The M2 Protein of the Influenza A Virus Interacts with PEX19 to Facilitate Virus Replication by Disrupting the Function of Peroxisome
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Yeast Two-Hybrid Assay
2.3. Plasmids
2.4. Antibodies
2.5. Co-Immunoprecipitation Assay
2.6. Western Blotting
2.7. Cell Viability Assay
2.8. siRNA Knockdown and Virus Infection
2.9. Generation of PEX19_KO A549 Cells and Virus Infection
2.10. Establishment of an A549 Stable Cell Line Overexpressing PEX19 and Virus Infection
2.11. Plaque Assay
2.12. Peroxisome and Reactive Oxygen Species (ROS) Detection
2.13. Confocal Microscopy
2.14. Quantitative Reverse Transcription PCR (RT-qPCR) Analysis
2.15. Statistical Analysis
3. Results
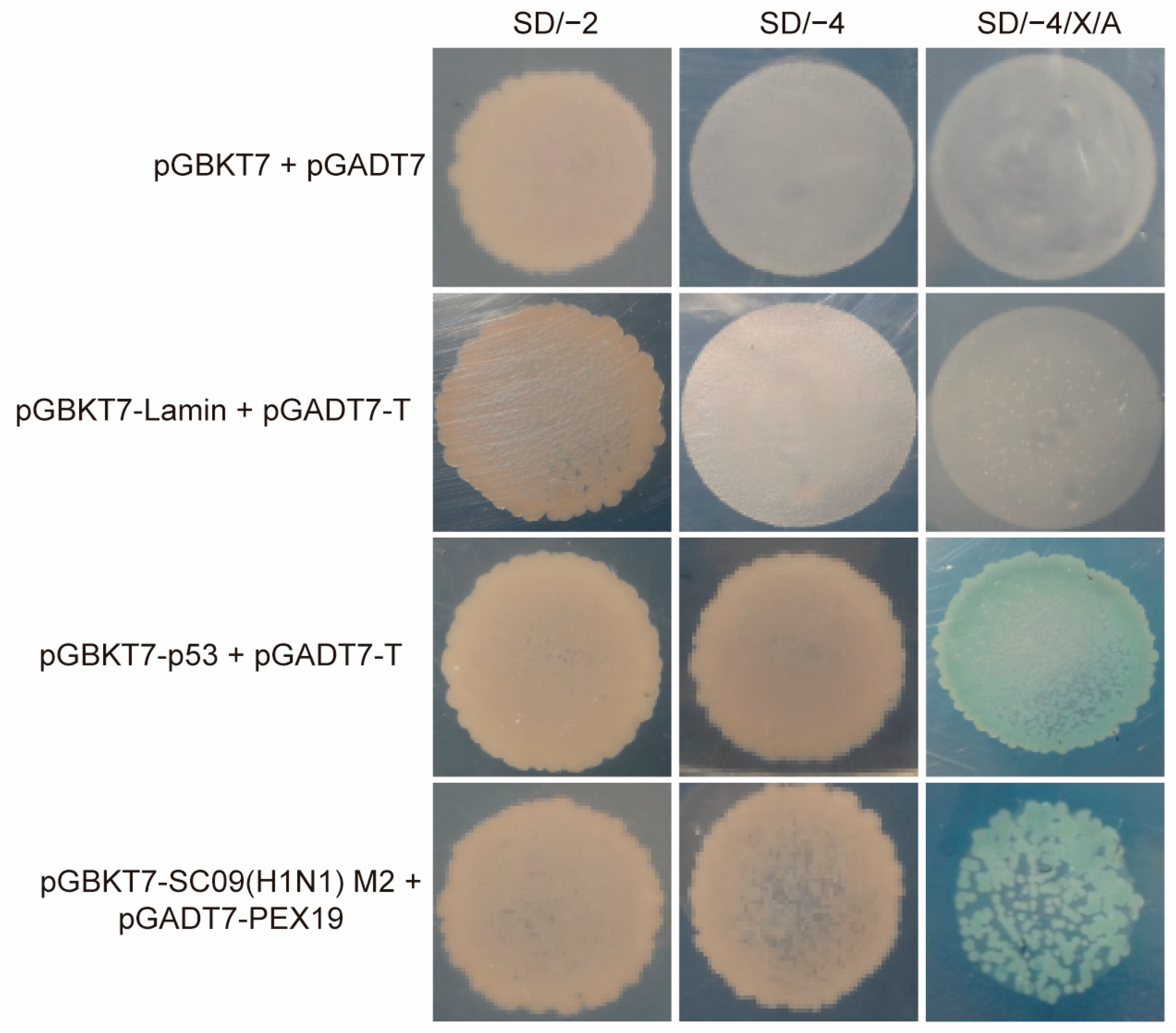
3.1. Identification of PEX19 as a Potential Interacting Host Factor of IAV M2 by the Yeast Two-Hybrid System
3.2. IAV M2 Interacts with PEX19 in Human Cells
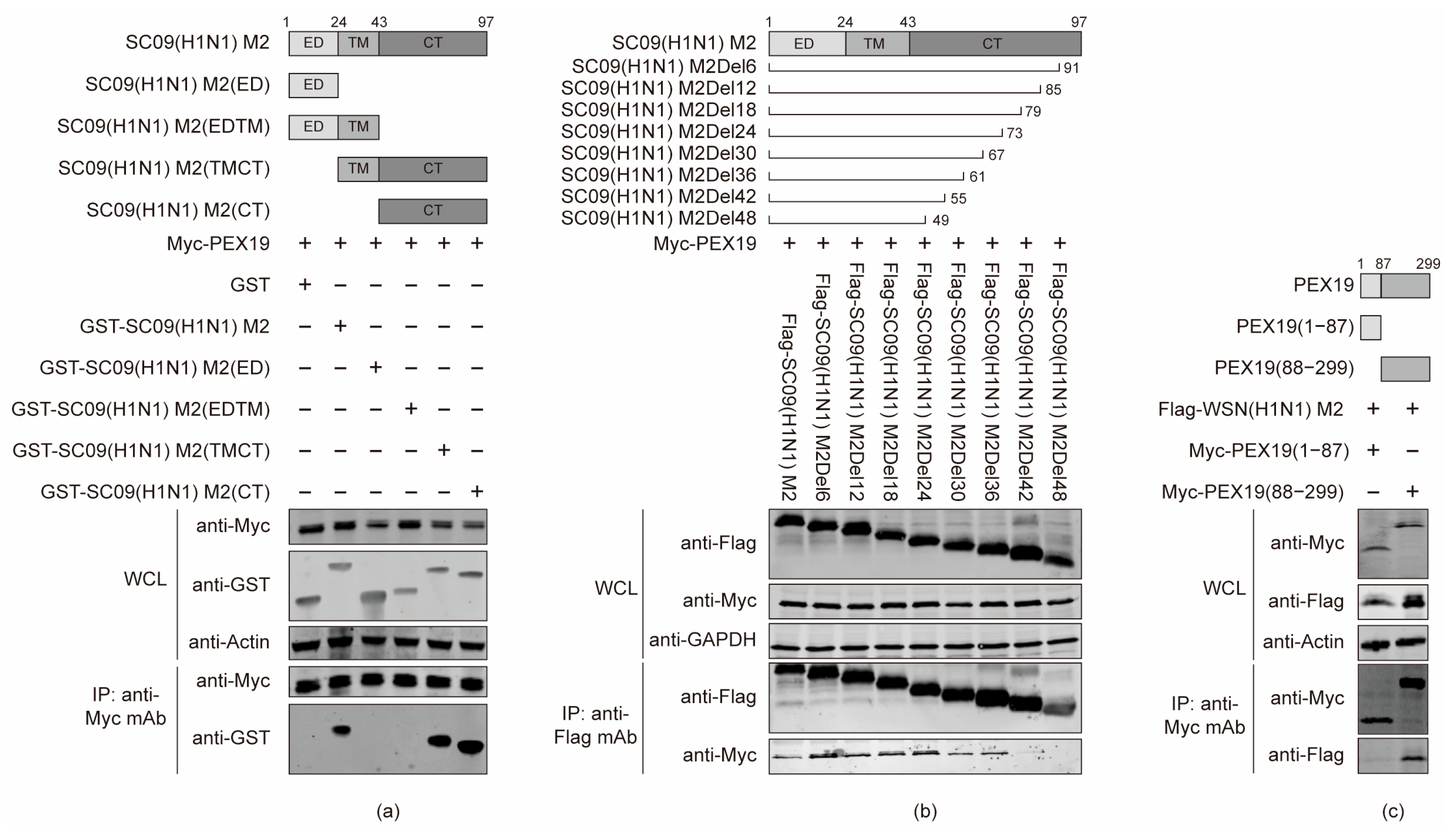
3.3. The Cytoplasmic Tail Domain of M2 and the C Terminus of PEX19 Are Key Regions of Interactions
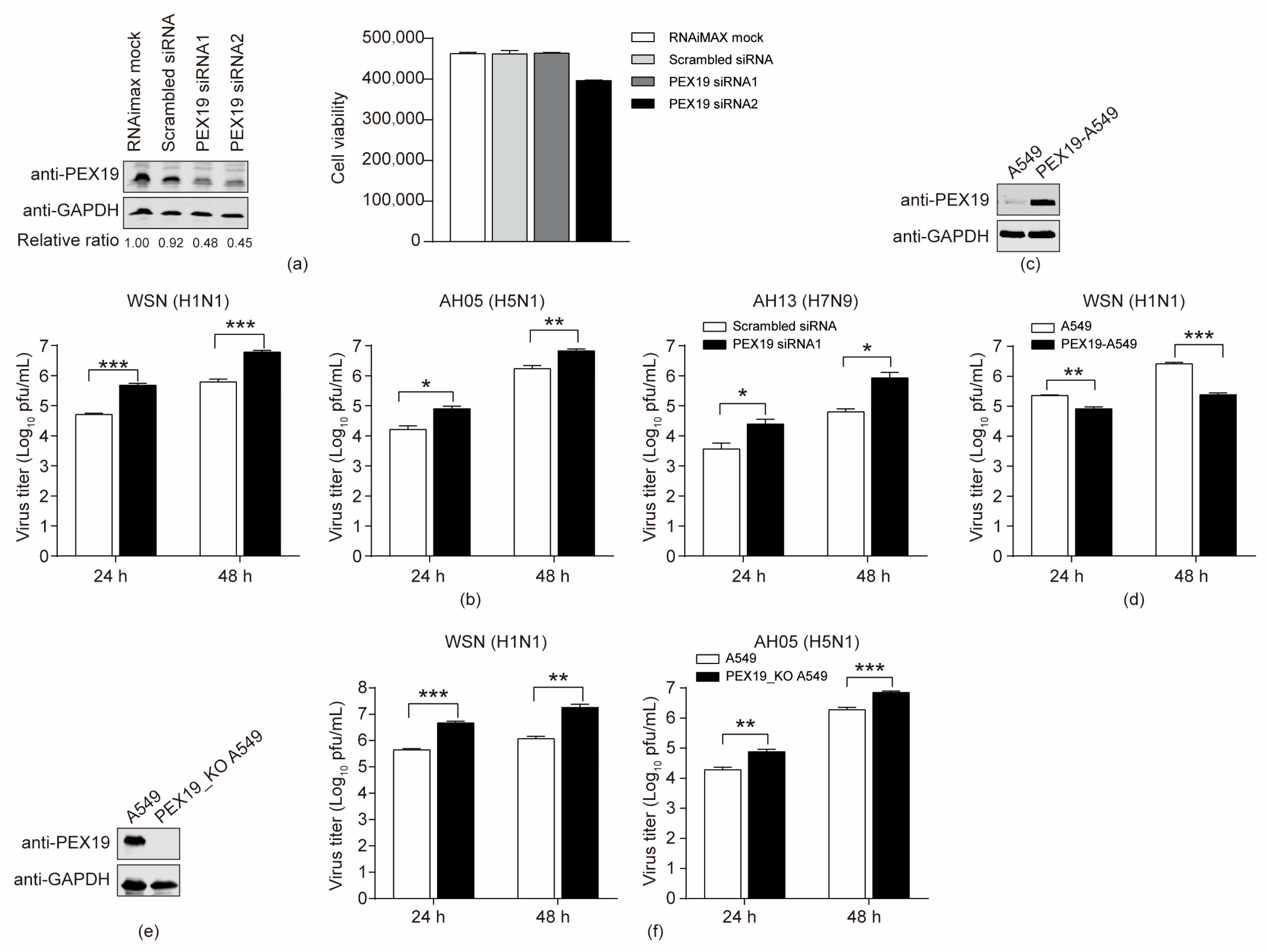
3.4. PEX19 Inhibits the Replication of IAV
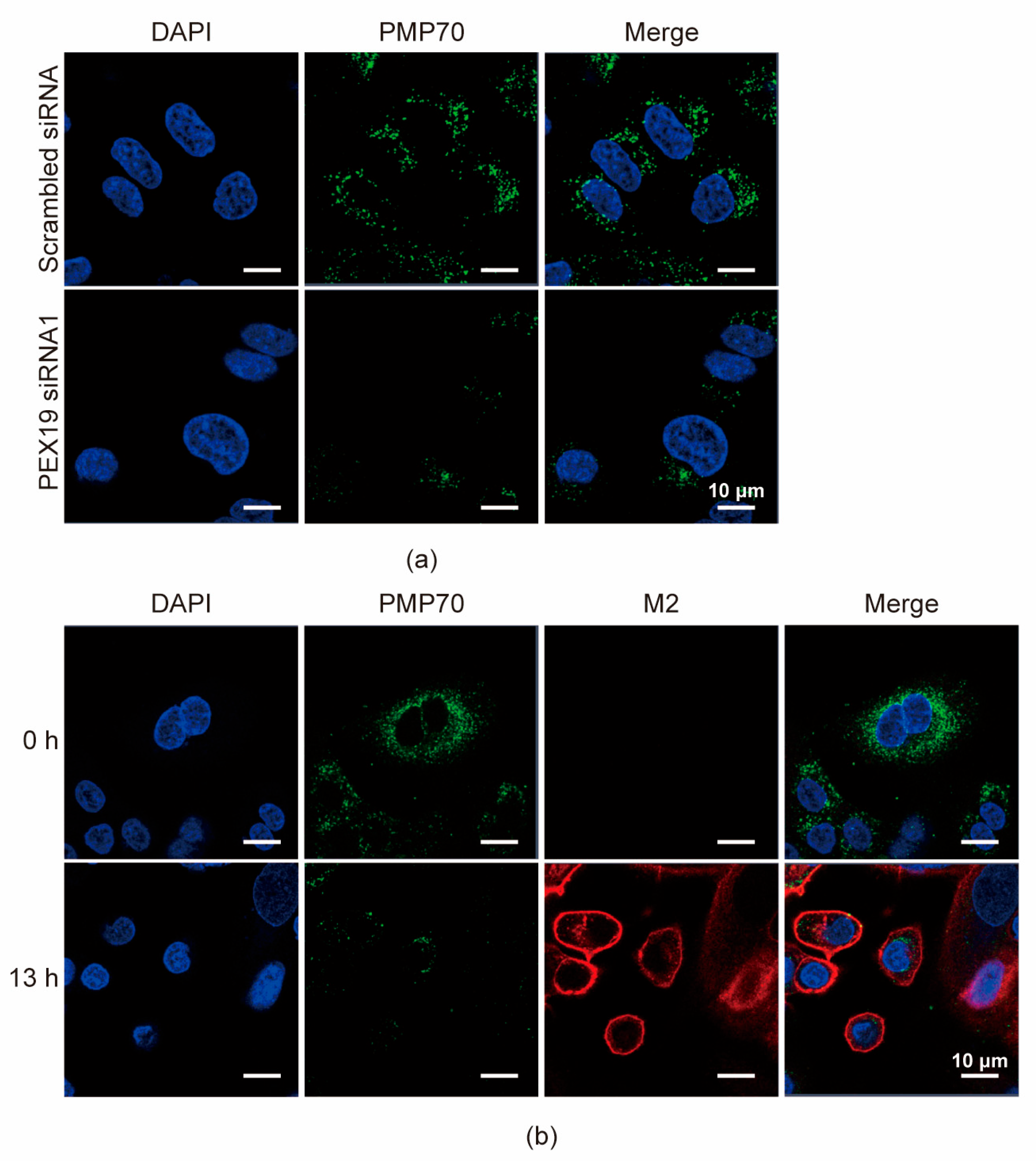
3.5. IAV Infection Decreases the Pool of Peroxisomes in A549 Cells
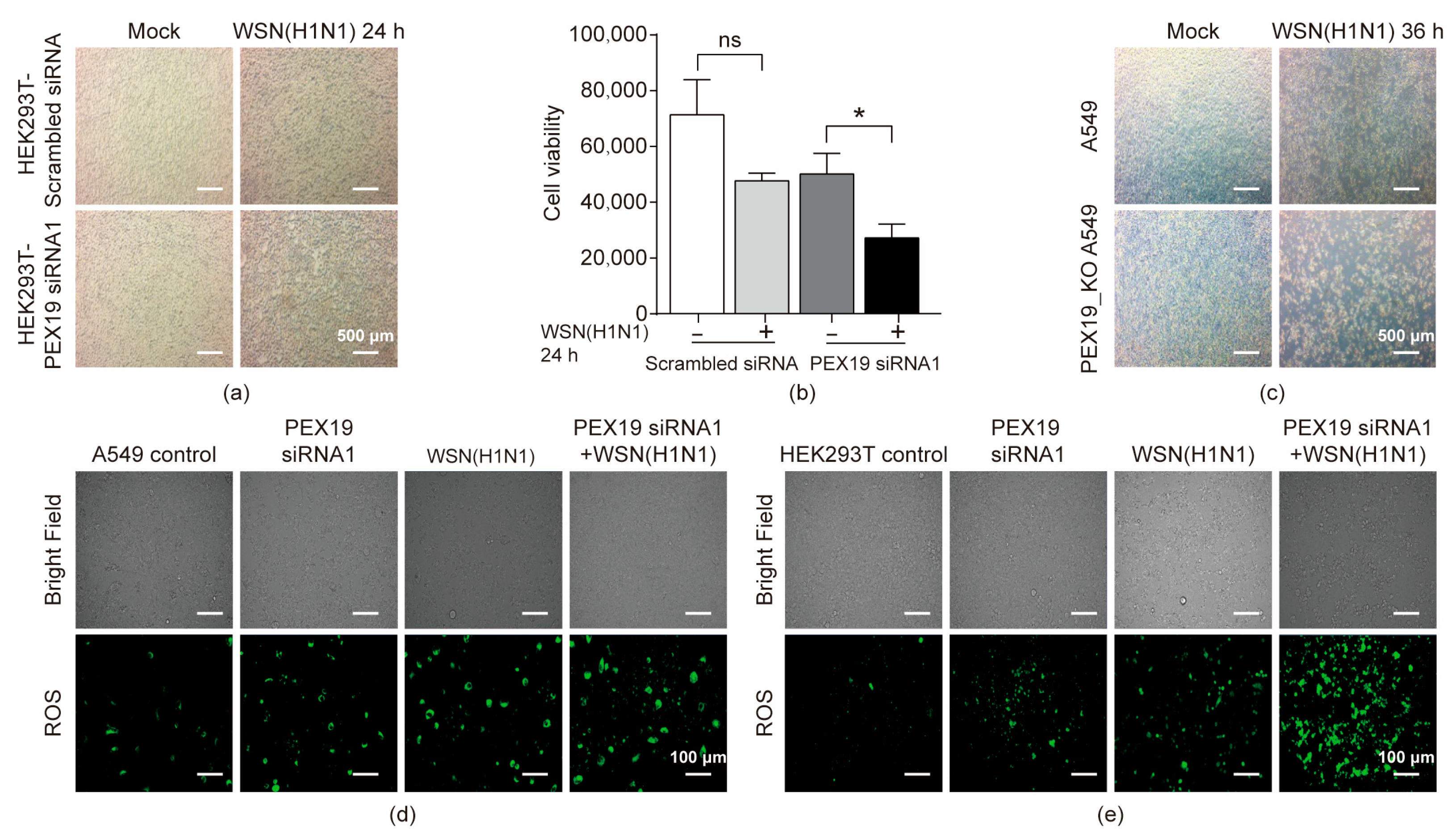
3.6. PEX19 Deficiency Promotes Cell Damage in IAV-Infected Cells by Inducing ROS and Compromising the ROS-Processing Function of Peroxisomes
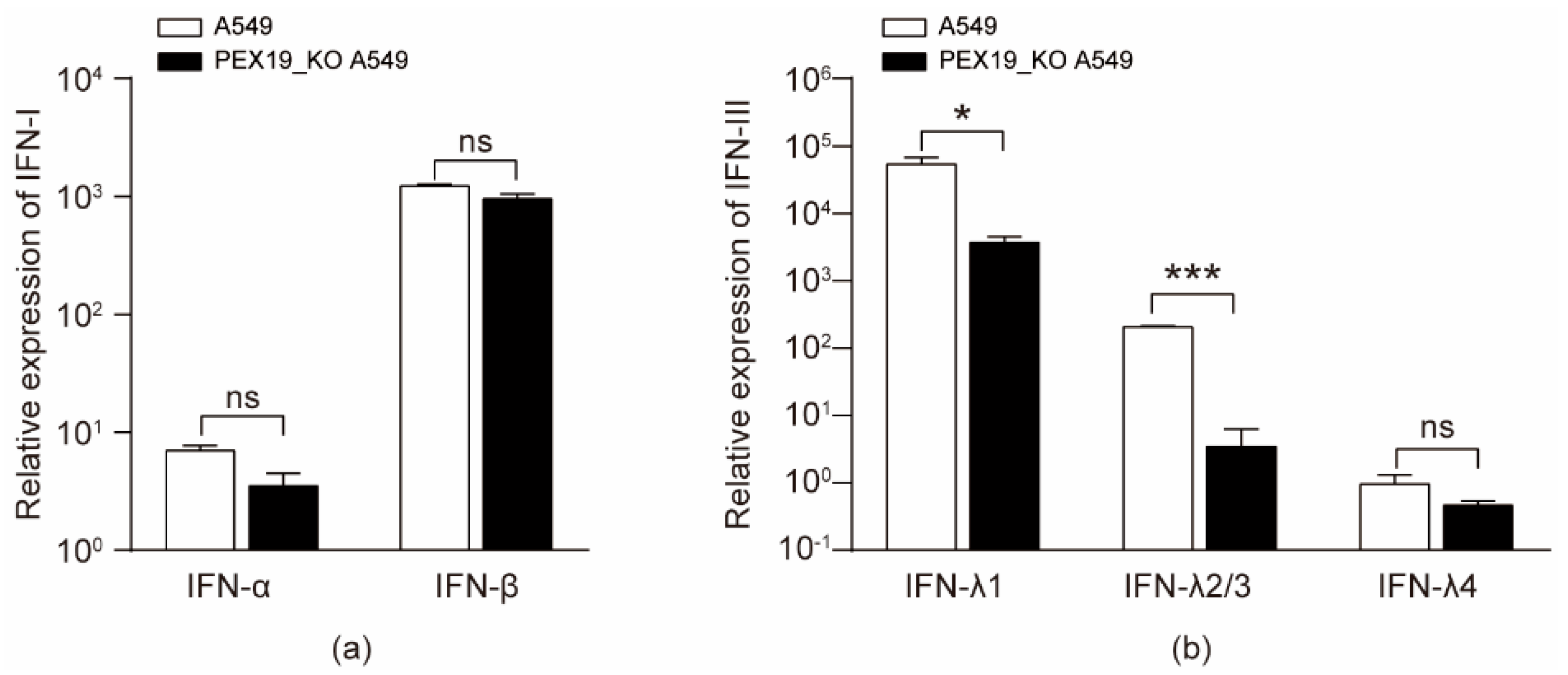
3.7. IAV Infection and PEX19 Knockout Reduce the Levels of Type III Interferons
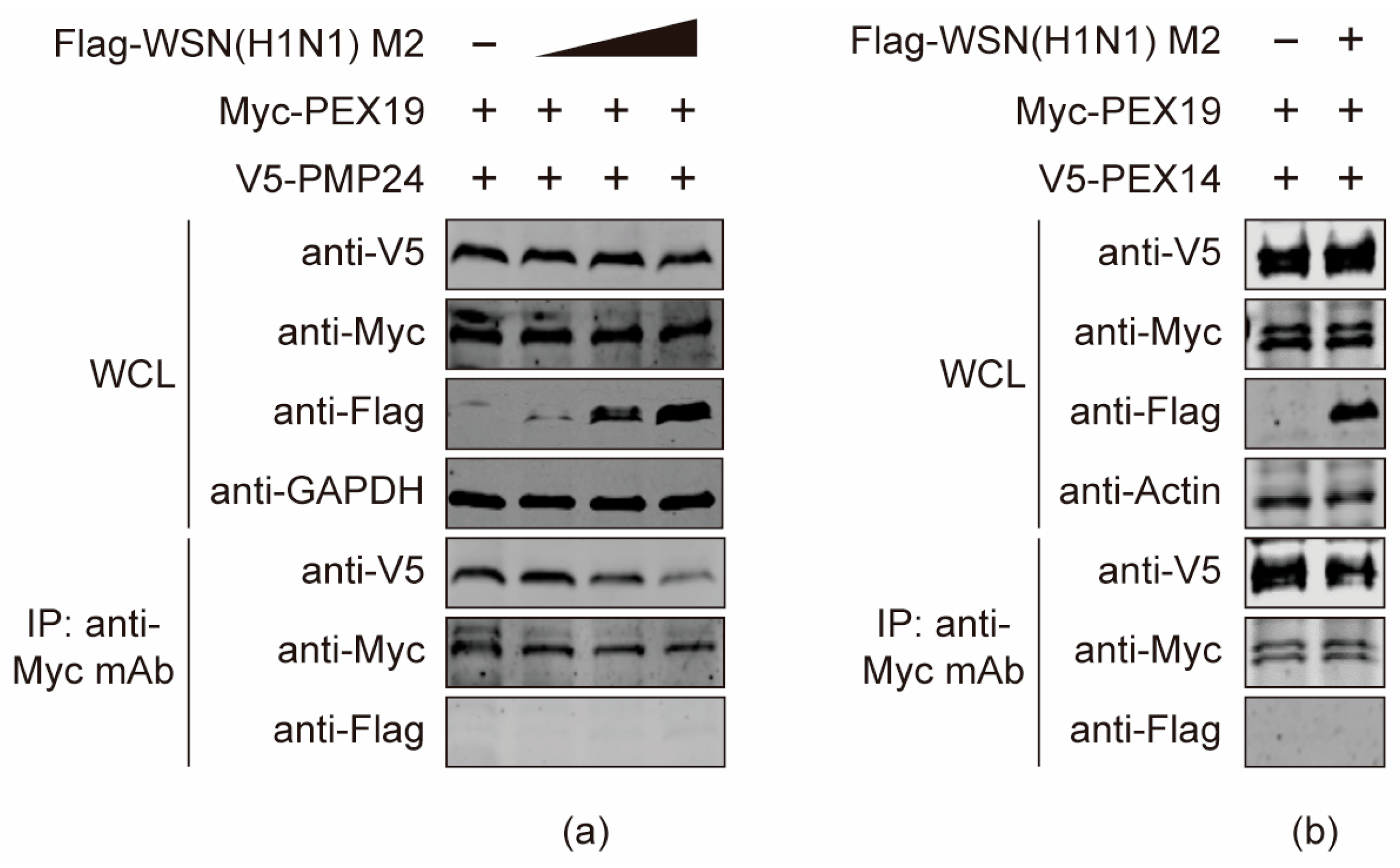
3.8. The M2 Protein Disturbs the Interactions between PEX19 and Peroxisome-Associated Factors
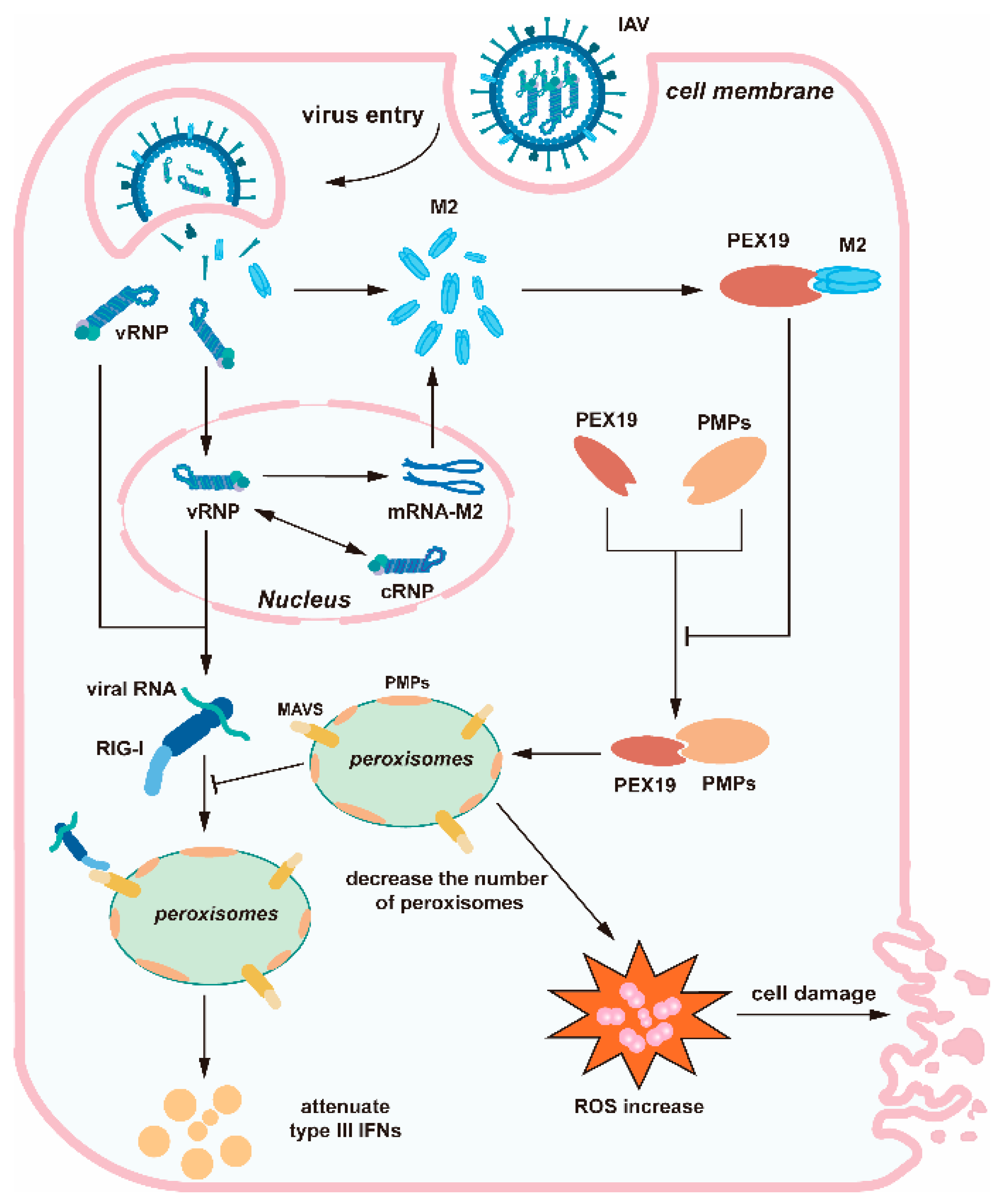
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Taubenberger, J.K.; Kash, J.C. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 2010, 7, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yuan, H.; Gao, R.; Zhang, J.; Wang, D.; Xiong, Y.; Fan, G.; Yang, F.; Li, X.; Zhou, J.; et al. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: A descriptive study. Lancet 2014, 383, 714–721. [Google Scholar] [CrossRef]
- Yang, R.; Sun, H.; Gao, F.; Luo, K.; Huang, Z.; Tong, Q.; Song, H.; Han, Q.; Liu, J.; Lan, Y.; et al. Human infection of avian influenza A H3N8 virus and the viral origins: A descriptive study. Lancet Microbe 2022, 3, e824–e834. [Google Scholar] [CrossRef] [PubMed]
- Carnaccini, S.; Perez, D.R. H9 Influenza Viruses: An Emerging Challenge. Cold Spring Harb. Perspect. Med. 2020, 10, a038588. [Google Scholar] [CrossRef]
- Shi, J.; Zeng, X.; Cui, P.; Yan, C.; Chen, H. Alarming situation of emerging H5 and H7 avian influenza and effective control strategies. Emerg. Microbes Infect. 2023, 12, 2155072. [Google Scholar] [CrossRef]
- Gu, W.; Shi, J.; Cui, P.; Yan, C.; Zhang, Y.; Wang, C.; Zhang, Y.; Xing, X.; Zeng, X.; Liu, L.; et al. Novel H5N6 reassortants bearing the clade 2.3.4.4b HA gene of H5N8 virus have been detected in poultry and caused multiple human infections in China. Emerg. Microbes Infect. 2022, 11, 1174–1185. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Cao, B.; Hu, Y.; Feng, Z.; Wang, D.; Hu, W.; Chen, J.; Jie, Z.; Qiu, H.; Xu, K.; et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 2013, 368, 1888–1897. [Google Scholar] [CrossRef] [PubMed]
- Hull, J.D.; Gilmore, R.; Lamb, R.A. Integration of a small integral membrane protein, M2, of influenza virus into the endoplasmic reticulum: Analysis of the internal signal-anchor domain of a protein with an ectoplasmic NH2 terminus. J. Cell Biol. 1988, 106, 1489–1498. [Google Scholar] [CrossRef]
- Lamb, R.A.; Zebedee, S.L.; Richardson, C.D. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 1985, 40, 627–633. [Google Scholar] [CrossRef]
- Pinto, L.H.; Lamb, R.A. The M2 proton channels of influenza A and B viruses. J. Biol. Chem. 2006, 281, 8997–9000. [Google Scholar] [CrossRef]
- Alvarado-Facundo, E.; Gao, Y.; Ribas-Aparicio, R.M.; Jimenez-Alberto, A.; Weiss, C.D.; Wang, W. Influenza virus M2 protein ion channel activity helps to maintain pandemic 2009 H1N1 virus hemagglutinin fusion competence during transport to the cell surface. J. Virol. 2015, 89, 1975–1985. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Lamb, R.A. Influenza virus M2 protein ion channel activity stabilizes the native form of fowl plague virus hemagglutinin during intracellular transport. J. Virol. 1994, 68, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Iwatsuki-Horimoto, K.; Horimoto, T.; Noda, T.; Kiso, M.; Maeda, J.; Watanabe, S.; Muramoto, Y.; Fujii, K.; Kawaoka, Y. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. J. Virol. 2006, 80, 5233–5240. [Google Scholar] [CrossRef]
- Rossman, J.S.; Jing, X.; Leser, G.P.; Lamb, R.A. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 2010, 142, 902–913. [Google Scholar] [CrossRef] [PubMed]
- Beale, R.; Wise, H.; Stuart, A.; Ravenhill, B.J.; Digard, P.; Randow, F. A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell Host Microbe 2014, 15, 239–247. [Google Scholar] [CrossRef]
- Gannage, M.; Dormann, D.; Albrecht, R.; Dengjel, J.; Torossi, T.; Ramer, P.C.; Lee, M.; Strowig, T.; Arrey, F.; Conenello, G.; et al. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 2009, 6, 367–380. [Google Scholar] [CrossRef]
- Ma, H.; Kien, F.; Maniere, M.; Zhang, Y.; Lagarde, N.; Tse, K.S.; Poon, L.L.; Nal, B. Human annexin A6 interacts with influenza a virus protein M2 and negatively modulates infection. J. Virol. 2012, 86, 1789–1801. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Li, Y.; Yan, J.; Gao, G.F. Na+/K+-ATPase beta1 subunit interacts with M2 proteins of influenza A and B viruses and affects the virus replication. Sci. China Life Sci. 2010, 53, 1098–1105. [Google Scholar] [CrossRef]
- Tripathi, S.; Pohl, M.O.; Zhou, Y.; Rodriguez-Frandsen, A.; Wang, G.; Stein, D.A.; Moulton, H.M.; DeJesus, P.; Che, J.; Mulder, L.C.; et al. Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe 2015, 18, 723–735. [Google Scholar] [CrossRef]
- Zhu, P.; Liang, L.; Shao, X.; Luo, W.; Jiang, S.; Zhao, Q.; Sun, N.; Zhao, Y.; Li, J.; Wang, J.; et al. Host Cellular Protein TRAPPC6ADelta Interacts with Influenza A Virus M2 Protein and Regulates Viral Propagation by Modulating M2 Trafficking. J. Virol. 2017, 91, e01757-16. [Google Scholar] [CrossRef]
- Kammerer, S.; Arnold, N.; Gutensohn, W.; Mewes, H.W.; Kunau, W.H.; Hofler, G.; Roscher, A.A.; Braun, A. Genomic organization and molecular characterization of a gene encoding HsPXF, a human peroxisomal farnesylated protein. Genomics 1997, 45, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Sacksteder, K.A.; Jones, J.M.; South, S.T.; Li, X.; Liu, Y.; Gould, S.J. PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J. Cell Biol. 2000, 148, 931–944. [Google Scholar] [CrossRef] [PubMed]
- Matsuzono, Y.; Kinoshita, N.; Tamura, S.; Shimozawa, N.; Hamasaki, M.; Ghaedi, K.; Wanders, R.J.; Suzuki, Y.; Kondo, N.; Fujiki, Y. Human PEX19: cDNA cloning by functional complementation, mutation analysis in a patient with Zellweger syndrome, and potential role in peroxisomal membrane assembly. Proc. Natl. Acad. Sci. USA 1999, 96, 2116–2121. [Google Scholar] [CrossRef]
- Jansen, R.L.M.; van der Klei, I.J. The peroxisome biogenesis factors Pex3 and Pex19: Multitasking proteins with disputed functions. FEBS Lett. 2019, 593, 457–474. [Google Scholar] [CrossRef] [PubMed]
- Schueller, N.; Holton, S.J.; Fodor, K.; Milewski, M.; Konarev, P.; Stanley, W.A.; Wolf, J.; Erdmann, R.; Schliebs, W.; Song, Y.H.; et al. The peroxisomal receptor Pex19p forms a helical mPTS recognition domain. EMBO J. 2010, 29, 2491–2500. [Google Scholar] [CrossRef] [PubMed]
- Indari, O.; Rani, A.; Baral, B.; Ergun, S.; Bala, K.; Karnati, S.; Jha, H.C. Modulation of peroxisomal compartment by Epstein-Barr virus. Microb. Pathog. 2023, 174, 105946. [Google Scholar] [CrossRef]
- Magalhaes, A.C.; Ferreira, A.R.; Gomes, S.; Vieira, M.; Gouveia, A.; Valenca, I.; Islinger, M.; Nascimento, R.; Schrader, M.; Kagan, J.C.; et al. Peroxisomes are platforms for cytomegalovirus’ evasion from the cellular immune response. Sci. Rep. 2016, 6, 26028. [Google Scholar] [CrossRef]
- You, J.; Hou, S.; Malik-Soni, N.; Xu, Z.; Kumar, A.; Rachubinski, R.A.; Frappier, L.; Hobman, T.C. Flavivirus Infection Impairs Peroxisome Biogenesis and Early Antiviral Signaling. J. Virol. 2015, 89, 12349–12361. [Google Scholar] [CrossRef]
- Choi, Y.B.; Choi, Y.; Harhaj, E.W. Peroxisomes support human herpesvirus 8 latency by stabilizing the viral oncogenic protein vFLIP via the MAVS-TRAF complex. PLoS Pathog. 2018, 14, e1007058. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, L.; Wang, G.; Song, Y.; Shan, Z.; Wang, X.; Deng, G.; Shi, J.; Tian, G.; Zeng, X.; et al. M6PR interacts with the HA2 subunit of influenza A virus to facilitate the fusion of viral and endosomal membranes. Sci. China Life Sci. 2024, 67, 579–595. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, L.; Wang, G.; Shi, W.; Hu, Y.; Wang, B.; Zeng, X.; Tian, G.; Deng, G.; Shi, J.; et al. Influenza A virus use of BinCARD1 to facilitate the binding of viral NP to importin alpha7 is counteracted by TBK1-p62 axis-mediated autophagy. Cell Mol. Immunol. 2022, 19, 1168–1184. [Google Scholar] [CrossRef]
- Vlahos, R.; Stambas, J.; Selemidis, S. Suppressing production of reactive oxygen species (ROS) for influenza A virus therapy. Trends Pharmacol. Sci. 2012, 33, 3–8. [Google Scholar] [CrossRef]
- Sugiura, A.; Mattie, S.; Prudent, J.; McBride, H.M. Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature 2017, 542, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Lodhi, I.J.; Semenkovich, C.F. Peroxisomes: A nexus for lipid metabolism and cellular signaling. Cell Metab. 2014, 19, 380–392. [Google Scholar] [CrossRef]
- Malik, G.; Zhou, Y. Innate Immune Sensing of Influenza A Virus. Viruses 2020, 12, 755. [Google Scholar] [CrossRef] [PubMed]
- Bender, S.; Reuter, A.; Eberle, F.; Einhorn, E.; Binder, M.; Bartenschlager, R. Activation of Type I and III Interferon Response by Mitochondrial and Peroxisomal MAVS and Inhibition by Hepatitis C Virus. PLoS Pathog. 2015, 11, e1005264. [Google Scholar] [CrossRef] [PubMed]
- Odendall, C.; Dixit, E.; Stavru, F.; Bierne, H.; Franz, K.M.; Durbin, A.F.; Boulant, S.; Gehrke, L.; Cossart, P.; Kagan, J.C. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 2014, 15, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, G.; Shang, H.H.; Xia, Z.J.; Subramani, S. Functional regions of the peroxin Pex19 necessary for peroxisome biogenesis. J. Biol. Chem. 2017, 292, 11547–11560. [Google Scholar] [CrossRef]
- Emmanouilidis, L.; Schutz, U.; Tripsianes, K.; Madl, T.; Radke, J.; Rucktaschel, R.; Wilmanns, M.; Schliebs, W.; Erdmann, R.; Sattler, M. Allosteric modulation of peroxisomal membrane protein recognition by farnesylation of the peroxisomal import receptor PEX19. Nat. Commun. 2017, 8, 14635. [Google Scholar] [CrossRef]
- Albertini, M.; Rehling, P.; Erdmann, R.; Girzalsky, W.; Kiel, J.A.; Veenhuis, M.; Kunau, W.H. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell 1997, 89, 83–92. [Google Scholar] [CrossRef]
- Emmanouilidis, L.; Gopalswamy, M.; Passon, D.M.; Wilmanns, M.; Sattler, M. Structural biology of the import pathways of peroxisomal matrix proteins. Biochim. Biophys. Acta 2016, 1863, 804–813. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Anderson-Baron, M.N.; Haskins, J.; Hughes, S.C.; Simmonds, A.J. Recruitment of Peroxin 14 to lipid droplets affects lipid storage in Drosophila. J. Cell Sci. 2022, 135, jcs259092. [Google Scholar] [CrossRef] [PubMed]
- Blankestijn, M.; Bloks, V.W.; Struik, D.; Huijkman, N.; Kloosterhuis, N.; Wolters, J.C.; Wanders, R.J.A.; Vaz, F.M.; Islinger, M.; Kuipers, F.; et al. Mice with a deficiency in Peroxisomal Membrane Protein 4 (PXMP4) display mild changes in hepatic lipid metabolism. Sci. Rep. 2022, 12, 2512. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.; Liang, L.; Zhao, P.; Lin, W.; Zhuang, Y.; Jiang, L.; Chen, H.; Li, C. The M2 Protein of the Influenza A Virus Interacts with PEX19 to Facilitate Virus Replication by Disrupting the Function of Peroxisome. Viruses 2024, 16, 1309. https://doi.org/10.3390/v16081309
Liu T, Liang L, Zhao P, Lin W, Zhuang Y, Jiang L, Chen H, Li C. The M2 Protein of the Influenza A Virus Interacts with PEX19 to Facilitate Virus Replication by Disrupting the Function of Peroxisome. Viruses. 2024; 16(8):1309. https://doi.org/10.3390/v16081309
Chicago/Turabian StyleLiu, Tanbin, Libin Liang, Pu Zhao, Weipeng Lin, Yichao Zhuang, Li Jiang, Hualan Chen, and Chengjun Li. 2024. "The M2 Protein of the Influenza A Virus Interacts with PEX19 to Facilitate Virus Replication by Disrupting the Function of Peroxisome" Viruses 16, no. 8: 1309. https://doi.org/10.3390/v16081309
APA StyleLiu, T., Liang, L., Zhao, P., Lin, W., Zhuang, Y., Jiang, L., Chen, H., & Li, C. (2024). The M2 Protein of the Influenza A Virus Interacts with PEX19 to Facilitate Virus Replication by Disrupting the Function of Peroxisome. Viruses, 16(8), 1309. https://doi.org/10.3390/v16081309






