TRIM5α: A Protean Architect of Viral Recognition and Innate Immunity
Abstract
:1. Introduction
2. TRIM5α in Antiviral Defense: CA Recognition, Immune Signaling, and Restriction
3. SPRYing into Evolution
3.1. Retroviral Battles Shape Genomic Adaptations
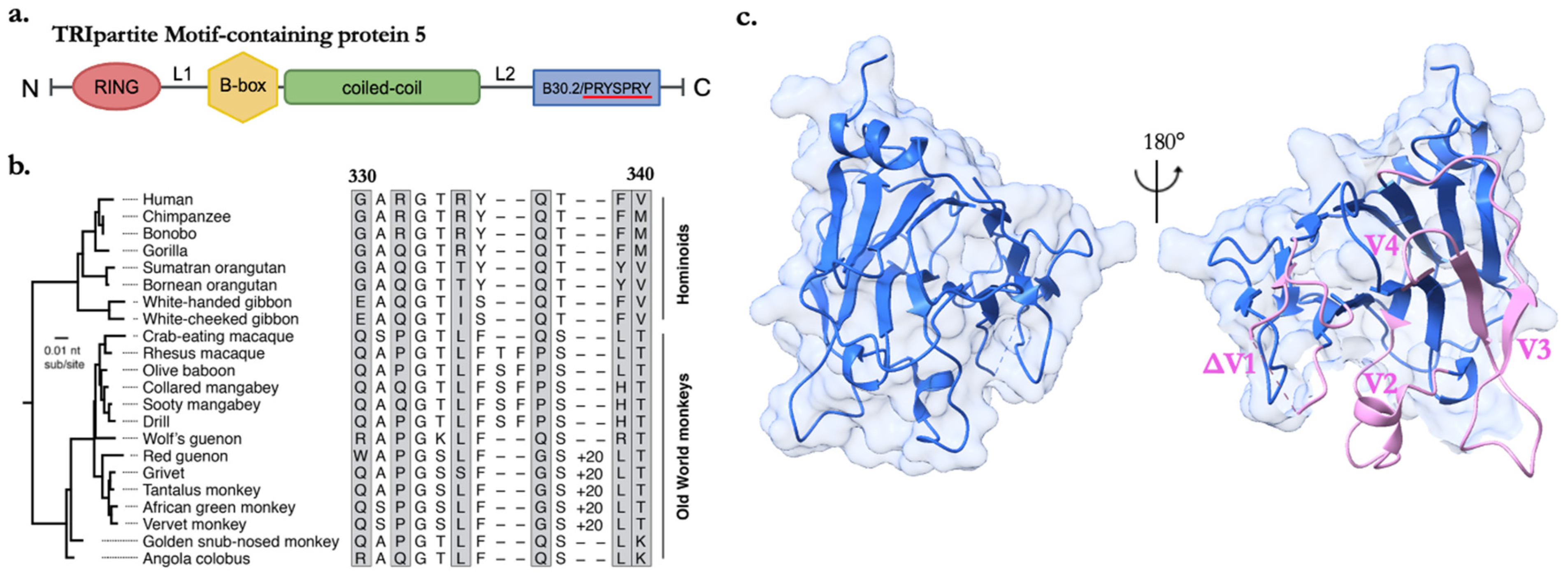
3.2. The Structural Dynamics and Evolutionary Resilience of the SPRY Domain’s v1 Loop
4. Strategies of Molecular Interactions
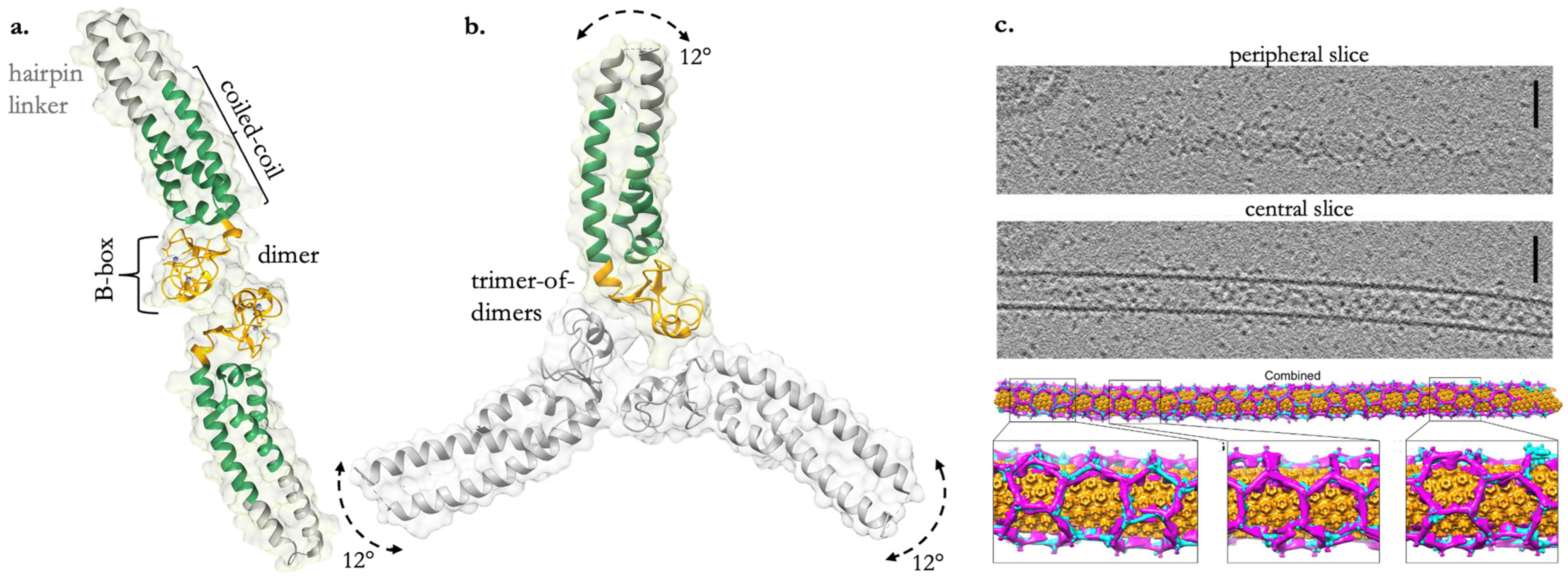
5. Exploring the Multivalency of the Coiled-Coil and B-Box Domains
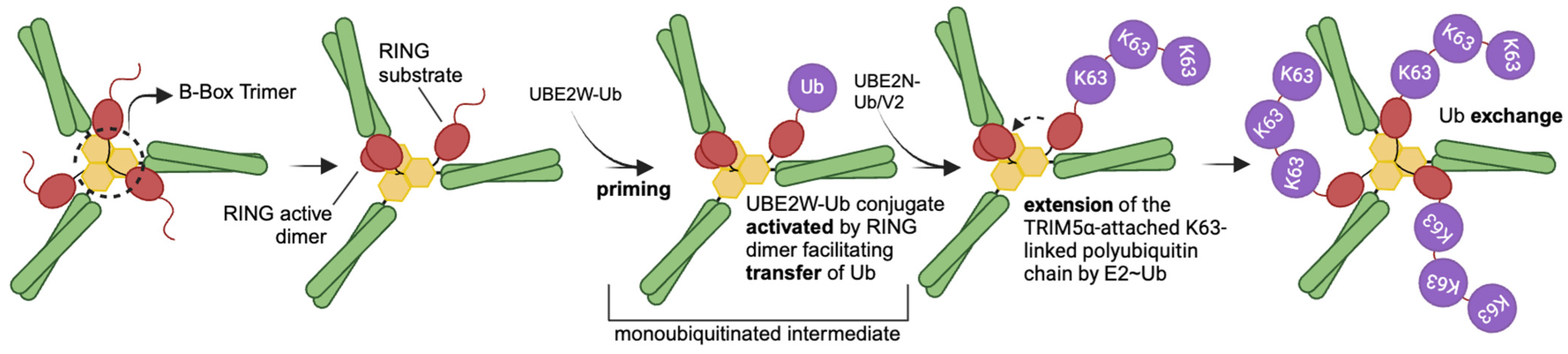
6. The RING Domain as an Effector of Restriction
7. Beyond Retroviruses: Novel Targets and Antiviral Mechanisms
8. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Colomer-Lluch, M.; Ruiz, A.; Moris, A.; Prado, J.G. Restriction Factors: From Intrinsic Viral Restriction to Shaping Cellular Immunity Against HIV-1. Front. Immunol. 2018, 9, 2876. [Google Scholar] [CrossRef]
- Hotter, D.; Kirchhoff, F. Interferons and beyond: Induction of antiretroviral restriction factors. J. Leukoc. Biol. 2018, 103, 465–477. [Google Scholar] [CrossRef]
- Boso, G.; Kozak, C.A. Retroviral Restriction Factors and Their Viral Targets: Restriction Strategies and Evolutionary Adaptations. Microorganisms 2020, 8, 1965. [Google Scholar] [CrossRef]
- Park, A.; Iwasaki, A. Type I and Type III Interferons—Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe 2020, 27, 870–878. [Google Scholar] [CrossRef]
- Sauter, D.; Kirchhoff, F. Evolutionary conflicts and adverse effects of antiviral factors. eLife 2021, 10, e65243. [Google Scholar] [CrossRef]
- Lilly, F. Susceptibility to two strains of Friend leukemia virus in mice. Science 1967, 155, 461–462. [Google Scholar] [CrossRef]
- Yap, M.W.; Colbeck, E.; Ellis, S.A.; Stoye, J.P. Evolution of the retroviral restriction gene Fv1: Inhibition of non-MLV retroviruses. PLoS Pathog. 2014, 10, e1003968. [Google Scholar] [CrossRef]
- Fernandes, A.P.; Agueda-Pinto, A.; Pinheiro, A.; Rebelo, H.; Esteves, P.J. Evolution of TRIM5 and TRIM22 in Bats Reveals a Complex Duplication Process. Viruses 2022, 14, 345. [Google Scholar] [CrossRef]
- Diaz-Griffero, F.; Kar, A.; Perron, M.; Xiang, S.H.; Javanbakht, H.; Li, X.; Sodroski, J. Modulation of retroviral restriction and proteasome inhibitor-resistant turnover by changes in the TRIM5alpha B-box 2 domain. J. Virol. 2007, 81, 10362–10378. [Google Scholar] [CrossRef]
- Hatziioannou, T.; Perez-Caballero, D.; Yang, A.; Cowan, S.; Bieniasz, P.D. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc. Natl. Acad. Sci. USA 2004, 101, 10774–10779. [Google Scholar] [CrossRef]
- Keckesova, Z.; Ylinen, L.M.; Towers, G.J. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 2004, 101, 10780–10785. [Google Scholar] [CrossRef]
- Perron, M.J.; Stremlau, M.; Song, B.; Ulm, W.; Mulligan, R.C.; Sodroski, J. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 2004, 101, 11827–11832. [Google Scholar] [CrossRef]
- Stremlau, M.; Owens, C.M.; Perron, M.J.; Kiessling, M.; Autissier, P.; Sodroski, J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 2004, 427, 848–853. [Google Scholar] [CrossRef]
- Stremlau, M.; Perron, M.; Lee, M.; Li, Y.; Song, B.; Javanbakht, H.; Diaz-Griffero, F.; Anderson, D.J.; Sundquist, W.I.; Sodroski, J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. USA 2006, 103, 5514–5519. [Google Scholar] [CrossRef]
- Wilson, S.J.; Webb, B.L.; Maplanka, C.; Newman, R.M.; Verschoor, E.J.; Heeney, J.L.; Towers, G.J. Rhesus macaque TRIM5 alleles have divergent antiretroviral specificities. J. Virol. 2008, 82, 7243–7247. [Google Scholar] [CrossRef]
- Yap, M.W.; Nisole, S.; Lynch, C.; Stoye, J.P. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 2004, 101, 10786–10791. [Google Scholar] [CrossRef]
- Ylinen, L.M.; Keckesova, Z.; Wilson, S.J.; Ranasinghe, S.; Towers, G.J. Differential restriction of human immunodeficiency virus type 2 and simian immunodeficiency virus SIVmac by TRIM5alpha alleles. J. Virol. 2005, 79, 11580–11587. [Google Scholar] [CrossRef]
- Han, K.; Lou, D.I.; Sawyer, S.L. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet. 2011, 7, e1002388. [Google Scholar] [CrossRef]
- Jaworska, A.M.; Wlodarczyk, N.A.; Mackiewicz, A.; Czerwinska, P. The role of TRIM family proteins in the regulation of cancer stem cell self-renewal. Stem Cells 2020, 38, 165–173. [Google Scholar] [CrossRef]
- van Gent, M.; Sparrer, K.M.J.; Gack, M.U. TRIM Proteins and Their Roles in Antiviral Host Defenses. Annu. Rev. Virol. 2018, 5, 385–405. [Google Scholar] [CrossRef]
- Ozato, K.; Shin, D.M.; Chang, T.H.; Morse, H.C., 3rd. TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 2008, 8, 849–860. [Google Scholar] [CrossRef]
- Diaz-Griffero, F.; Qin, X.R.; Hayashi, F.; Kigawa, T.; Finzi, A.; Sarnak, Z.; Lienlaf, M.; Yokoyama, S.; Sodroski, J. A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction. J. Virol. 2009, 83, 10737–10751. [Google Scholar] [CrossRef]
- Ganser-Pornillos, B.K.; Chandrasekaran, V.; Pornillos, O.; Sodroski, J.G.; Sundquist, W.I.; Yeager, M. Hexagonal assembly of a restricting TRIM5alpha protein. Proc. Natl. Acad. Sci. USA 2011, 108, 534–539. [Google Scholar] [CrossRef]
- Li, X.; Sodroski, J. The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J. Virol. 2008, 82, 11495–11502. [Google Scholar] [CrossRef]
- Li, X.; Yeung, D.F.; Fiegen, A.M.; Sodroski, J. Determinants of the higher order association of the restriction factor TRIM5alpha and other tripartite motif (TRIM) proteins. J. Biol. Chem. 2011, 286, 27959–27970. [Google Scholar] [CrossRef]
- Ganser-Pornillos, B.K.; Pornillos, O. Restriction of HIV-1 and other retroviruses by TRIM5. Nat. Rev. Microbiol. 2019, 17, 546–556. [Google Scholar] [CrossRef]
- Song, B.; Gold, B.; O’Huigin, C.; Javanbakht, H.; Li, X.; Stremlau, M.; Winkler, C.; Dean, M.; Sodroski, J. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates. J. Virol. 2005, 79, 6111–6121. [Google Scholar] [CrossRef]
- Zuliani-Alvarez, L.; Govasli, M.L.; Rasaiyaah, J.; Monit, C.; Perry, S.O.; Sumner, R.P.; McAlpine-Scott, S.; Dickson, C.; Rifat Faysal, K.M.; Hilditch, L.; et al. Evasion of cGAS and TRIM5 defines pandemic HIV. Nat. Microbiol. 2022, 7, 1762–1776. [Google Scholar] [CrossRef]
- Sawyer, S.L.; Wu, L.I.; Emerman, M.; Malik, H.S. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 2005, 102, 2832–2837. [Google Scholar] [CrossRef]
- Ganser, B.K.; Li, S.; Klishko, V.Y.; Finch, J.T.; Sundquist, W.I. Assembly and analysis of conical models for the HIV-1 core. Science 1999, 283, 80–83. [Google Scholar] [CrossRef]
- Mattei, S.; Glass, B.; Hagen, W.J.; Krausslich, H.G.; Briggs, J.A. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 2016, 354, 1434–1437. [Google Scholar] [CrossRef]
- Zhao, G.; Perilla, J.R.; Yufenyuy, E.L.; Meng, X.; Chen, B.; Ning, J.; Ahn, J.; Gronenborn, A.M.; Schulten, K.; Aiken, C.; et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 2013, 497, 643–646. [Google Scholar] [CrossRef]
- Volkmann, B.; Wittmann, S.; Lagisquet, J.; Deutschmann, J.; Eissmann, K.; Ross, J.J.; Biesinger, B.; Gramberg, T. Human TRIM5alpha senses and restricts LINE-1 elements. Proc. Natl. Acad. Sci. USA 2020, 117, 17965–17976. [Google Scholar] [CrossRef]
- Chiramel, A.I.; Meyerson, N.R.; McNally, K.L.; Broeckel, R.M.; Montoya, V.R.; Mendez-Solis, O.; Robertson, S.J.; Sturdevant, G.L.; Lubick, K.J.; Nair, V.; et al. TRIM5alpha Restricts Flavivirus Replication by Targeting the Viral Protease for Proteasomal Degradation. Cell Rep. 2019, 27, 3269–3283.e6. [Google Scholar] [CrossRef]
- Li, H.; Saucedo-Cuevas, L.; Yuan, L.; Ross, D.; Johansen, A.; Sands, D.; Stanley, V.; Guemez-Gamboa, A.; Gregor, A.; Evans, T.; et al. Zika Virus Protease Cleavage of Host Protein Septin-2 Mediates Mitotic Defects in Neural Progenitors. Neuron 2019, 101, 1089–1098.e4. [Google Scholar] [CrossRef]
- Stabell, A.C.; Meyerson, N.R.; Gullberg, R.C.; Gilchrist, A.R.; Webb, K.J.; Old, W.M.; Perera, R.; Sawyer, S.L. Dengue viruses cleave STING in humans but not in nonhuman primates, their presumed natural reservoir. eLife 2018, 7, e31919. [Google Scholar] [CrossRef]
- Yu, C.Y.; Liang, J.J.; Li, J.K.; Lee, Y.L.; Chang, B.L.; Su, C.I.; Huang, W.J.; Lai, M.M.; Lin, Y.L. Dengue Virus Impairs Mitochondrial Fusion by Cleaving Mitofusins. PLoS Pathog. 2015, 11, e1005350. [Google Scholar] [CrossRef]
- Zhao, Y.; Lu, Y.; Richardson, S.; Sreekumar, M.; Albarnaz, J.D.; Smith, G.L. TRIM5alpha restricts poxviruses and is antagonized by CypA and the viral protein C6. Nature 2023, 620, 873–880. [Google Scholar] [CrossRef]
- Li, Y.L.; Chandrasekaran, V.; Carter, S.D.; Woodward, C.L.; Christensen, D.E.; Dryden, K.A.; Pornillos, O.; Yeager, M.; Ganser-Pornillos, B.K.; Jensen, G.J.; et al. Primate TRIM5 proteins form hexagonal nets on HIV-1 capsids. eLife 2016, 5, e16269. [Google Scholar] [CrossRef]
- Pertel, T.; Hausmann, S.; Morger, D.; Zuger, S.; Guerra, J.; Lascano, J.; Reinhard, C.; Santoni, F.A.; Uchil, P.D.; Chatel, L.; et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 2011, 472, 361–365. [Google Scholar] [CrossRef]
- Sebastian, S.; Luban, J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology 2005, 2, 40. [Google Scholar] [CrossRef]
- Skorupka, K.A.; Roganowicz, M.D.; Christensen, D.E.; Wan, Y.; Pornillos, O.; Ganser-Pornillos, B.K. Hierarchical assembly governs TRIM5alpha recognition of HIV-1 and retroviral capsids. Sci. Adv. 2019, 5, eaaw3631. [Google Scholar] [CrossRef]
- Campbell, E.M.; Weingart, J.; Sette, P.; Opp, S.; Sastri, J.; O’Connor, S.K.; Talley, S.; Diaz-Griffero, F.; Hirsch, V.; Bouamr, F. TRIM5alpha-Mediated Ubiquitin Chain Conjugation Is Required for Inhibition of HIV-1 Reverse Transcription and Capsid Destabilization. J. Virol. 2016, 90, 1849–1857. [Google Scholar] [CrossRef]
- Lukic, Z.; Hausmann, S.; Sebastian, S.; Rucci, J.; Sastri, J.; Robia, S.L.; Luban, J.; Campbell, E.M. TRIM5alpha associates with proteasomal subunits in cells while in complex with HIV-1 virions. Retrovirology 2011, 8, 93. [Google Scholar] [CrossRef]
- Wu, X.; Anderson, J.L.; Campbell, E.M.; Joseph, A.M.; Hope, T.J. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc. Natl. Acad. Sci. USA 2006, 103, 7465–7470. [Google Scholar] [CrossRef]
- Fletcher, A.J.; Vaysburd, M.; Maslen, S.; Zeng, J.; Skehel, J.M.; Towers, G.J.; James, L.C. Trivalent RING Assembly on Retroviral Capsids Activates TRIM5 Ubiquitination and Innate Immune Signaling. Cell Host Microbe 2018, 24, 761–775.e6. [Google Scholar] [CrossRef]
- Towers, G.; Bock, M.; Martin, S.; Takeuchi, Y.; Stoye, J.P.; Danos, O. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 2000, 97, 12295–12299. [Google Scholar] [CrossRef]
- Hofmann, W.; Schubert, D.; LaBonte, J.; Munson, L.; Gibson, S.; Scammell, J.; Ferrigno, P.; Sodroski, J. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 1999, 73, 10020–10028. [Google Scholar] [CrossRef]
- Fernandes, A.P.; OhAinle, M.; Esteves, P.J. Patterns of Evolution of TRIM Genes Highlight the Evolutionary Plasticity of Antiviral Effectors in Mammals. Genome Biol. Evol. 2023, 15, evad209. [Google Scholar] [CrossRef]
- Sawyer, S.L.; Emerman, M.; Malik, H.S. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog. 2007, 3, e197. [Google Scholar] [CrossRef]
- Yang, H.; Ji, X.; Zhao, G.; Ning, J.; Zhao, Q.; Aiken, C.; Gronenborn, A.M.; Zhang, P.; Xiong, Y. Structural insight into HIV-1 capsid recognition by rhesus TRIM5alpha. Proc. Natl. Acad. Sci. USA 2012, 109, 18372–18377. [Google Scholar] [CrossRef]
- Newman, R.M.; Hall, L.; Kirmaier, A.; Pozzi, L.A.; Pery, E.; Farzan, M.; O’Neil, S.P.; Johnson, W. Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 2008, 4, e1000003. [Google Scholar] [CrossRef]
- Sayah, D.M.; Sokolskaja, E.; Berthoux, L.; Luban, J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 2004, 430, 569–573. [Google Scholar] [CrossRef]
- Yu, A.; Skorupka, K.A.; Pak, A.J.; Ganser-Pornillos, B.K.; Pornillos, O.; Voth, G.A. TRIM5alpha self-assembly and compartmentalization of the HIV-1 viral capsid. Nat. Commun. 2020, 11, 1307. [Google Scholar] [CrossRef]
- Liu, C.; Perilla, J.R.; Ning, J.; Lu, M.; Hou, G.; Ramalho, R.; Himes, B.A.; Zhao, G.; Bedwell, G.J.; Byeon, I.J.; et al. Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site. Nat. Commun. 2016, 7, 10714. [Google Scholar] [CrossRef]
- Ni, T.; Gerard, S.; Zhao, G.; Dent, K.; Ning, J.; Zhou, J.; Shi, J.; Anderson-Daniels, J.; Li, W.; Jang, S.; et al. Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A. Nat. Struct. Mol. Biol. 2020, 27, 855–862. [Google Scholar] [CrossRef]
- Kim, K.; Dauphin, A.; Komurlu, S.; McCauley, S.M.; Yurkovetskiy, L.; Carbone, C.; Diehl, W.E.; Strambio-De-Castillia, C.; Campbell, E.M.; Luban, J. Cyclophilin A protects HIV-1 from restriction by human TRIM5alpha. Nat. Microbiol. 2019, 4, 2044–2051. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Stremlau, M.; Lee, M.; Sodroski, J. Removal of arginine 332 allows human TRIM5alpha to bind human immunodeficiency virus capsids and to restrict infection. J. Virol. 2006, 80, 6738–6744. [Google Scholar] [CrossRef]
- Pham, Q.T.; Bouchard, A.; Grutter, M.G.; Berthoux, L. Generation of human TRIM5alpha mutants with high HIV-1 restriction activity. Gene Ther. 2010, 17, 859–871. [Google Scholar] [CrossRef] [PubMed]
- Yap, M.W.; Nisole, S.; Stoye, J.P. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr. Biol. 2005, 15, 73–78. [Google Scholar] [CrossRef]
- Tenthorey, J.L.; Young, C.; Sodeinde, A.; Emerman, M.; Malik, H.S. Mutational resilience of antiviral restriction favors primate TRIM5alpha in host-virus evolutionary arms races. eLife 2020, 9, e59988. [Google Scholar] [CrossRef] [PubMed]
- Tenthorey, J.L.; Del Banco, S.; Ramzan, I.; Klingenberg, H.; Liu, C.; Emerman, M.; Malik, H.S. Indels allow antiviral proteins to evolve functional novelty inaccessible by missense mutations. bioRxiv 2024. [Google Scholar] [CrossRef] [PubMed]
- Twentyman, J.; Khalifeh, A.; Felton, A.L.; Emerman, M.; Ohainle, M. Primate TRIM34 is a broadly-acting, TRIM5-dependent lentiviral restriction factor. Retrovirology 2023, 20, 15. [Google Scholar] [CrossRef] [PubMed]
- Bozek, K.; Nakayama, E.E.; Kono, K.; Shioda, T. Electrostatic potential of human immunodeficiency virus type 2 and rhesus macaque simian immunodeficiency virus capsid proteins. Front. Microbiol. 2012, 3, 206. [Google Scholar] [CrossRef]
- Keown, J.R.; Black, M.M.; Ferron, A.; Yap, M.; Barnett, M.J.; Pearce, F.G.; Stoye, J.P.; Goldstone, D.C. A helical LC3-interacting region mediates the interaction between the retroviral restriction factor Trim5alpha and mammalian autophagy-related ATG8 proteins. J. Biol. Chem. 2018, 293, 18378–18386. [Google Scholar] [CrossRef] [PubMed]
- Javanbakht, H.; Yuan, W.; Yeung, D.F.; Song, B.; Diaz-Griffero, F.; Li, Y.; Li, X.; Stremlau, M.; Sodroski, J. Characterization of TRIM5alpha trimerization and its contribution to human immunodeficiency virus capsid binding. Virology 2006, 353, 234–246. [Google Scholar] [CrossRef]
- Sanchez, J.G.; Okreglicka, K.; Chandrasekaran, V.; Welker, J.M.; Sundquist, W.I.; Pornillos, O. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. Proc. Natl. Acad. Sci. USA 2014, 111, 2494–2499. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.M.; Roganowicz, M.D.; Skorupka, K.; Alam, S.L.; Christensen, D.; Doss, G.; Wan, Y.; Frank, G.A.; Ganser-Pornillos, B.K.; Sundquist, W.I.; et al. Mechanism of B-box 2 domain-mediated higher-order assembly of the retroviral restriction factor TRIM5alpha. eLife 2016, 5, e16309. [Google Scholar] [CrossRef]
- Roganowicz, M.D.; Komurlu, S.; Mukherjee, S.; Plewka, J.; Alam, S.L.; Skorupka, K.A.; Wan, Y.; Dawidowski, D.; Cafiso, D.S.; Ganser-Pornillos, B.K.; et al. TRIM5alpha SPRY/coiled-coil interactions optimize avid retroviral capsid recognition. PLoS Pathog. 2017, 13, e1006686. [Google Scholar] [CrossRef]
- Maillard, P.V.; Ecco, G.; Ortiz, M.; Trono, D. The specificity of TRIM5 alpha-mediated restriction is influenced by its coiled-coil domain. J. Virol. 2010, 84, 5790–5801. [Google Scholar] [CrossRef]
- Yudina, Z.; Roa, A.; Johnson, R.; Biris, N.; de Souza Aranha Vieira, D.A.; Tsiperson, V.; Reszka, N.; Taylor, A.B.; Hart, P.J.; Demeler, B.; et al. RING Dimerization Links Higher-Order Assembly of TRIM5alpha to Synthesis of K63-Linked Polyubiquitin. Cell Rep. 2015, 12, 788–797. [Google Scholar] [CrossRef]
- Danielson, C.M.; Cianci, G.C.; Hope, T.J. Recruitment and dynamics of proteasome association with rhTRIM5alpha cytoplasmic complexes during HIV-1 infection. Traffic 2012, 13, 1206–1217. [Google Scholar] [CrossRef]
- Lienlaf, M.; Hayashi, F.; Di Nunzio, F.; Tochio, N.; Kigawa, T.; Yokoyama, S.; Diaz-Griffero, F. Contribution of E3-ubiquitin ligase activity to HIV-1 restriction by TRIM5alpha(rh): Structure of the RING domain of TRIM5alpha. J. Virol. 2011, 85, 8725–8737. [Google Scholar] [CrossRef]
- Rold, C.J.; Aiken, C. Proteasomal degradation of TRIM5alpha during retrovirus restriction. PLoS Pathog. 2008, 4, e1000074. [Google Scholar] [CrossRef]
- Perez-Caballero, D.; Hatziioannou, T.; Zhang, F.; Cowan, S.; Bieniasz, P.D. Restriction of human immunodeficiency virus type 1 by TRIM-CypA occurs with rapid kinetics and independently of cytoplasmic bodies, ubiquitin, and proteasome activity. J. Virol. 2005, 79, 15567–15572. [Google Scholar] [CrossRef]
- Roa, A.; Hayashi, F.; Yang, Y.; Lienlaf, M.; Zhou, J.; Shi, J.; Watanabe, S.; Kigawa, T.; Yokoyama, S.; Aiken, C.; et al. RING domain mutations uncouple TRIM5alpha restriction of HIV-1 from inhibition of reverse transcription and acceleration of uncoating. J. Virol. 2012, 86, 1717–1727. [Google Scholar] [CrossRef]
- Fletcher, A.J.; Christensen, D.E.; Nelson, C.; Tan, C.P.; Schaller, T.; Lehner, P.J.; Sundquist, W.I.; Towers, G.J. TRIM5alpha requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription. EMBO J. 2015, 34, 2078–2095. [Google Scholar] [CrossRef]
- Herkules, F.; Yu, C.H.; Taylor, A.B.; Dougherty, V.; Weintraub, S.T.; Ivanov, D.N. Structural and functional asymmetry of RING trimerization controls priming and extension events in TRIM5alpha autoubiquitylation. Nat. Commun. 2022, 13, 7104. [Google Scholar] [CrossRef]
- Brennan, G.; Kozyrev, Y.; Hu, S.L. TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc. Natl. Acad. Sci. USA 2008, 105, 3569–3574. [Google Scholar] [CrossRef]
- Virgen, C.A.; Hatziioannou, T. Antiretroviral activity and Vif sensitivity of rhesus macaque APOBEC3 proteins. J. Virol. 2007, 81, 13932–13937. [Google Scholar] [CrossRef]
- Battivelli, E.; Lecossier, D.; Matsuoka, S.; Migraine, J.; Clavel, F.; Hance, A.J. Strain-specific differences in the impact of human TRIM5alpha, different TRIM5alpha alleles, and the inhibition of capsid-cyclophilin A interactions on the infectivity of HIV-1. J. Virol. 2010, 84, 11010–11019. [Google Scholar] [CrossRef] [PubMed]
- Battivelli, E.; Migraine, J.; Lecossier, D.; Matsuoka, S.; Perez-Bercoff, D.; Saragosti, S.; Clavel, F.; Hance, A.J. Modulation of TRIM5alpha activity in human cells by alternatively spliced TRIM5 isoforms. J. Virol. 2011, 85, 7828–7835. [Google Scholar] [CrossRef] [PubMed]
- Berube, J.; Bouchard, A.; Berthoux, L. Both TRIM5alpha and TRIMCyp have only weak antiviral activity in canine D17 cells. Retrovirology 2007, 4, 68. [Google Scholar] [CrossRef] [PubMed]
- Merindol, N.; El-Far, M.; Sylla, M.; Masroori, N.; Dufour, C.; Li, J.X.; Cherry, P.; Plourde, M.B.; Tremblay, C.; Berthoux, L. HIV-1 capsids from B27/B57+ elite controllers escape Mx2 but are targeted by TRIM5alpha, leading to the induction of an antiviral state. PLoS Pathog. 2018, 14, e1007398. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, C.M.; Sarrami-Forooshani, R.; Setiawan, L.C.; Zijlstra-Willems, E.M.; van Hamme, J.L.; Tigchelaar, W.; van der Wel, N.N.; Kootstra, N.A.; Gringhuis, S.I.; Geijtenbeek, T.B. Receptor usage dictates HIV-1 restriction by human TRIM5alpha in dendritic cell subsets. Nature 2016, 540, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Battivelli, E.; Migraine, J.; Lecossier, D.; Yeni, P.; Clavel, F.; Hance, A.J. Gag cytotoxic T lymphocyte escape mutations can increase sensitivity of HIV-1 to human TRIM5alpha, linking intrinsic and acquired immunity. J. Virol. 2011, 85, 11846–11854. [Google Scholar] [CrossRef] [PubMed]
- Granier, C.; Battivelli, E.; Lecuroux, C.; Venet, A.; Lambotte, O.; Schmitt-Boulanger, M.; Delaugerre, C.; Molina, J.M.; Chakrabarti, L.A.; Clavel, F.; et al. Pressure from TRIM5alpha contributes to control of HIV-1 replication by individuals expressing protective HLA-B alleles. J. Virol. 2013, 87, 10368–10380. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Guardeno, J.M.; Apolonia, L.; Betancor, G.; Malim, M.H. Immunoproteasome activation enables human TRIM5alpha restriction of HIV-1. Nat. Microbiol. 2019, 4, 933–940. [Google Scholar] [CrossRef]
- van Manen, D.; Rits, M.A.; Beugeling, C.; van Dort, K.; Schuitemaker, H.; Kootstra, N.A. The effect of Trim5 polymorphisms on the clinical course of HIV-1 infection. PLoS Pathog. 2008, 4, e18. [Google Scholar] [CrossRef]
- Pierson, T.C.; Diamond, M.S. The continued threat of emerging flaviviruses. Nat. Microbiol. 2020, 5, 796–812. [Google Scholar] [CrossRef]
- Qing, M.; Yang, F.; Zhang, B.; Zou, G.; Robida, J.M.; Yuan, Z.; Tang, H.; Shi, P.Y. Cyclosporine inhibits flavivirus replication through blocking the interaction between host cyclophilins and viral NS5 protein. Antimicrob. Agents Chemother. 2009, 53, 3226–3235. [Google Scholar] [CrossRef] [PubMed]
- Vidotto, A.; Morais, A.T.; Ribeiro, M.R.; Pacca, C.C.; Terzian, A.C.; Gil, L.H.; Mohana-Borges, R.; Gallay, P.; Nogueira, M.L. Systems Biology Reveals NS4B-Cyclophilin A Interaction: A New Target to Inhibit YFV Replication. J. Proteome Res. 2017, 16, 1542–1555. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.L. Vaccinia Virus Protein C6: A Multifunctional Interferon Antagonist. Adv. Exp. Med. Biol. 2018, 1052, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Koepke, L.; Gack, M.U.; Sparrer, K.M. The antiviral activities of TRIM proteins. Curr. Opin. Microbiol. 2021, 59, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Mandell, M.A.; Kimura, T.; Jain, A.; Johansen, T.; Deretic, V. TRIM proteins regulate autophagy: TRIM5 is a selective autophagy receptor mediating HIV-1 restriction. Autophagy 2014, 10, 2387–2388. [Google Scholar] [CrossRef] [PubMed]
- Blanchet, F.P.; Moris, A.; Nikolic, D.S.; Lehmann, M.; Cardinaud, S.; Stalder, R.; Garcia, E.; Dinkins, C.; Leuba, F.; Wu, L.; et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity 2010, 32, 654–669. [Google Scholar] [CrossRef]
- Cloherty, A.P.M.; Rader, A.G.; Compeer, B.; Ribeiro, C.M.S. Human TRIM5alpha: Autophagy Connects Cell-Intrinsic HIV-1 Restriction and Innate Immune Sensor Functioning. Viruses 2021, 13, 320. [Google Scholar] [CrossRef]
| TRIM5α | HIV-1 | SIVmac | SIVagm | N-MLV | B-MLV | EIAV |
|---|---|---|---|---|---|---|
| human | weak | - | - | + | - | + |
| rhesus macaque | + | - | + | weak | - | + |
| TRIM5α Characteristics | HIV-1 | Orthoflaviviruses | Orthopoxviruses |
|---|---|---|---|
| Viral target | Capsid lattice (structural) | Viral protease (nonstructural) | Capsid protein L3 (structural) |
| RING | + | + | + |
| SPRY/B30.2 | + | + | + |
| Oligomerization | + | + | + |
| TRIM-CypA | + | - | ? |
| Proteasome | +/- | + degradative | ? |
| Ubiquitination of target | - | + (K48) | + ? |
| Human TRIM5α | +/- | ++ | ++ |
| Innate immune signaling | + | - | +/- (with recognition of L3) |
| Antagonist(s) | CypA | ? | CypA; C6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spada, S.J.; Grigg, M.E.; Bouamr, F.; Best, S.M.; Zhang, P. TRIM5α: A Protean Architect of Viral Recognition and Innate Immunity. Viruses 2024, 16, 997. https://doi.org/10.3390/v16070997
Spada SJ, Grigg ME, Bouamr F, Best SM, Zhang P. TRIM5α: A Protean Architect of Viral Recognition and Innate Immunity. Viruses. 2024; 16(7):997. https://doi.org/10.3390/v16070997
Chicago/Turabian StyleSpada, Stephanie J., Michael E. Grigg, Fadila Bouamr, Sonja M. Best, and Peijun Zhang. 2024. "TRIM5α: A Protean Architect of Viral Recognition and Innate Immunity" Viruses 16, no. 7: 997. https://doi.org/10.3390/v16070997
APA StyleSpada, S. J., Grigg, M. E., Bouamr, F., Best, S. M., & Zhang, P. (2024). TRIM5α: A Protean Architect of Viral Recognition and Innate Immunity. Viruses, 16(7), 997. https://doi.org/10.3390/v16070997






