization of a novel insect-specific flavivirus from Brazil: Potential for inhibition of infection of arthropod cells with medically important flaviviruses. J. Gen. Virol. 2014, 95 Pt 12, 2796–2808. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bruenn, J.A. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2003, 31, 1821–1829. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Charon, J.; Buchmann, J.P.; Sadiq, S.; Holmes, E.C. RdRp-scan: A bioinformatic resource to identify and annotate divergent RNA viruses in metagenomic sequence data. Virus Evol. 2022, 8, veac082. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Monttinen, H.A.M.; Ravantti, J.J.; Poranen, M.M. Structure Unveils Relationships between RNA Virus Polymerases. Viruses 2021, 13, 313. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nguyen, P.T.T.; Culverwell, C.L.; Suvanto, M.T.; Korhonen, E.M.; Uusitalo, R.; Vapalahti, O.; Smura, T.; Huhtamo, E. Characterisation of the RNA Virome of Nine Ochlerotatus Species in Finland. Viruses 2022, 14, 1489. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, Z.; Zhao, H.; Li, Z.; Huang, M.; Si, N.; Zhao, H.; Wei, X.; Sun, B.; Gao, G.F.; Xu, Z.; et al. First Discovery of Phenuiviruses within Diverse RNA Viromes of Asiatic Toad (Bufo gargarizans) by Metagenomics Sequencing. Viruses 2023, 15, 750. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- French, R.K.; Anderson, S.H.; Cain, K.E.; Greene, T.C.; Minor, M.; Miskelly, C.M.; Montoya, J.M.; Wille, M.; Muller, C.G.; Taylor, M.W.; et al. Host phylogeny shapes viral transmission networks in an island ecosystem. Nat. Ecol. Evol. 2023, 7, 1834–1843. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Geoghegan, J.L.; Di Giallonardo, F.; Wille, M.; Ortiz-Baez, A.S.; Costa, V.A.; Ghaly, T.; Mifsud, J.C.O.; Turnbull, O.M.H.; Bellwood, D.R.; E Williamson, J.; et al. Virome composition in marine fish revealed by meta-transcriptomics. Virus Evol. 2021, 7, veab005. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ramirez-Martinez, M.M.; Bennett, A.J.; Dunn, C.D.; Yuill, T.M.; Goldberg, T.L. Bat Flies of the Family Streblidae (Diptera: Hippoboscoidea) Host Relatives of Medically and Agriculturally Important “Bat-Associated” Viruses. Viruses 2021, 13, 860. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Waldron, F.M.; Stone, G.N.; Obbard, D.J. Metagenomic sequencing suggests a diversity of RNA interference-like responses to viruses across multicellular eukaryotes. PLoS Genet. 2018, 14, e1007533. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| Virus | Proposed Taxonomic Classification | 1 Amount of Genome Sequenced (nt. or bp) | 2 Closest Known Relative Based on Amino Acid Sequence Alignments | % Amino Acid Identity (% Coverage) [Translation Product(s)] |
|---|---|---|---|---|
| Chihuahua culicoides nodavirus 1 | Nodaviridae | 996 | Riboviria sp. (QJI53480.1) | 34.5 (98) [CP] |
| Chihuahua culicoides partitivirus 1 | Partitiviridae | 1543 | Hubei partiti-like virus 56 (APG78242.1) | 67.8 (99) [RdRp] |
| Chihuahua culicoides partitivirus 2 | Partitiviridae | 1703 | Riboviria sp. (WKV33652.1) | 62.3 (95) [RdRp] |
| Chihuahua culicoides solemo-like virus 1 | Solemoviridae? | 1159 [segment 1] | Erysiphe necator associated sobemo-like virus 3 (QKN22638.1) | 57.4 (99) [RdRp] |
| 1480 [segment 2] | Turkana Sobemo-like virus (UCW41649.1) | 59.0 (100) [CP] | ||
| Chihuahua culicoides tombusvirus 1 | Tombusviridae | 327 | Hubei tombus-like virus 8 (YP_009336791.1) | 50.5 (87) [CP] |
| Chihuahua culicoides totivirus 1 | Totiviridae | 774 | Mute swan feces associated toti-like virus 1 (QUS52816.1) | 40.0 (100) [RdRp] |
| Chihuahua culicoides virus 1 | Unclassified | 1849 | Leuven wasp-associated virus 1 (QZZ63336.1, QZZ63337.1) | 25.2 (94) [HP] 36.8 (100) [RdRp] |
| Virus | a No. Pools Positive | b Minimal Infection Rate |
|---|---|---|
| Chihuahua culicoides nodavirus 1 | 1 | 0.22 |
| Chihuahua culicoides partitivirus 1 | 5 | 1.09 |
| Chihuahua culicoides partitivirus 2 | 3 | 0.66 |
| Chihuahua culicoides solemo-like virus 1 | 5 | 1.09 |
| c Chihuahua culicoides tombusvirus 1 | 1 | 0.22 |
| Chihuahua culicoides totivirus 1 | 3 | 0.66 |
| Chihuahua culicoides virus 1 | 1 | 0.22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laredo-Tiscareño, S.V.; Garza-Hernandez, J.A.; Tangudu, C.S.; Dankaona, W.; Rodríguez-Alarcón, C.A.; Adame-Gallegos, J.R.; De Luna Santillana, E.J.; Huerta, H.; Gonzalez-Peña, R.; Rivera-Martínez, A.; et al. Discovery of Novel Viruses in Culicoides Biting Midges in Chihuahua, Mexico. Viruses 2024, 16, 1160. https://doi.org/10.3390/v16071160
Laredo-Tiscareño SV, Garza-Hernandez JA, Tangudu CS, Dankaona W, Rodríguez-Alarcón CA, Adame-Gallegos JR, De Luna Santillana EJ, Huerta H, Gonzalez-Peña R, Rivera-Martínez A, et al. Discovery of Novel Viruses in Culicoides Biting Midges in Chihuahua, Mexico. Viruses. 2024; 16(7):1160. https://doi.org/10.3390/v16071160
Chicago/Turabian StyleLaredo-Tiscareño, S. Viridiana, Javier A. Garza-Hernandez, Chandra S. Tangudu, Wichan Dankaona, Carlos A. Rodríguez-Alarcón, Jaime R. Adame-Gallegos, Erick J. De Luna Santillana, Herón Huerta, Rodolfo Gonzalez-Peña, Alejandra Rivera-Martínez, and et al. 2024. "Discovery of Novel Viruses in Culicoides Biting Midges in Chihuahua, Mexico" Viruses 16, no. 7: 1160. https://doi.org/10.3390/v16071160
APA StyleLaredo-Tiscareño, S. V., Garza-Hernandez, J. A., Tangudu, C. S., Dankaona, W., Rodríguez-Alarcón, C. A., Adame-Gallegos, J. R., De Luna Santillana, E. J., Huerta, H., Gonzalez-Peña, R., Rivera-Martínez, A., Rubio-Tabares, E., Beristain-Ruiz, D. M., & Blitvich, B. J. (2024). Discovery of Novel Viruses in Culicoides Biting Midges in Chihuahua, Mexico. Viruses, 16(7), 1160. https://doi.org/10.3390/v16071160






