SARS-CoV-2 Modulation of HIV Latency Reversal in a Myeloid Cell Line: Direct and Bystander Effects
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines, Primary Cells, and Reagents
- the unstimulated group that was not exposed to reactivating stimuli (negative control);
- the stimulated group incubated with phorbol 12-myristate 13-acetate (PMA; P8139, Merck, St. Louis, MO, USA) at 30 ng/mL to induce proviral reactivation from latency (positive control);
- the study group exposed to different candidate stimuli.
2.2. SARS-CoV-2 Variants, Viral RNA Quantification, and MDM Infection
2.3. SARS-CoV-2 Inactivation via UVC Irradiation
2.4. Flow cytometry Analysis
2.5. ELISA for TNF-α, IL-6, and IL-1β Detection
2.6. TNF-α Neutralization
2.7. Statistical Analysis
2.8. Ethical Approval
3. Results
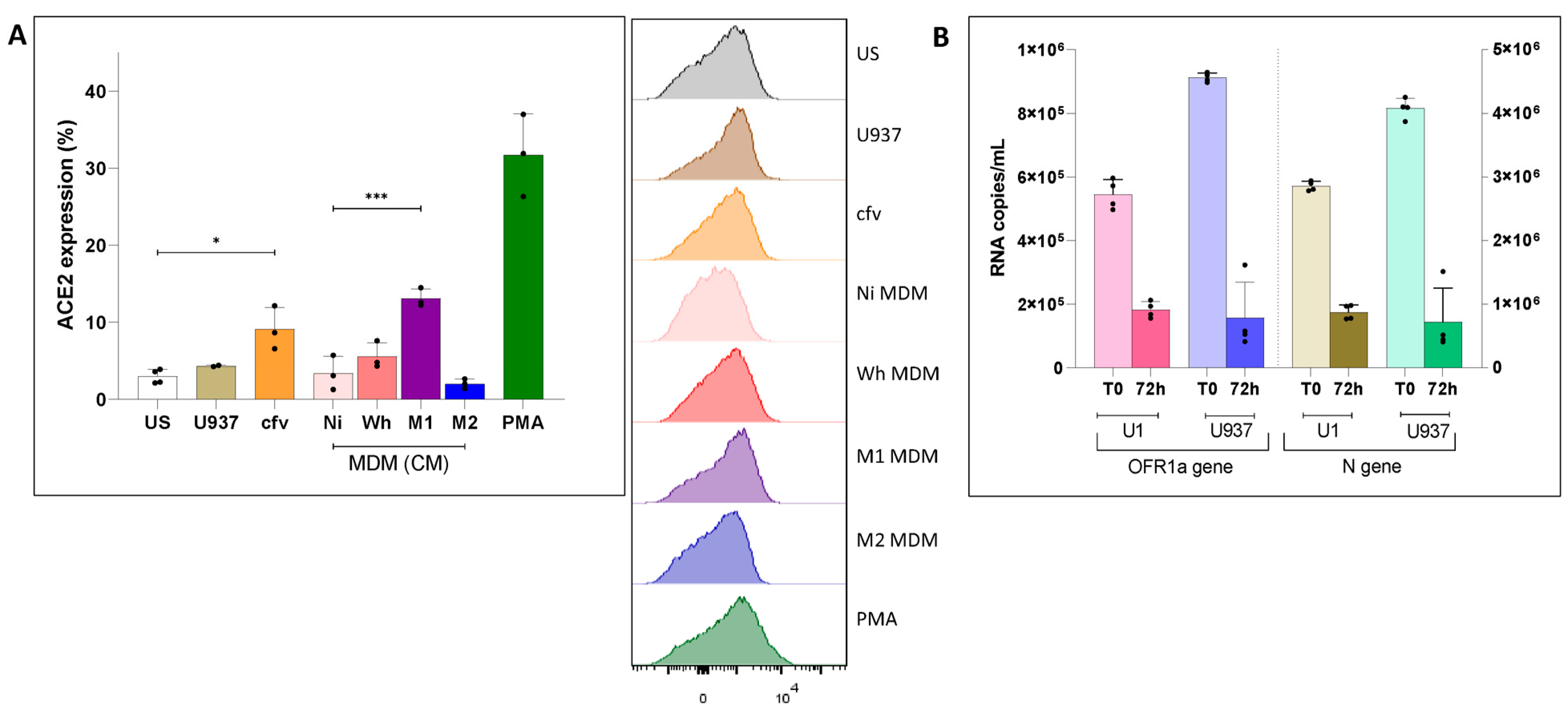
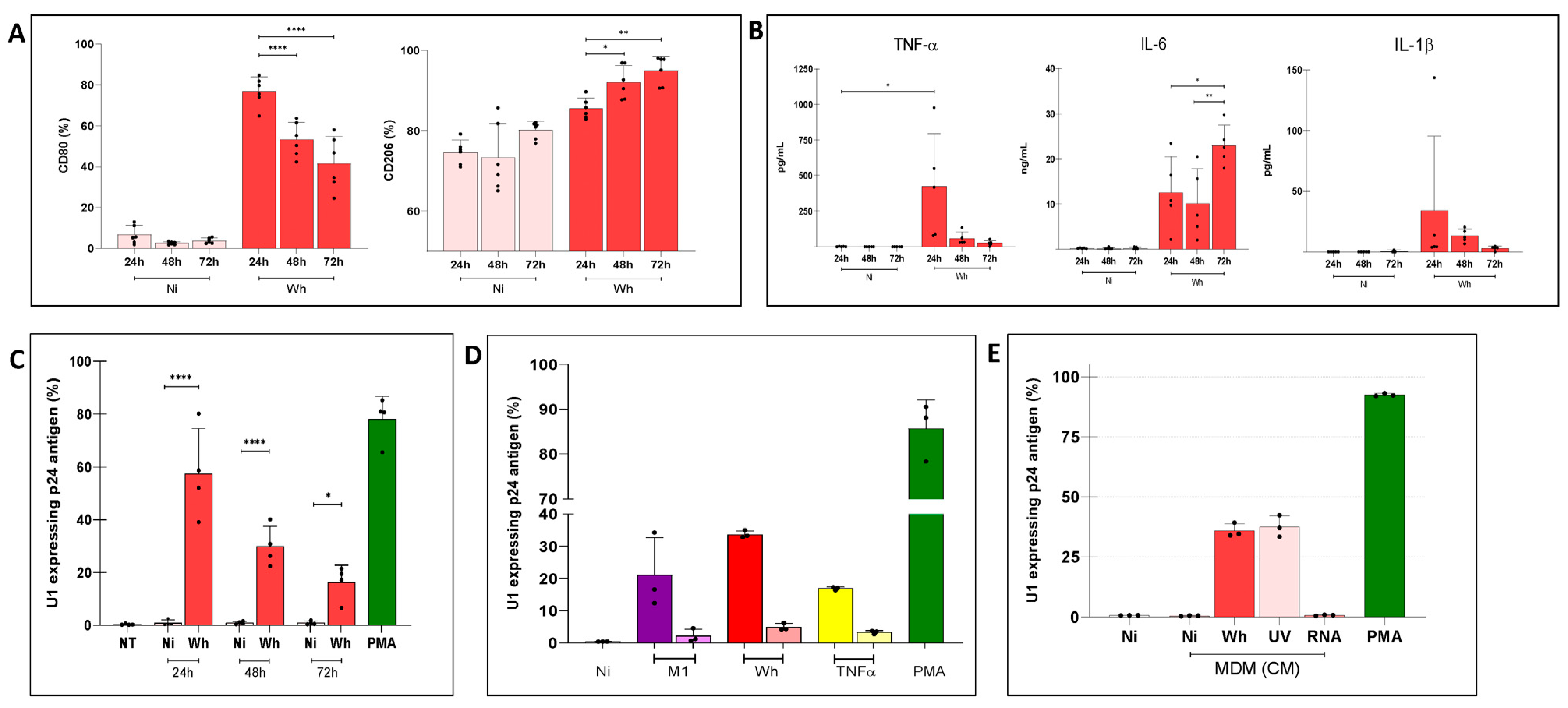
3.1. Modulation of ACE2 Surface Expression in Uninfected Cells and in Latently HIV-Infected U1 Cells
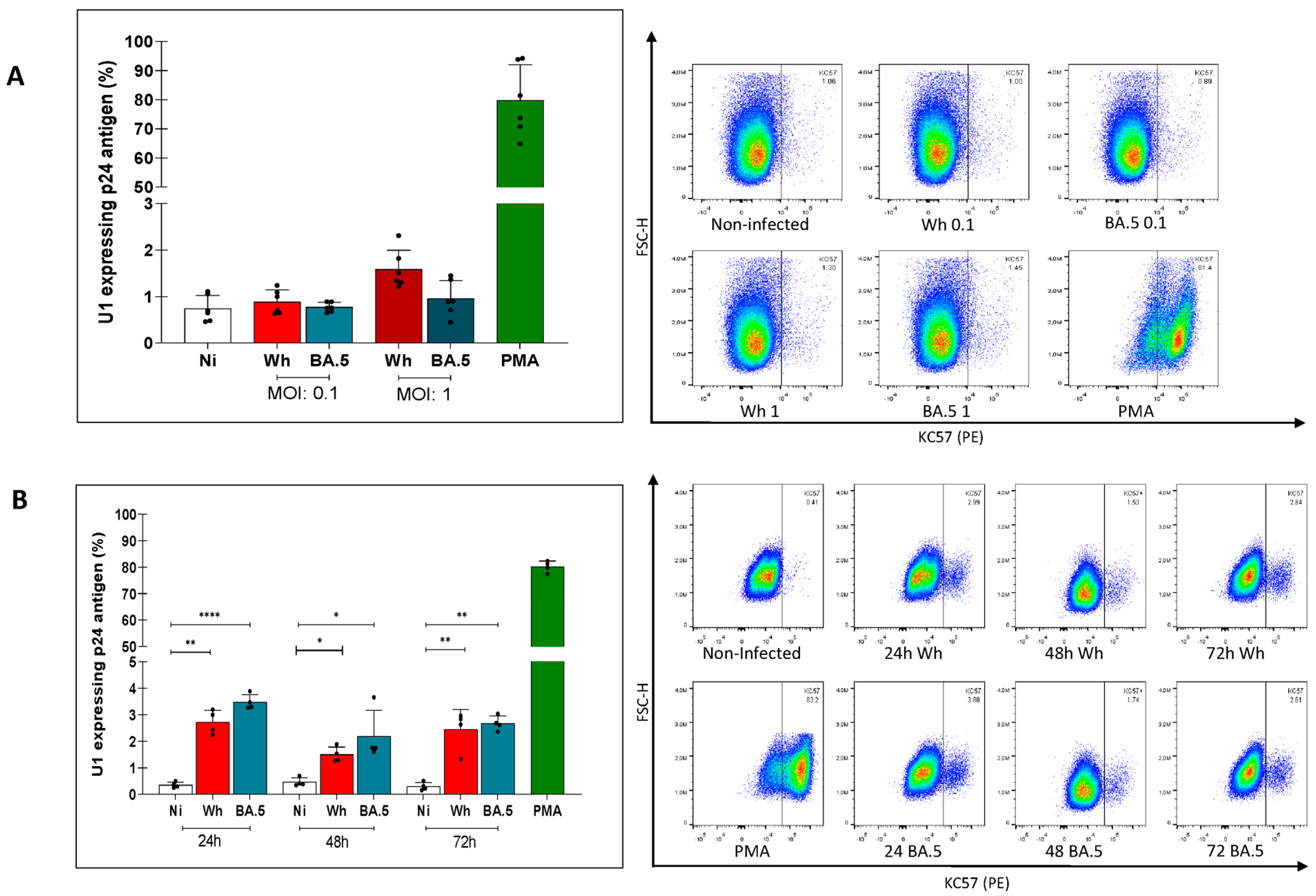
3.2. Modest Reversal of HIV-1 Latently Infected U1 Cells by Cell-Free SARS-CoV-2
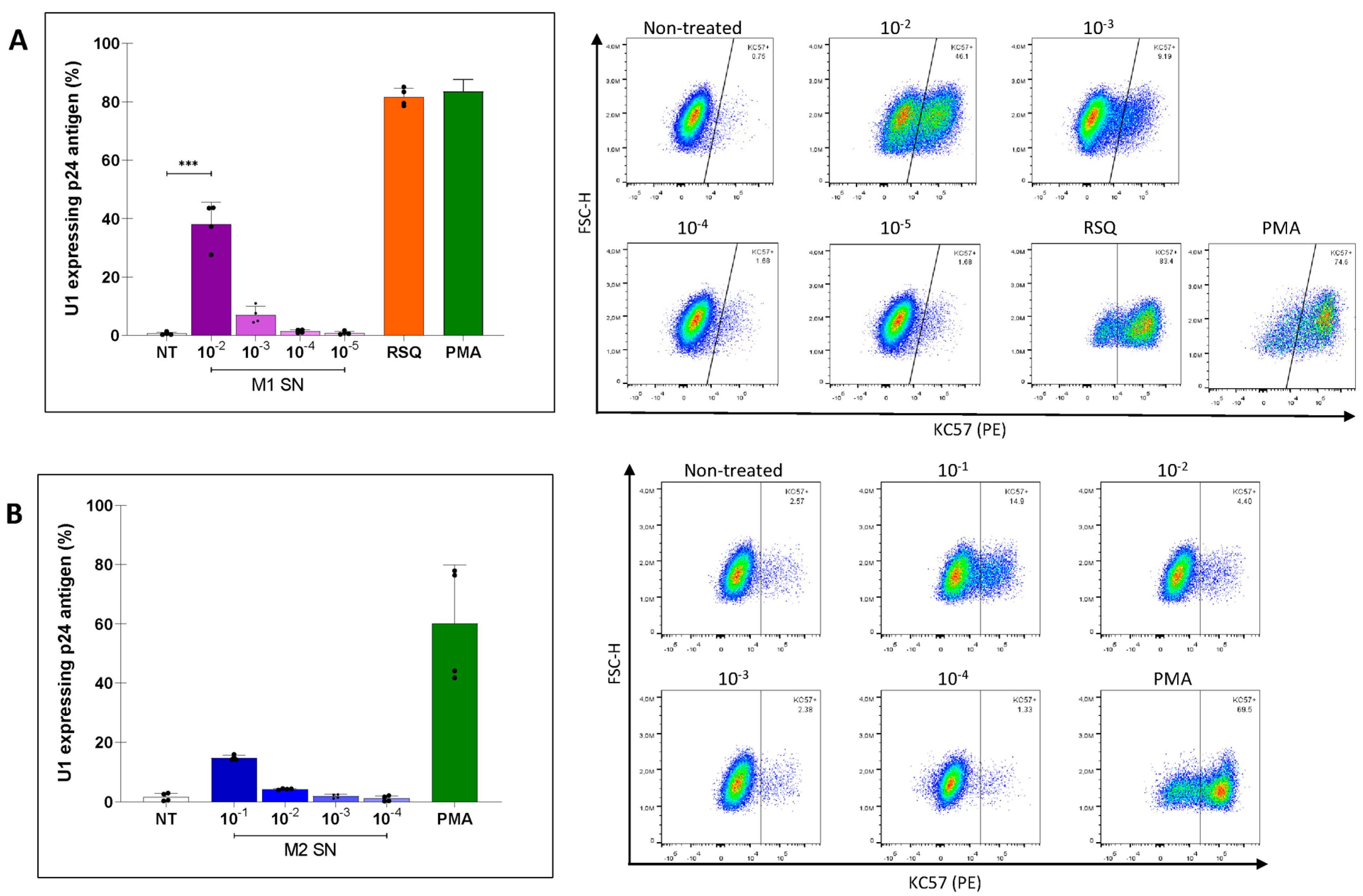
3.3. Modulation of HIV Latency in U1 Cells by Soluble Factors Released by M1 or M2-Polarized MDMs
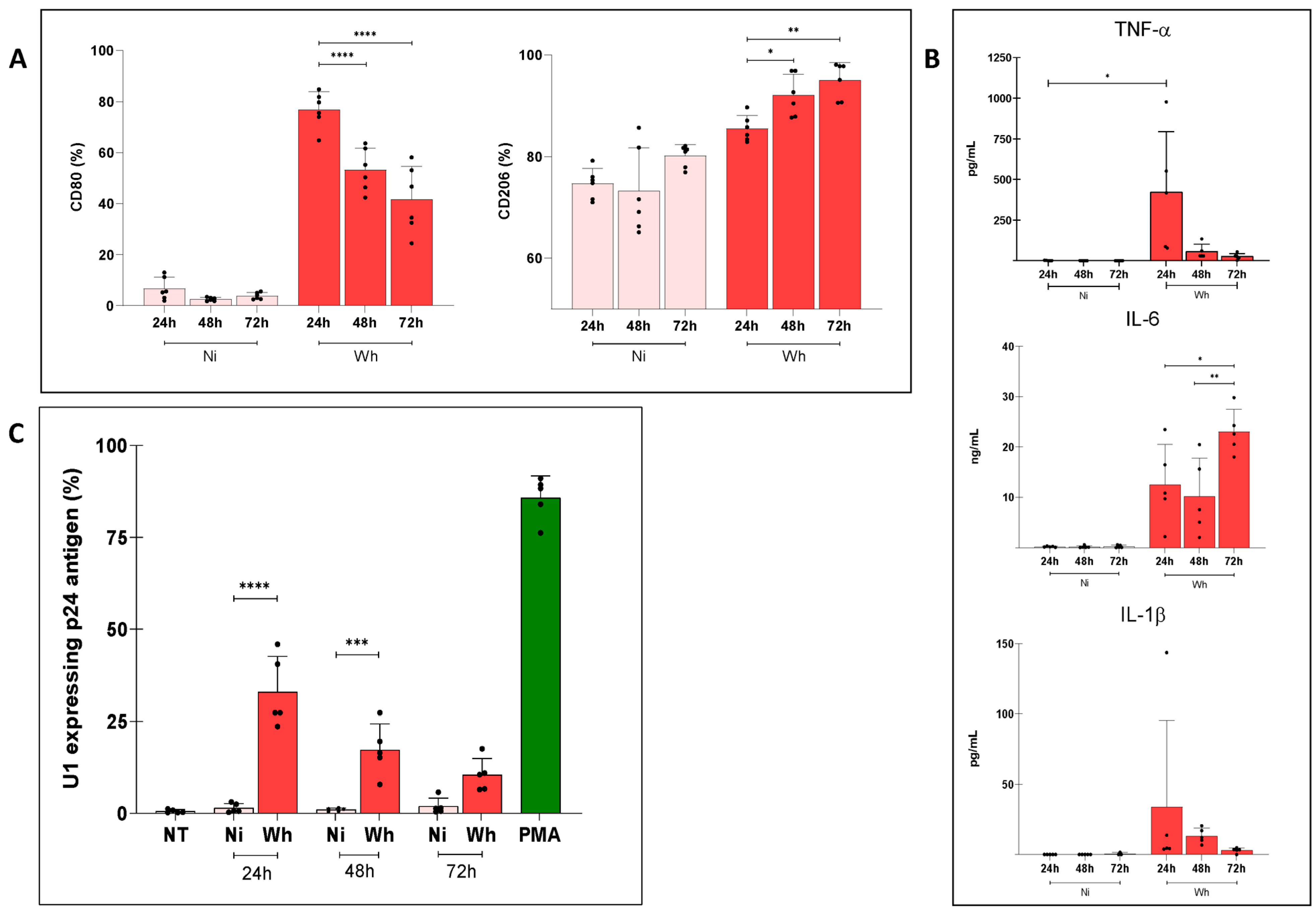
3.4. Soluble Factors Released from SARS-CoV-2-Exposed MDMs Reactivated Proviral Expression in U1 Cells
3.5. U1 Cells Exposed to Soluble Factors from Infectious and UV-Inactivated SARS-CoV-2-Exposed MDMs over Prolonged Time
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spinelli, M.A.; Jones, B.L.H.; Gandhi, M. COVID-19 Outcomes and Risk Factors Among People Living with HIV. Curr. HIV/AIDS Rep. 2022, 19, 425–432. [Google Scholar] [CrossRef]
- Garcia, F.; Leon, A.; Gatell, J.M.; Plana, M.; Gallart, T. Therapeutic vaccines against HIV infection. Hum. Vaccines Immunother. 2012, 8, 569–581. [Google Scholar] [CrossRef]
- Finzi, D.; Hermankova, M.; Pierson, T.; Carruth, L.M.; Buck, C.; Chaisson, R.E.; Quinn, T.C.; Chadwick, K.; Margolick, J.; Brookmeyer, R.; et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997, 278, 1295–1300. [Google Scholar] [CrossRef] [PubMed]
- Chun, T.W.; Stuyver, L.; Mizell, S.B.; Ehler, L.A.; Mican, J.A.; Baseler, M.; Lloyd, A.L.; Nowak, M.A.; Fauci, A.S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 1997, 94, 13193–13197. [Google Scholar] [CrossRef]
- Le Douce, V.; Herbein, G.; Rohr, O.; Schwartz, C. Molecular mechanisms of HIV-1 persistence in the monocyte-macrophage lineage. Retrovirology 2010, 7, 32. [Google Scholar] [CrossRef]
- Zhu, T. HIV-1 in peripheral blood monocytes: An underrated viral source. J. Antimicrob. Chemother. 2002, 50, 309–311. [Google Scholar] [CrossRef]
- Costiniuk, C.T.; Jenabian, M.A. The lungs as anatomical reservoirs of HIV infection. Rev. Med. Virol. 2014, 24, 35–54. [Google Scholar] [CrossRef] [PubMed]
- Banga, R.; Perreau, M. The multifaceted nature of HIV tissue reservoirs. Curr. Opin. HIV AIDS 2024, 19, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Kalada, W.; Cory, T.J. The Importance of Tissue Sanctuaries and Cellular Reservoirs of HIV-1. Curr. HIV Res. 2022, 20, 102–110. [Google Scholar]
- Gaskill, P.J.; Zandonatti, M.; Gilmartin, T.; Head, S.R.; Fox, H.S. Macrophage-derived simian immunodeficiency virus exhibits enhanced infectivity by comparison with T-cell-derived virus. J. Virol. 2008, 82, 1615–1621. [Google Scholar] [CrossRef]
- Devadas, K.; Hardegen, N.J.; Wahl, L.M.; Hewlett, I.K.; Clouse, K.A.; Yamada, K.M.; Dhawan, S. Mechanisms for macrophage-mediated HIV-1 induction. J. Immunol. 2004, 173, 6735–6744. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Julg, B.; Mohandas, S.; Bradfute, S.B.; Force, R.M.P.T. Viral persistence, reactivation, and mechanisms of long COVID. eLife 2023, 12, e86015. [Google Scholar] [CrossRef] [PubMed]
- Vojdani, A.; Vojdani, E.; Saidara, E.; Maes, M. Persistent SARS-CoV-2 Infection, EBV, HHV-6 and Other Factors May Contribute to Inflammation and Autoimmunity in Long COVID. Viruses 2023, 15, 400. [Google Scholar] [CrossRef] [PubMed]
- Peluso, M.J.; Deveau, T.M.; Munter, S.E.; Ryder, D.; Buck, A.; Beck-Engeser, G.; Chan, F.; Lu, S.; Goldberg, S.A.; Hoh, R.; et al. Chronic viral coinfections differentially affect the likelihood of developing long COVID. J. Clin. Investig. 2023, 133, e163669. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, E.M.; Terry, S.; Copertino, D.; Leyre, L.; Danesh, A.; Weiler, J.; Ward, A.R.; Khadka, P.; McNeil, E.; Bernard, K.; et al. SARS CoV-2 mRNA vaccination exposes latent HIV to Nef-specific CD8+ T-cells. Nat. Commun. 2022, 13, 4888. [Google Scholar] [CrossRef] [PubMed]
- Qu, M.M.; Song, B.; Yang, B.P.; Wang, Z.; Yu, M.; Zhang, Y.; Zhang, C.; Song, J.W.; Fan, X.; Xu, R.; et al. Effect of SARS-CoV-2 Breakthrough Infection on HIV Reservoirs and T-Cell Immune Recovery in 3-Dose Vaccinated People Living with HIV. Viruses 2023, 15, 2427. [Google Scholar] [CrossRef] [PubMed]
- Hamidzadeh, K.; Christensen, S.M.; Dalby, E.; Chandrasekaran, P.; Mosser, D.M. Macrophages and the Recovery from Acute and Chronic Inflammation. Annu. Rev. Physiol. 2017, 79, 567–592. [Google Scholar] [CrossRef] [PubMed]
- Felkle, D.; Zieba, K.; Kaleta, K.; Czaja, J.; Zyzdorf, A.; Sobocinska, W.; Jarczynski, M.; Bryniarski, K.; Nazimek, K. Overreactive macrophages in SARS-CoV-2 infection: The effects of ACEI. Int. Immunopharmacol. 2023, 124, 110858. [Google Scholar] [CrossRef]
- Garcia-Nicolas, O.; Godel, A.; Zimmer, G.; Summerfield, A. Macrophage phagocytosis of SARS-CoV-2-infected cells mediates potent plasmacytoid dendritic cell activation. Cell Mol. Immunol. 2023, 20, 835–849. [Google Scholar] [CrossRef]
- Grant, R.A.; Morales-Nebreda, L.; Markov, N.S.; Swaminathan, S.; Querrey, M.; Guzman, E.R.; Abbott, D.A.; Donnelly, H.K.; Donayre, A.; Goldberg, I.A.; et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature 2021, 590, 635–641. [Google Scholar] [CrossRef]
- Han, Y.; Wind-Rotolo, M.; Yang, H.C.; Siliciano, J.D.; Siliciano, R.F. Experimental approaches to the study of HIV-1 latency. Nat. Rev. Microbiol. 2007, 5, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Folks, T.M.; Justement, J.; Kinter, A.; Dinarello, C.A.; Fauci, A.S. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science 1987, 238, 800–802. [Google Scholar] [CrossRef]
- Jordan, A.; Bisgrove, D.; Verdin, E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003, 22, 1868–1877. [Google Scholar] [CrossRef] [PubMed]
- Folks, T.M.; Justement, J.; Kinter, A.; Schnittman, S.; Orenstein, J.; Poli, G.; Fauci, A.S. Characterization of a promonocyte clone chronically infected with HIV and inducible by 13-phorbol-12-myristate acetate. J. Immunol. 1988, 140, 1117–1122. [Google Scholar] [CrossRef]
- Sviercz, F.; Jarmoluk, P.; Godoy Coto, J.; Cevallos, C.; Freiberger, R.N.; Lopez, C.A.M.; Ennis, I.L.; Delpino, M.V.; Quarleri, J. The abortive SARS-CoV-2 infection of osteoclast precursors promotes their differentiation into osteoclasts. J. Med. Virol. 2024, 96, e29597. [Google Scholar] [CrossRef] [PubMed]
- Sviercz, F.A.; Jarmoluk, P.; Cevallos, C.G.; Lopez, C.A.M.; Freiberger, R.N.; Guano, A.; Adamczyk, A.; Ostrowski, M.; Delpino, M.V.; Quarleri, J. Massively HIV-1-infected macrophages exhibit a severely hampered ability to differentiate into osteoclasts. Front. Immunol. 2023, 14, 1206099. [Google Scholar] [CrossRef] [PubMed]
- Biasin, M.; Bianco, A.; Pareschi, G.; Cavalleri, A.; Cavatorta, C.; Fenizia, C.; Galli, P.; Lessio, L.; Lualdi, M.; Tombetti, E.; et al. UV-C irradiation is highly effective in inactivating SARS-CoV-2 replication. Sci. Rep. 2021, 11, 6260. [Google Scholar] [CrossRef]
- Song, X.; Hu, W.; Yu, H.; Zhao, L.; Zhao, Y.; Zhao, X.; Xue, H.H.; Zhao, Y. Little to no expression of angiotensin-converting enzyme-2 on most human peripheral blood immune cells but highly expressed on tissue macrophages. Cytometry A 2023, 103, 136–145. [Google Scholar] [CrossRef]
- Chau, C.W.; To, A.; Au-Yeung, R.K.H.; Tang, K.; Xiang, Y.; Ruan, D.; Zhang, L.; Wong, H.; Zhang, S.; Au, M.T.; et al. SARS-CoV-2 infection activates inflammatory macrophages in vascular immune organoids. Sci. Rep. 2024, 14, 8781. [Google Scholar] [CrossRef]
- Labzin, L.I.; Chew, K.Y.; Eschke, K.; Wang, X.; Esposito, T.; Stocks, C.J.; Rae, J.; Patrick, R.; Mostafavi, H.; Hill, B.; et al. Macrophage ACE2 is necessary for SARS-CoV-2 replication and subsequent cytokine responses that restrict continued virion release. Sci. Signal. 2023, 16, eabq1366. [Google Scholar] [CrossRef]
- Kinter, A.L.; Biswas, P.; Alfano, M.; Justement, J.S.; Mantelli, B.; Rizzi, C.; Gatti, A.R.; Vicenzi, E.; Bressler, P.; Poli, G. Interleukin-6 and glucocorticoids synergistically induce human immunodeficiency virus type-1 expression in chronically infected U1 cells by a long terminal repeat independent post-transcriptional mechanism. Mol. Med. 2001, 7, 668–678. [Google Scholar] [CrossRef]
- Bertani, F.R.; Mozetic, P.; Fioramonti, M.; Iuliani, M.; Ribelli, G.; Pantano, F.; Santini, D.; Tonini, G.; Trombetta, M.; Businaro, L.; et al. Classification of M1/M2-polarized human macrophages by label-free hyperspectral reflectance confocal microscopy and multivariate analysis. Sci. Rep. 2017, 7, 8965. [Google Scholar] [CrossRef] [PubMed]
- Rodell, C.B.; Arlauckas, S.P.; Cuccarese, M.F.; Garris, C.S.; Li, R.; Ahmed, M.S.; Kohler, R.H.; Pittet, M.J.; Weissleder, R. TLR7/8-agonist-loaded nanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat. Biomed. Eng. 2018, 2, 578–588. [Google Scholar] [CrossRef] [PubMed]
- Lian, Q.; Zhang, K.; Zhang, Z.; Duan, F.; Guo, L.; Luo, W.; Mok, B.W.; Thakur, A.; Ke, X.; Motallebnejad, P.; et al. Differential effects of macrophage subtypes on SARS-CoV-2 infection in a human pluripotent stem cell-derived model. Nat. Commun. 2022, 13, 2028. [Google Scholar] [CrossRef]
- Li, X.; Cai, H.; Wu, W.; Si, S.; Zhu, M. Exposure duration of ambient fine particulate matter determines the polarization of macrophages. Cent. Eur. J. Immunol. 2023, 48, 219–227. [Google Scholar] [CrossRef]
- Feng, H.; Yin, Y.; Ren, Y.; Li, M.; Zhang, D.; Xu, M.; Cai, X.; Kang, J. Effect of CSE on M1/M2 polarization in alveolar and peritoneal macrophages at different concentrations and exposure in vitro. In Vitro Cell Dev. Biol. Anim. 2020, 56, 154–164. [Google Scholar] [CrossRef]
- Kawasuji, H.; Morinaga, Y.; Tani, H.; Yoshida, Y.; Takegoshi, Y.; Kaneda, M.; Murai, Y.; Kimoto, K.; Ueno, A.; Miyajima, Y.; et al. SARS-CoV-2 RNAemia with a higher nasopharyngeal viral load is strongly associated with disease severity and mortality in patients with COVID-19. J. Med. Virol. 2022, 94, 147–153. [Google Scholar] [CrossRef]
- Gracheva, A.V.; Korchevaya, E.R.; Ammour, Y.I.; Smirnova, D.I.; Sokolova, O.S.; Glukhov, G.S.; Moiseenko, A.V.; Zubarev, I.V.; Samoilikov, R.V.; Leneva, I.A.; et al. Immunogenic properties of SARS-CoV-2 inactivated by ultraviolet light. Arch. Virol. 2022, 167, 2181–2191. [Google Scholar] [CrossRef]
- Lo, C.W.; Matsuura, R.; Iimura, K.; Wada, S.; Shinjo, A.; Benno, Y.; Nakagawa, M.; Takei, M.; Aida, Y. UVC disinfects SARS-CoV-2 by induction of viral genome damage without apparent effects on viral morphology and proteins. Sci. Rep. 2021, 11, 13804. [Google Scholar] [CrossRef] [PubMed]
- Dufour, C.; Gantner, P.; Fromentin, R.; Chomont, N. Correction: The multifaceted nature of HIV latency. J. Clin. Investig. 2020, 130, 3381–3390, Erratum in J. Clin. Investig. 2021, 131, e151380. [Google Scholar] [CrossRef]
- Symons, J.; Chopra, A.; Malatinkova, E.; De Spiegelaere, W.; Leary, S.; Cooper, D.; Abana, C.O.; Rhodes, A.; Rezaei, S.D.; Vandekerckhove, L.; et al. HIV integration sites in latently infected cell lines: Evidence of ongoing replication. Retrovirology 2017, 14, 2. [Google Scholar] [CrossRef] [PubMed]
- Poli, G. U1 and OM10.1. Myeloid Cell Lines as Surrogate Models of Reversible Proviral Latency. Methods Mol. Biol. 2022, 2407, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Rodari, A.; Poli, G.; Van Lint, C. Jurkat-Derived (J-Lat, J1.1, and Jurkat E4) and CEM-Derived T Cell Lines (8E5 and ACH-2) as Models of Reversible Proviral Latency. Methods Mol. Biol. 2022, 2407, 3–15. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Ziegler, C.G.K.; Allon, S.J.; Nyquist, S.K.; Mbano, I.M.; Miao, V.N.; Tzouanas, C.N.; Cao, Y.; Yousif, A.S.; Bals, J.; Hauser, B.M.; et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020, 181, 1016–1035.e19. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Dong, Y.; Bo, X.; Cheng, Y.; Cheng, J. Large Screening Identifies ACE2 Positively Correlates With NF-kappaB Signaling Activity and Targeting NF-kappaB Signaling Drugs Suppress ACE2 Levels. Front. Pharmacol. 2021, 12, 771555. [Google Scholar] [CrossRef]
- Shen, X.R.; Geng, R.; Li, Q.; Chen, Y.; Li, S.F.; Wang, Q.; Min, J.; Yang, Y.; Li, B.; Jiang, R.D.; et al. ACE2-independent infection of T lymphocytes by SARS-CoV-2. Signal Transduct. Target. Ther. 2022, 7, 83. [Google Scholar] [CrossRef]
- Poli, G.; Kinter, A.; Justement, J.S.; Kehrl, J.H.; Bressler, P.; Stanley, S.; Fauci, A.S. Tumor necrosis factor alpha functions in an autocrine manner in the induction of human immunodeficiency virus expression. Proc. Natl. Acad. Sci. USA 1990, 87, 782–785. [Google Scholar] [CrossRef]
- Kosyreva, A.; Dzhalilova, D.; Lokhonina, A.; Vishnyakova, P.; Fatkhudinov, T. The Role of Macrophages in the Pathogenesis of SARS-CoV-2-Associated Acute Respiratory Distress Syndrome. Front. Immunol. 2021, 12, 682871. [Google Scholar] [CrossRef]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef]
- Zhang, D.; Guo, R.; Lei, L.; Liu, H.; Wang, Y.; Wang, Y.; Qian, H.; Dai, T.; Zhang, T.; Lai, Y.; et al. Frontline Science: COVID-19 infection induces readily detectable morphologic and inflammation-related phenotypic changes in peripheral blood monocytes. J. Leukoc. Biol. 2021, 109, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Pantazi, I.; Al-Qahtani, A.A.; Alhamlan, F.S.; Alothaid, H.; Matou-Nasri, S.; Sourvinos, G.; Vergadi, E.; Tsatsanis, C. SARS-CoV-2/ACE2 Interaction Suppresses IRAK-M Expression and Promotes Pro-Inflammatory Cytokine Production in Macrophages. Front. Immunol. 2021, 12, 683800. [Google Scholar] [CrossRef] [PubMed]
- Poli, G.; Kinter, A.L.; Justement, J.S.; Bressler, P.; Kehrl, J.H.; Fauci, A.S. Retinoic acid mimics transforming growth factor beta in the regulation of human immunodeficiency virus expression in monocytic cells. Proc. Natl. Acad. Sci. USA 1992, 89, 2689–2693. [Google Scholar] [CrossRef] [PubMed]
- Goletti, D.; Kinter, A.L.; Hardy, E.C.; Poli, G.; Fauci, A.S. Modulation of endogenous IL-1 beta and IL-1 receptor antagonist results in opposing effects on HIV expression in chronically infected monocytic cells. J. Immunol. 1996, 156, 3501–3508. [Google Scholar] [CrossRef] [PubMed]
- Grebennikov, D.; Kholodareva, E.; Sazonov, I.; Karsonova, A.; Meyerhans, A.; Bocharov, G. Intracellular Life Cycle Kinetics of SARS-CoV-2 Predicted Using Mathematical Modelling. Viruses 2021, 13, 1735. [Google Scholar] [CrossRef]
- Ponde, N.O.; Shoger, K.E.; Khatun, M.S.; Sarkar, M.K.; Dey, I.; Taylor, T.C.; Cisney, R.N.; Arunkumar, S.P.; Gudjonsson, J.E.; Kolls, J.K.; et al. SARS-CoV-2 ORF8 Mediates Signals in Macrophages and Monocytes through MyD88 Independently of the IL-17 Receptor. J. Immunol. 2023, 211, 252–260. [Google Scholar] [CrossRef]
- Hakre, S.; Chavez, L.; Shirakawa, K.; Verdin, E. HIV latency: Experimental systems and molecular models. FEMS Microbiol. Rev. 2012, 36, 706–716. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jarmoluk, P.; Sviercz, F.A.; Cevallos, C.; Freiberger, R.N.; López, C.A.; Poli, G.; Delpino, M.V.; Quarleri, J. SARS-CoV-2 Modulation of HIV Latency Reversal in a Myeloid Cell Line: Direct and Bystander Effects. Viruses 2024, 16, 1310. https://doi.org/10.3390/v16081310
Jarmoluk P, Sviercz FA, Cevallos C, Freiberger RN, López CA, Poli G, Delpino MV, Quarleri J. SARS-CoV-2 Modulation of HIV Latency Reversal in a Myeloid Cell Line: Direct and Bystander Effects. Viruses. 2024; 16(8):1310. https://doi.org/10.3390/v16081310
Chicago/Turabian StyleJarmoluk, Patricio, Franco Agustín Sviercz, Cintia Cevallos, Rosa Nicole Freiberger, Cynthia Alicia López, Guido Poli, M. Victoria Delpino, and Jorge Quarleri. 2024. "SARS-CoV-2 Modulation of HIV Latency Reversal in a Myeloid Cell Line: Direct and Bystander Effects" Viruses 16, no. 8: 1310. https://doi.org/10.3390/v16081310
APA StyleJarmoluk, P., Sviercz, F. A., Cevallos, C., Freiberger, R. N., López, C. A., Poli, G., Delpino, M. V., & Quarleri, J. (2024). SARS-CoV-2 Modulation of HIV Latency Reversal in a Myeloid Cell Line: Direct and Bystander Effects. Viruses, 16(8), 1310. https://doi.org/10.3390/v16081310








