Genetic Characterization of African Swine Fever Italian Clusters in the 2022–2023 Epidemic Wave by a Multi-Gene Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimens
2.2. Molecular Marker Analysis: Conventional PCR, Sanger Sequencing and Phylogenetic Analysis
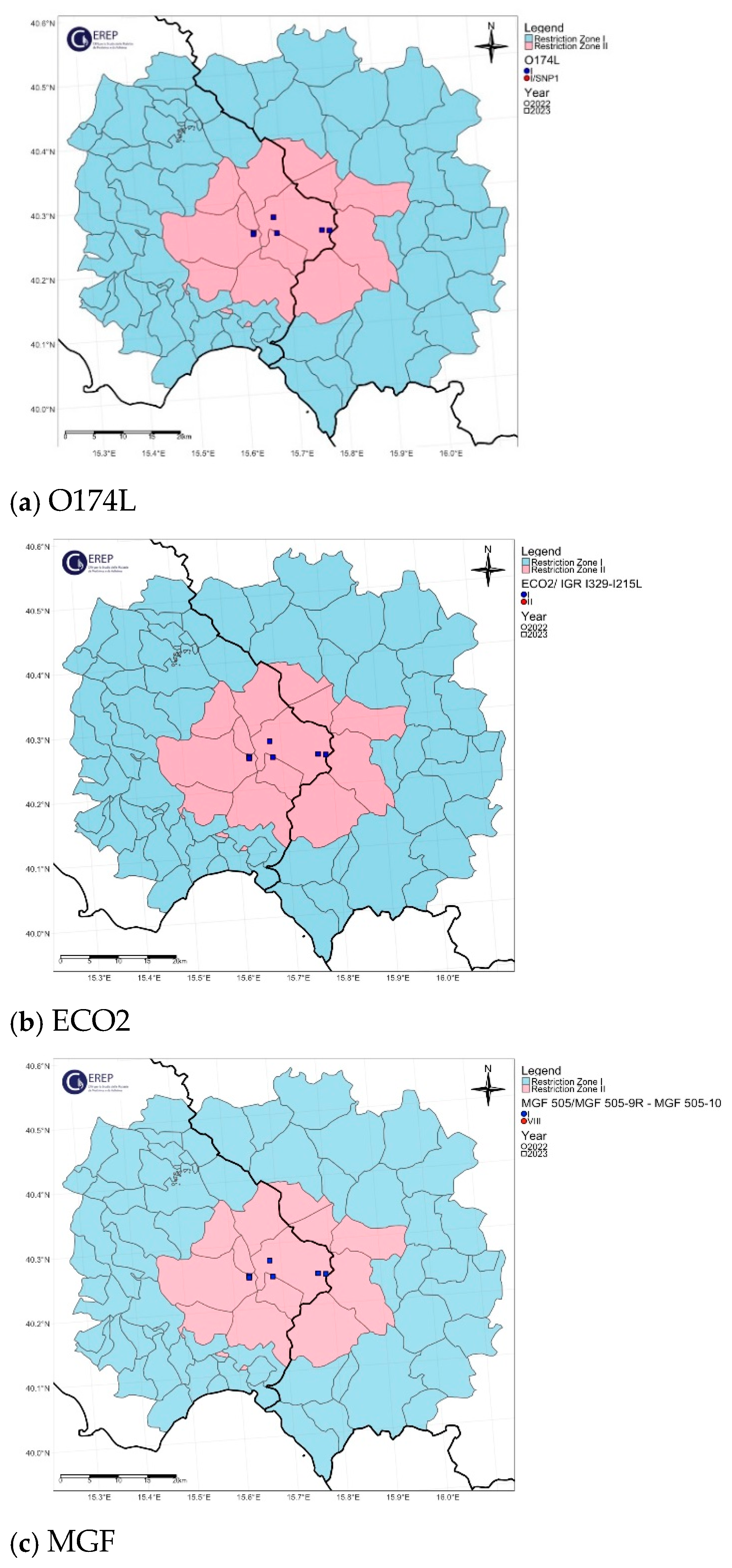
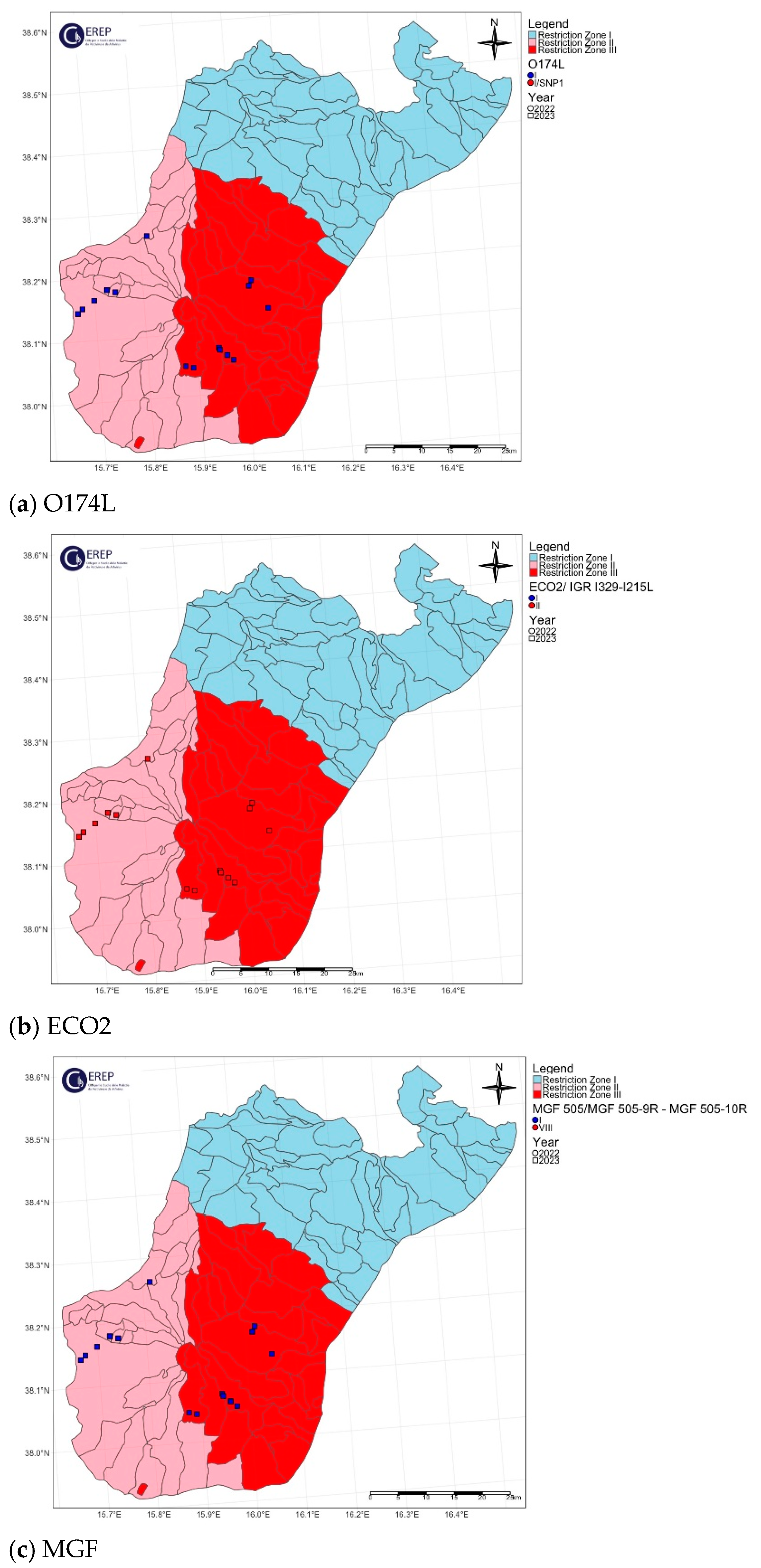
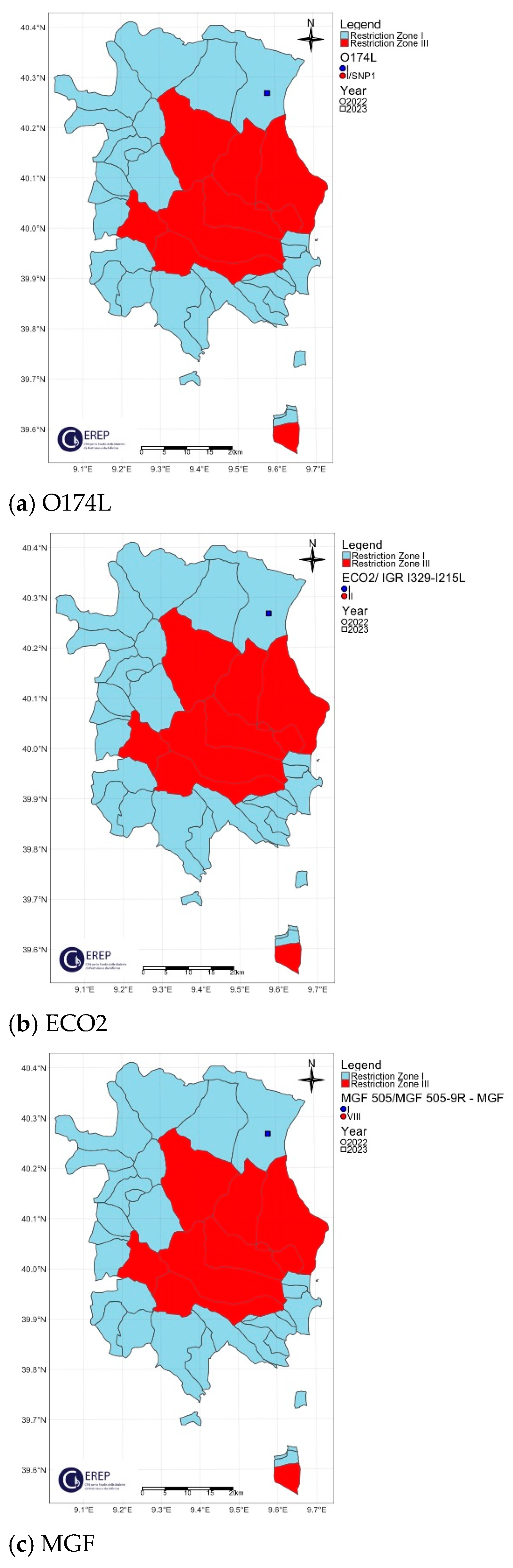
3. Results
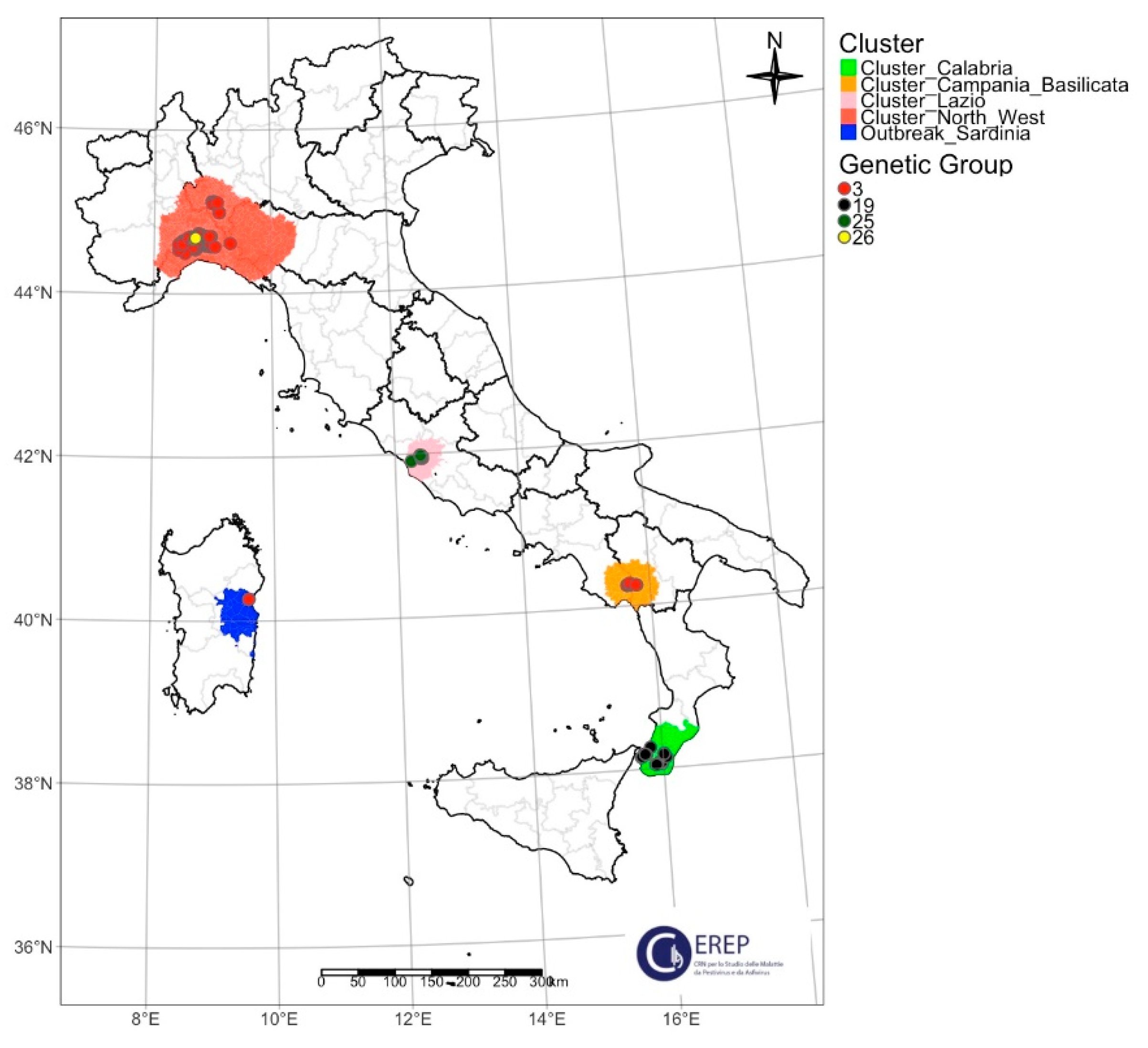
3.1. Phylogenetic Analysis
3.2. Genetic Variants of the Molecular Markers
3.3. Classification of Italian ASFV Strains in Four Different Genetic Groups
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rowlands, R.J.; Michaud, V.; Heath, L.; Hutchings, G.; Oura, C.; Vosloo, W.; Dwarka, R.; Onashvili, T.; Albina, E.; Dixon, L.K. African Swine Fever Virus Isolate, Georgia, 2007. Emerg. Infect. Dis. 2008, 14, 1870. [Google Scholar] [CrossRef]
- Bastos, A.D.S.; Penrith, M.-L.; Macome, F.; Pinto, F.; Thomson, G.R. Co-Circulation of Two Genetically Distinct Viruses in an Outbreak of African Swine Fever in Mozambique: No Evidence for Individual Co-Infection. Vet. Microbiol. 2004, 103, 169–182. [Google Scholar] [CrossRef]
- Lubisi, B.A.; Dwarka, R.M.; Meenowa, D.; Jaumally, R. An Investigation into the First Outbreak of African Swine Fever in the Republic of Mauritius. Transbound. Emerg. Dis. 2009, 56, 178–188. [Google Scholar] [CrossRef]
- Penrith, M.-L.; Van Heerden, J.; Heath, L.; Abworo, E.O.; Bastos, A.D.S. Review of the Pig-Adapted African Swine Fever Viruses in and Outside Africa. Pathogens 2022, 11, 1190. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Fernández-Pinero, J.; Pelayo, V.; Gazaev, I.; Markowska-Daniel, I.; Pridotkas, G.; Nieto, R.; Fernández-Pacheco, P.; Bokhan, S.; Nevolko, O.; et al. Genetic Variation among African Swine Fever Genotype II Viruses, Eastern and Central Europe. Emerg. Infect. Dis. 2014, 20, 1544–1547. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cordón, P.J.; Montoya, M.; Reis, A.L.; Dixon, L.K. African Swine Fever: A Re-Emerging Viral Disease Threatening the Global Pig Industry. Vet. J. 2018, 233, 41–48. [Google Scholar] [CrossRef]
- Gaudreault, N.N.; Madden, D.W.; Wilson, W.C.; Trujillo, J.D.; Richt, J.A. African Swine Fever Virus: An Emerging DNA Arbovirus. Front. Vet. Sci. 2020, 7, 215. [Google Scholar] [CrossRef] [PubMed]
- Forth, J.H.; Tignon, M.; Cay, A.B.; Forth, L.F.; Höper, D.; Blome, S.; Beer, M. Comparative Analysis of Whole-Genome Sequence of African Swine Fever Virus Belgium 2018/1. Emerg. Infect. Dis. 2019, 25, 1249–1252. [Google Scholar] [CrossRef] [PubMed]
- Sauter-Louis, C.; Forth, J.H.; Probst, C.; Staubach, C.; Hlinak, A.; Rudovsky, A.; Holland, D.; Schlieben, P.; Göldner, M.; Schatz, J.; et al. Joining the Club: First Detection of African Swine Fever in Wild Boar in Germany. Transbound. Emerg. Dis. 2021, 68, 1744–1752. [Google Scholar] [CrossRef]
- Ramirez-Medina, E.; O’Donnell, V.; Silva, E.; Espinoza, N.; Velazquez-Salinas, L.; Moran, K.; Daite, D.A.; Barrette, R.; Faburay, B.; Holland, R.; et al. Experimental Infection of Domestic Pigs with an African Swine Fever Virus Field Strain Isolated in 2021 from the Dominican Republic. Viruses 2022, 14, 1090. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/home/en (accessed on 29 March 2024).
- Rajukumar, K.; Senthilkumar, D.; Venkatesh, G.; Singh, F.; Patil, V.P.; Kombiah, S.; Tosh, C.; Dubey, C.K.; Sen, A.; Barman, N.N.; et al. Genetic Characterization of African Swine Fever Virus from Domestic Pigs in India. Transbound. Emerg. Dis. 2021, 68, 2687–2692. [Google Scholar] [CrossRef] [PubMed]
- WAHIS: World Animal Health Information System. Available online: https://wahis.woah.org/#/home (accessed on 29 March 2024).
- Iscaro, C.; Dondo, A.; Ruocco, L.; Masoero, L.; Giammarioli, M.; Zoppi, S.; Guberti, V.; Feliziani, F. January 2022: Index Case of New African Swine Fever Incursion in Mainland Italy. Transbound. Emerg. Dis. 2022, 69, 1707–1711. [Google Scholar] [CrossRef] [PubMed]
- Giammarioli, M.; Alessandro, D.; Cammà, C.; Masoero, L.; Torresi, C.; Marcacci, M.; Zoppi, S.; Curini, V.; Rinaldi, A.; Rossi, E.; et al. Molecular Characterization of the First African Swine Fever Virus Genotype II Strains Identified from Mainland Italy, 2022. Pathogens 2023, 12, 372. [Google Scholar] [CrossRef] [PubMed]
- Rajala, E.; Gröndal, H.; Sternberg Lewerin, S. The First Outbreak of African Swine Fever in Sweden: A Survey of Pig Farmers’ Perceptions of Information Received, Risks, Biosecurity Measures and Future Prospects. Acta Vet. Scand. 2023, 65, 58. [Google Scholar] [CrossRef] [PubMed]
- Giammarioli, M.; Marcacci, M.; Scicluna, M.T.; Cersini, A.; Torresi, C.; Curini, V.; Ancora, M.; Rinaldi, A.; Sala, M.G.; Rossi, E.; et al. Complete Genome of African Swine Fever Virus Genotype II in Central Italy. Microbiol. Resour. Announc. 2023, 12, e0136422. [Google Scholar] [CrossRef]
- Dei Giudici, S.; Loi, F.; Ghisu, S.; Angioi, P.P.; Zinellu, S.; Fiori, M.S.; Carusillo, F.; Brundu, D.; Franzoni, G.; Zidda, G.M.; et al. The Long-Jumping of African Swine Fever: First Genotype II Notified in Sardinia, Italy. Viruses 2023, 16, 32. [Google Scholar] [CrossRef] [PubMed]
- Penrith, M.L.; Gavin, R.; Thomson, A.B. Infectious Diseases of Livestock, 2nd ed.; Oxford University Press: Oxford, UK, 2004. [Google Scholar]
- Dixon, L.K.; Abrams, C.C.; Bowick, G.; Goatley, L.C.; Kay-Jackson, P.C.; Chapman, D.; Liverani, E.; Nix, R.; Silk, R.; Zhang, F. African Swine Fever Virus Proteins Involved in Evading Host Defence Systems. Vet. Immunol. Immunopathol. 2004, 100, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Dixon, L.K.; Chapman, D.A.G.; Netherton, C.L.; Upton, C. African Swine Fever Virus Replication and Genomics. Virus Res. 2013, 173, 3–14. [Google Scholar] [CrossRef]
- Gallardo, C.; Mwaengo, D.M.; MacHaria, J.M.; Arias, M.; Taracha, E.A.; Soler, A.; Okoth, E.; Martín, E.; Kasiti, J.; Bishop, R.P. Enhanced Discrimination of African Swine Fever Virus Isolates through Nucleotide Sequencing of the P54, P72, and PB602L (CVR) Genes. Virus Genes 2009, 38, 85–95. [Google Scholar] [CrossRef]
- Gallardo, C.; Nieto, R.; Soler, A.; Pelayo, V.; Fernández-Pinero, J.; Markowska-Daniel, I.; Pridotkas, G.; Nurmoja, I.; Granta, R.; Simón, A.; et al. Assessment of African Swine Fever Diagnostic Techniques as a Response to the Epidemic Outbreaks in Eastern European Union Countries: How to Improve Surveillance and Control Programs. J. Clin. Microbiol. 2015, 53, 2555–2565. [Google Scholar] [CrossRef]
- Bastos, A.D.S.; Penrith, M.L.; Crucière, C.; Edrich, J.L.; Hutchings, G.; Roger, F.; Couacy-Hymann, E.; Thomson, G.R. Genotyping Field Strains of African Swine Fever Virus by Partial P72 Gene Characterisation. Arch. Virol. 2003, 148, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Lubisi, B.A.; Bastos, A.D.S.; Dwarka, R.M.; Vosloo, W. Molecular Epidemiology of African Swine Fever in East Africa. Arch. Virol. 2005, 150, 2439–2452. [Google Scholar] [CrossRef] [PubMed]
- Boshoff, C.I.; Bastos, A.D.S.; Gerber, L.J.; Vosloo, W. Genetic Characterisation of African Swine Fever Viruses from Outbreaks in Southern Africa (1973–1999). Vet. Microbiol. 2007, 121, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Achenbach, J.E.; Gallardo, C.; Nieto-Pelegrín, E.; Rivera-Arroyo, B.; Degefa-Negi, T.; Arias, M.; Jenberie, S.; Mulisa, D.D.; Gizaw, D.; Gelaye, E.; et al. Identification of a New Genotype of African Swine Fever Virus in Domestic Pigs from Ethiopia. Transbound. Emerg. Dis. 2017, 64, 1393–1404. [Google Scholar] [CrossRef] [PubMed]
- Quembo, C.J.; Jori, F.; Vosloo, W.; Heath, L. Genetic Characterization of African Swine Fever Virus Isolates from Soft Ticks at the Wildlife/Domestic Interface in Mozambique and Identification of a Novel Genotype. Transbound. Emerg. Dis. 2018, 65, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, W.; Moreno, C.; Duran, U.; Henao, N.; Bencosme, M.; Lora, P.; Reyes, R.; Núñez, R.; De Gracia, A.; Perez, A.M. African Swine Fever in the Dominican Republic. Transbound. Emerg. Dis. 2021, 68, 3018–3019. [Google Scholar] [CrossRef]
- Xiong, D.; Zhang, X.; Xiong, J.; Yu, J.; Wei, H. Rapid Genome-Wide Sequence Typing of African Swine Fever Virus Based on Alleles. Virus Res. 2021, 297, 198357. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Okoth, E.; Pelayo, V.; Anchuelo, R.; Martín, E.; Simón, A.; Llorente, A.; Nieto, R.; Soler, A.; Martín, R.; et al. African Swine Fever Viruses with Two Different Genotypes, Both of Which Occur in Domestic Pigs, Are Associated with Ticks and Adult Warthogs, Respectively, at a Single Geographical Site. J. Gen. Virol. 2011, 92, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Giammarioli, M.; Gallardo, C.; Oggiano, A.; Iscaro, C.; Nieto, R.; Pellegrini, C.; Dei Giudici, S.; Arias, M.; De Mia, G.M. Genetic Characterisation of African Swine Fever Viruses from Recent and Historical Outbreaks in Sardinia (1978–2009). Virus Genes 2011, 42, 377–387. [Google Scholar] [CrossRef]
- Lubisi, B.A.; Bastos, A.D.S.; Dwarka, R.M.; Vosloo, W. Intra-Genotypic Resolution of African Swine Fever Viruses from an East African Domestic Pig Cycle: A Combined P72-CVR Approach. Virus Genes 2007, 35, 729–735. [Google Scholar] [CrossRef]
- Nix, R.J.; Gallardo, C.; Hutchings, G.; Blanco, E.; Dixon, L.K. Molecular Epidemiology of African Swine Fever Virus Studied by Analysis of Four Variable Genome Regions. Arch. Virol. 2006, 151, 2475–2494. [Google Scholar] [CrossRef] [PubMed]
- Irusta, P.M.; Borca, M.V.; Kutish, G.F.; Lu, Z.; Caler, E.; Carrillo, C.; Rock, D.L. Amino Acid Tandem Repeats within a Late Viral Gene Define the Central Variable Region of African Swine Fever Virus. Virology 1996, 220, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Cobbold, C.; Windsor, M.; Wileman, T. A Virally Encoded Chaperone Specialized for Folding of the Major Capsid Protein of African Swine Fever Virus. J. Virol. 2001, 75, 7221–7229. [Google Scholar] [CrossRef] [PubMed]
- Mazloum, A.; van Schalkwyk, A.; Chernyshev, R.; Igolkin, A.; Heath, L.; Sprygin, A. A Guide to Molecular Characterization of Genotype II African Swine Fever Virus: Essential and Alternative Genome Markers. Microorganisms 2023, 11, 642. [Google Scholar] [CrossRef] [PubMed]
- Jia, N.; Ou, Y.; Pejsak, Z.; Zhang, Y.; Zhang, J. Roles of African Swine Fever Virus Structural Proteins in Viral Infection. J. Vet. Res. 2017, 61, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Meekins, D.A.; Trujillo, J.D.; Gaudreault, N.N.; Morozov, I.; Pérez-Núñez, D.; Revilla, Y.; Richt, J.A. Long Amplicon Sequencing for Improved Genetic Characterization of African Swine Fever Virus. J. Virol. Methods 2020, 285, 113946. [Google Scholar] [CrossRef]
- Farlow, J.; Donduashvili, M.; Kokhreidze, M.; Kotorashvili, A.; Vepkhvadze, N.G.; Kotaria, N.; Gulbani, A. Intra-Epidemic Genome Variation in Highly Pathogenic African Swine Fever Virus (ASFV) from the Country of Georgia. Virol. J. 2018, 15, 190. [Google Scholar] [CrossRef] [PubMed]
- Simulundu, E.; Lubaba, C.H.; van Heerden, J.; Kajihara, M.; Mataa, L.; Chambaro, H.M.; Sinkala, Y.; Munjita, S.M.; Munang’andu, H.M.; Nalubamba, K.S.; et al. The Epidemiology of African Swine Fever in “Nonendemic” Regions of Zambia (1989–2015): Implications for Disease Prevention and Control. Viruses 2017, 9, 236. [Google Scholar] [CrossRef] [PubMed]
- Sanna, G.; Giudici, S.D.; Bacciu, D.; Angioi, P.P.; Giammarioli, M.; De Mia, G.M.; Oggiano, A. Improved Strategy for Molecular Characterization of African Swine Fever Viruses from Sardinia, Based on Analysis of P30, CD2V and I73R/I329L Variable Regions. Transbound. Emerg. Dis. 2017, 64, 1280–1286. [Google Scholar] [CrossRef]
- Onzere, C.K.; Bastos, A.D.; Okoth, E.A.; Lichoti, J.K.; Bochere, E.N.; Owido, M.G.; Ndambuki, G.; Bronsvoort, M.; Bishop, R.P. Multi-Locus Sequence Typing of African Swine Fever Viruses from Endemic Regions of Kenya and Eastern Uganda (2011–2013) Reveals Rapid B602L Central Variable Region Evolution. Virus Genes 2018, 54, 111–123. [Google Scholar] [CrossRef]
- Mazur-Panasiuk, N.; Walczak, M.; Juszkiewicz, M.; Woźniakowski, G. The Spillover of African Swine Fever in Western Poland Revealed Its Estimated Origin on the Basis of O174L, K145R, MGF 505-5R and IGR I73R/I329L Genomic Sequences. Viruses 2020, 12, 1094. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Casado, N.; Soler, A.; Djadjovski, I.; Krivko, L.; Madueño, E.; Nieto, R.; Perez, C.; Simon, A.; Ivanova, E.; et al. A Multi Gene-Approach Genotyping Method Identifies 24 Genetic Clusters within the Genotype II-European African Swine Fever Viruses Circulating from 2007 to 2022. Front. Vet. Sci. 2023, 10, 1112850. [Google Scholar] [CrossRef] [PubMed]
- Wernike, K.; Hoffmann, B.; Kalthoff, D.; König, P.; Beer, M. Development and Validation of a Triplex Real-Time PCR Assay for the Rapid Detection and Differentiation of Wild-Type and Glycoprotein E-Deleted Vaccine Strains of Bovine Herpesvirus Type 1. J. Virol. Methods 2011, 174, 77–84. [Google Scholar] [CrossRef] [PubMed]
- King, D.P.; Reid, S.M.; Hutchings, G.H.; Grierson, S.S.; Wilkinson, P.J.; Dixon, L.K.; Bastos, A.D.S.; Drew, T.W. Development of a TaqMan® PCR Assay with Internal Amplification Control for the Detection of African Swine Fever Virus. J. Virol. Methods 2003, 107, 53–61. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health (WOAH). African Swine Fever. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; WOAH: Paris, France, 2021; Volume 2, Chapter 3.9.1; Available online: https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/3.09.01_ASF.pdf (accessed on 29 March 2024).
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v6: Recent Updates to the Phylogenetic Tree Display and Annotation Tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Burland, T.G. DNASTAR’s Lasergene Sequence Analysis Software. Methods Mol. Biol. 2000, 132, 71–91. [Google Scholar] [CrossRef]
- RStudio: Integrated Development for R. RStudio, PBC. 2020. Available online: https://www.rstudio.com/ (accessed on 10 April 2024).
- Tennekes, M. Tmap: Thematic Maps in R. J. Stat. Softw. 2018, 84, 1–39. [Google Scholar] [CrossRef]
- Ramirez-Medina, E.; Velazquez-Salinas, L.; Rai, A.; Espinoza, N.; Valladares, A.; Silva, E.; Burton, L.; Spinard, E.; Meyers, A.; Risatti, G.; et al. Evaluation of the Deletion of the African Swine Fever Virus Gene O174L from the Genome of the Georgia Isolate. Viruses 2023, 15, 2134. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cordón, P.J.; Nunez, A.; Neimanis, A.; Wikström-Lassa, E.; Montoya, M.; Crooke, H.; Gavier-Widén, D. African Swine Fever: Disease Dynamics in Wild Boar Experimentally Infected with ASFV Isolates Belonging to Genotype I and II. Viruses 2019, 11, 852. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health African Swine Fever (ASF)—Situation Report 26. Available online: https://www.woah.org/en/document/african-swine-fever-asf-situation-report-26 (accessed on 31 January 2023).
- EFSA Panel on Animal Health and Welfare (AHAW). Scientific Opinion on African Swine Fever. EFSA J. 2014, 12, 3628–3677. [Google Scholar] [CrossRef]
- Commission Implementing Regulation (EU) 2024/808 of 29 February 2024 Amending Annexes I and II to Implementing Regulation (EU) 2023/594 Laying down Special Control Measures for African Swine Fever. Available online: https://eur-lex.europa.eu/eli/reg_impl/2024/808/oj (accessed on 4 March 2024).
- Bora, M.; Bora, D.P.; Manu, M.; Barman, N.N.; Dutta, L.J.; Kumar, P.P.; Poovathikkal, S.; Suresh, K.P.; Nimmanapalli, R. Assessment of Risk Factors of African Swine Fever in India: Perspectives on Future Outbreaks and Control Strategies. Pathogens 2020, 9, 1044. [Google Scholar] [CrossRef] [PubMed]
- Forth, J.H.; Calvelage, S.; Fischer, M.; Hellert, J.; Sehl-Ewert, J.; Roszyk, H.; Deutschmann, P.; Reichold, A.; Lange, M.; Thulke, H.-H.; et al. African Swine Fever Virus—Variants on the Rise. Emerg. Microbes Infect. 2023, 12, 2146537. [Google Scholar] [CrossRef]
- GenBank, National Library of Medicine (US), National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/nucleotide/ (accessed on 5 June 2024).
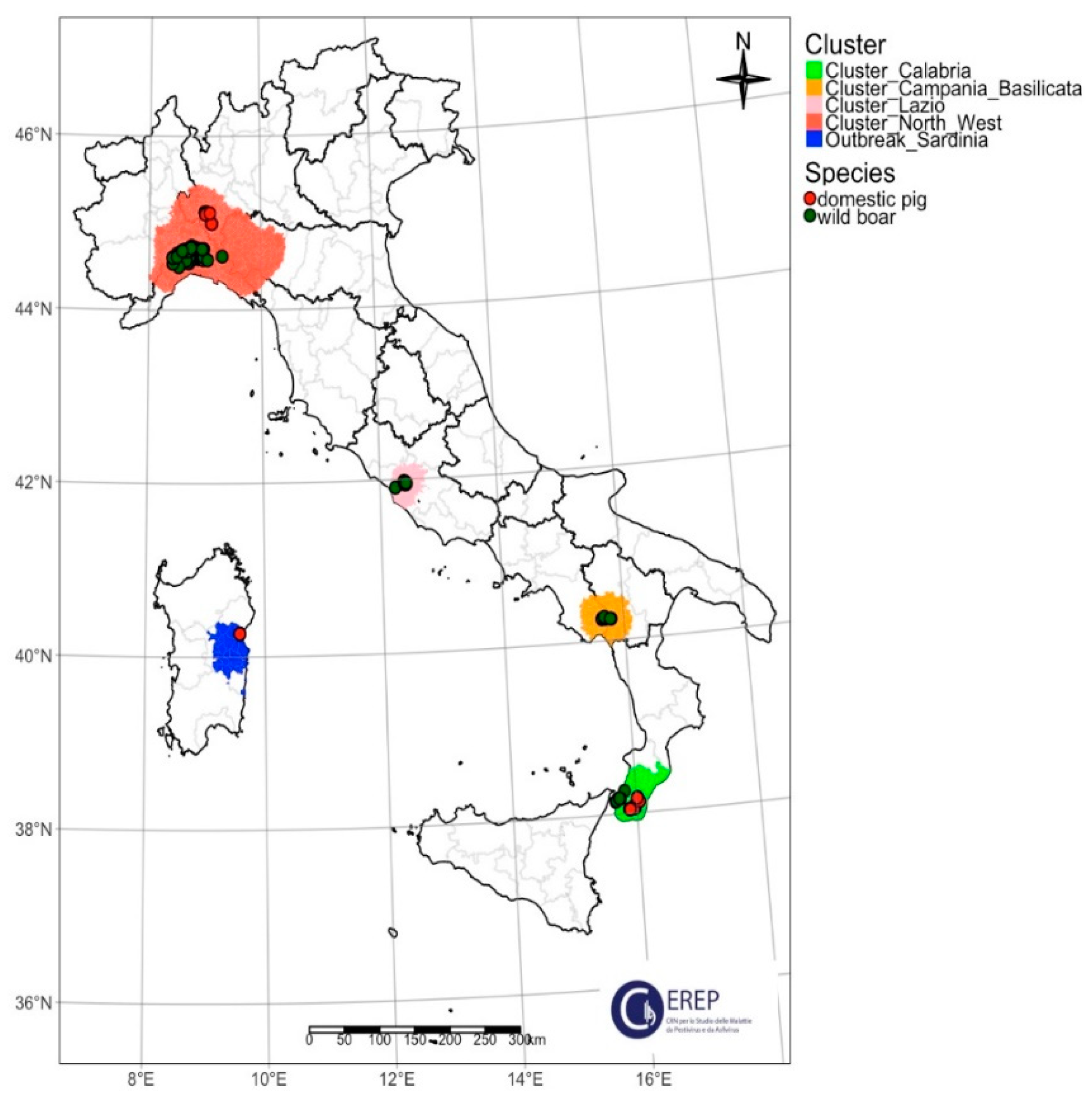
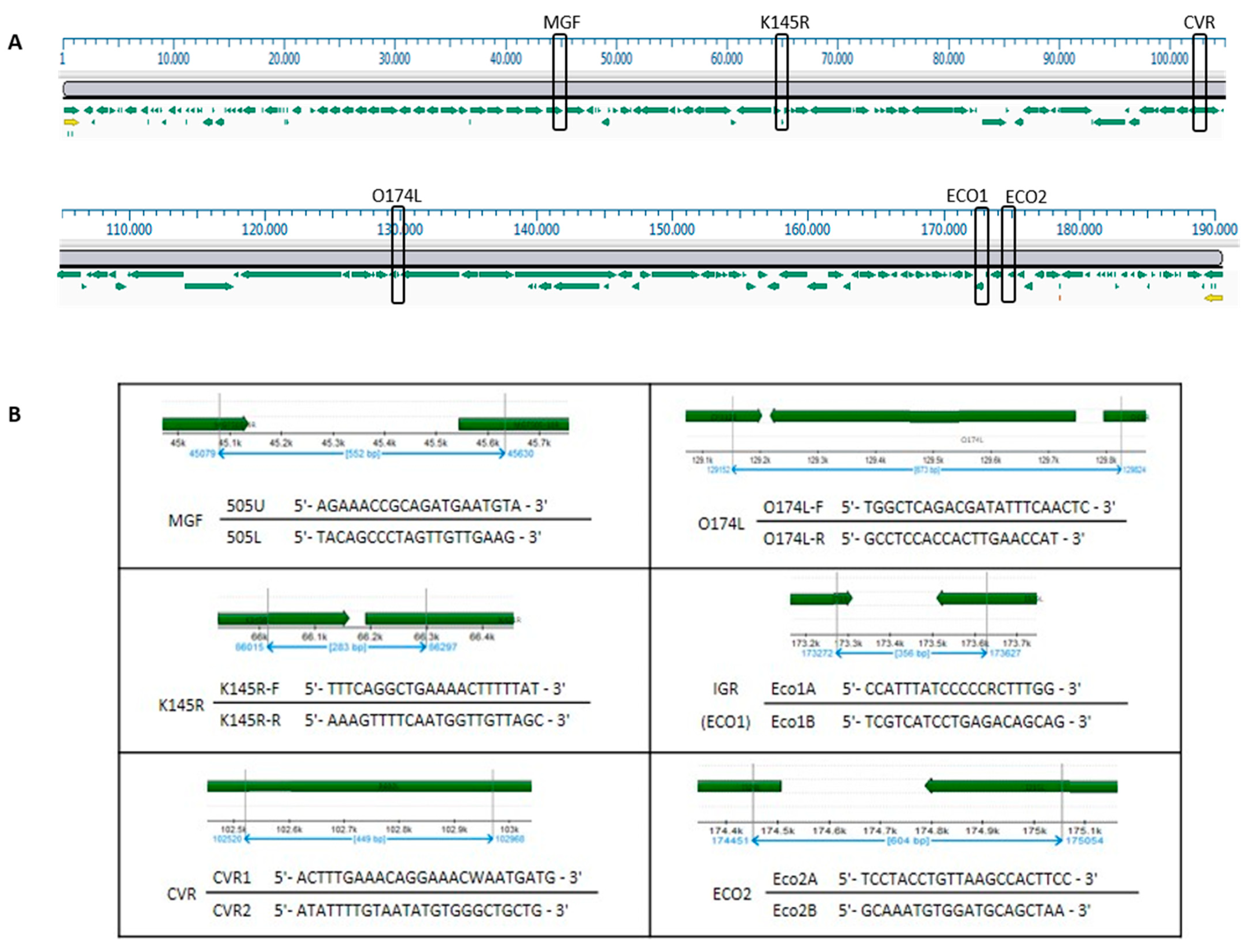
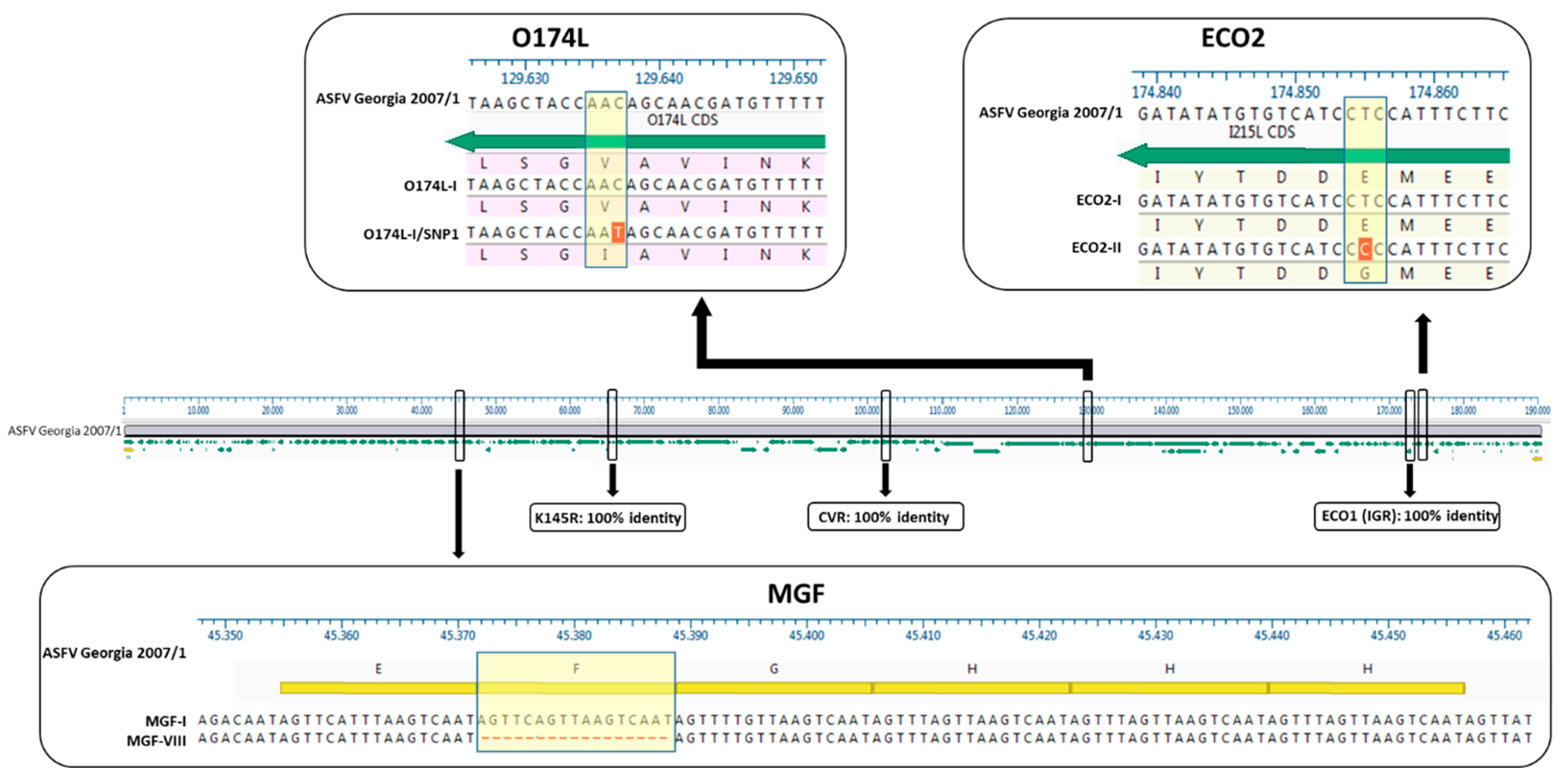

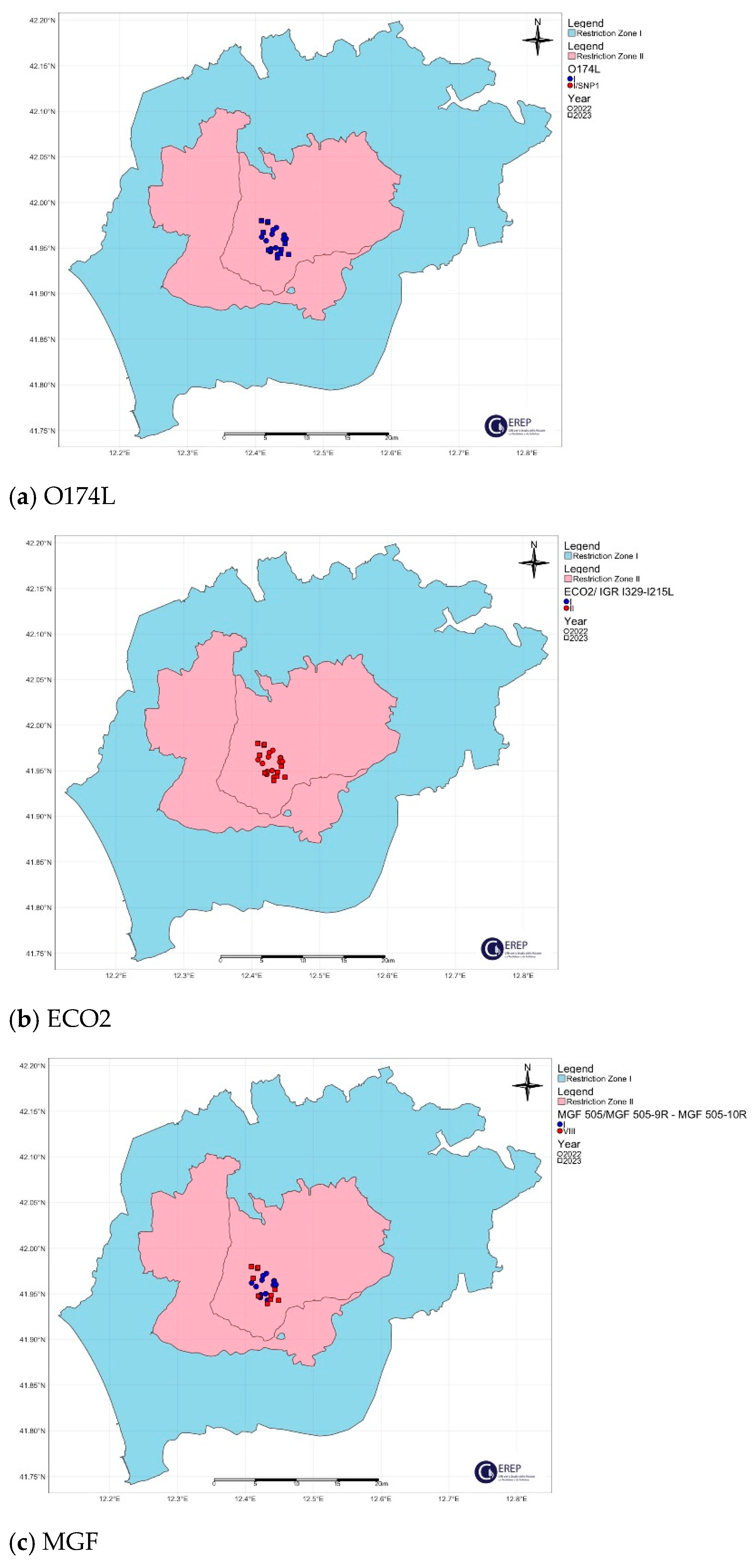
| Cluster/Outbreak | Region | Provinces | Collecting Date From–To (DD/MM/YYYY) | Host | Total | |
|---|---|---|---|---|---|---|
| Wild Boar | Domestic Pig | |||||
| Northwest | Piedmont | Alessandria | 07/01/2022–24/02/2023 | 46 | 46 | |
| Liguria | Genoa, Savona | 13/01/2022–22/12/2022 | 11 | 11 | ||
| Lombardy | Pavia | 16/08/2023–07/09/2023 | 1 | 7 | 8 | |
| Emilia Romagna | Piacenza | 16/11/2023 | 1 | 1 | ||
| Lazio | Lazio | Rome | 29/04/2022–11/07/2023 | 28 | 2 | 30 |
| Campania–Basilicata | Campania | Salerno | 22/05/2023–10/06/2023 | 9 | 9 | |
| Calabria | Reggio Calabria | Reggio Calabria | 03/05/2023–17/07/2023 | 7 | 17 | 24 |
| Sardinia | Sardinia | Nuoro | 19/09/2023 | 3 | 3 | |
| 103 | 29 | 132 | ||||
| Cluster/Outbreak | Provinces | CVR | IGR-ECO1 | O174L | K145R | MGF | ECO2 | Genetic Group |
|---|---|---|---|---|---|---|---|---|
| Northwest | Alessandria | I | II | I (91.3%) | I | I | I | 3 |
| I-SNP1 (8.7%) 1 | 26 | |||||||
| Genoa, Savona | I | II | I | I | I | I | 3 | |
| Pavia | I | II | I | I | I | I | 3 | |
| Piacenza | I | II | I | I | I | I | 3 | |
| Lazio | Rome | I | II | I | I | I (60%) 2 | II | 19 |
| VIII (40%) 3 | 25 | |||||||
| Campania–Basilicata | Salerno | I | II | I | I | I | I | 3 |
| Calabria | Reggio Calabria | I | II | I | I | I | II | 19 |
| Sardinia | Nuoro | I | II | I | I | I | I | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giammarioli, M.; Torresi, C.; Biccheri, R.; Cammà, C.; Marcacci, M.; Dondo, A.; Razzuoli, E.; Fusco, G.; Casalinuovo, F.; Scicluna, M.T.; et al. Genetic Characterization of African Swine Fever Italian Clusters in the 2022–2023 Epidemic Wave by a Multi-Gene Approach. Viruses 2024, 16, 1185. https://doi.org/10.3390/v16081185
Giammarioli M, Torresi C, Biccheri R, Cammà C, Marcacci M, Dondo A, Razzuoli E, Fusco G, Casalinuovo F, Scicluna MT, et al. Genetic Characterization of African Swine Fever Italian Clusters in the 2022–2023 Epidemic Wave by a Multi-Gene Approach. Viruses. 2024; 16(8):1185. https://doi.org/10.3390/v16081185
Chicago/Turabian StyleGiammarioli, Monica, Claudia Torresi, Roberta Biccheri, Cesare Cammà, Maurilia Marcacci, Alessandro Dondo, Elisabetta Razzuoli, Giovanna Fusco, Francesco Casalinuovo, Maria Teresa Scicluna, and et al. 2024. "Genetic Characterization of African Swine Fever Italian Clusters in the 2022–2023 Epidemic Wave by a Multi-Gene Approach" Viruses 16, no. 8: 1185. https://doi.org/10.3390/v16081185
APA StyleGiammarioli, M., Torresi, C., Biccheri, R., Cammà, C., Marcacci, M., Dondo, A., Razzuoli, E., Fusco, G., Casalinuovo, F., Scicluna, M. T., Dei Giudici, S., Martin, A. M. M., Rossi, E., Casciari, C., Pela, M., Iscaro, C., Gallardo, C., Marocco, G., Orrico, M., & Feliziani, F. (2024). Genetic Characterization of African Swine Fever Italian Clusters in the 2022–2023 Epidemic Wave by a Multi-Gene Approach. Viruses, 16(8), 1185. https://doi.org/10.3390/v16081185







