Kinetic Landscape of Single Virus-like Particles Highlights the Efficacy of SARS-CoV-2 Internalization
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Transfections
2.2. Transfection
2.3. Time-Lapse Live-Cell Imaging
2.4. Electron Microscopy
2.5. Virus-like Particle Preparation
2.6. Antibody Inhibition of SASR-CoV-2 VLPs
2.7. VLP Tracking
2.8. VLP Analysis
2.9. Statistical Analysis
3. Results
3.1. Visualization of VLP Internalization
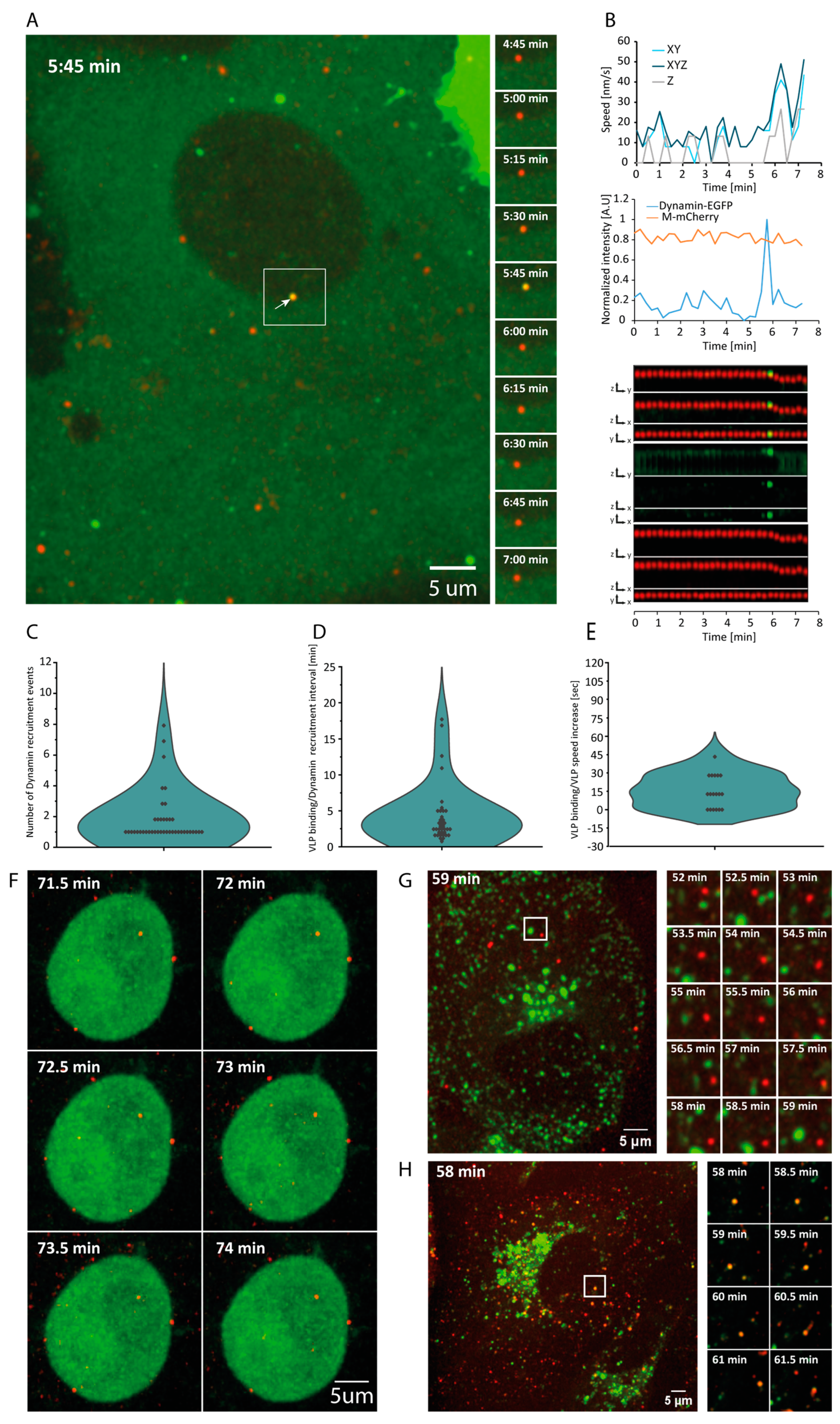
3.2. Dynamin-Dependent VLP Entry
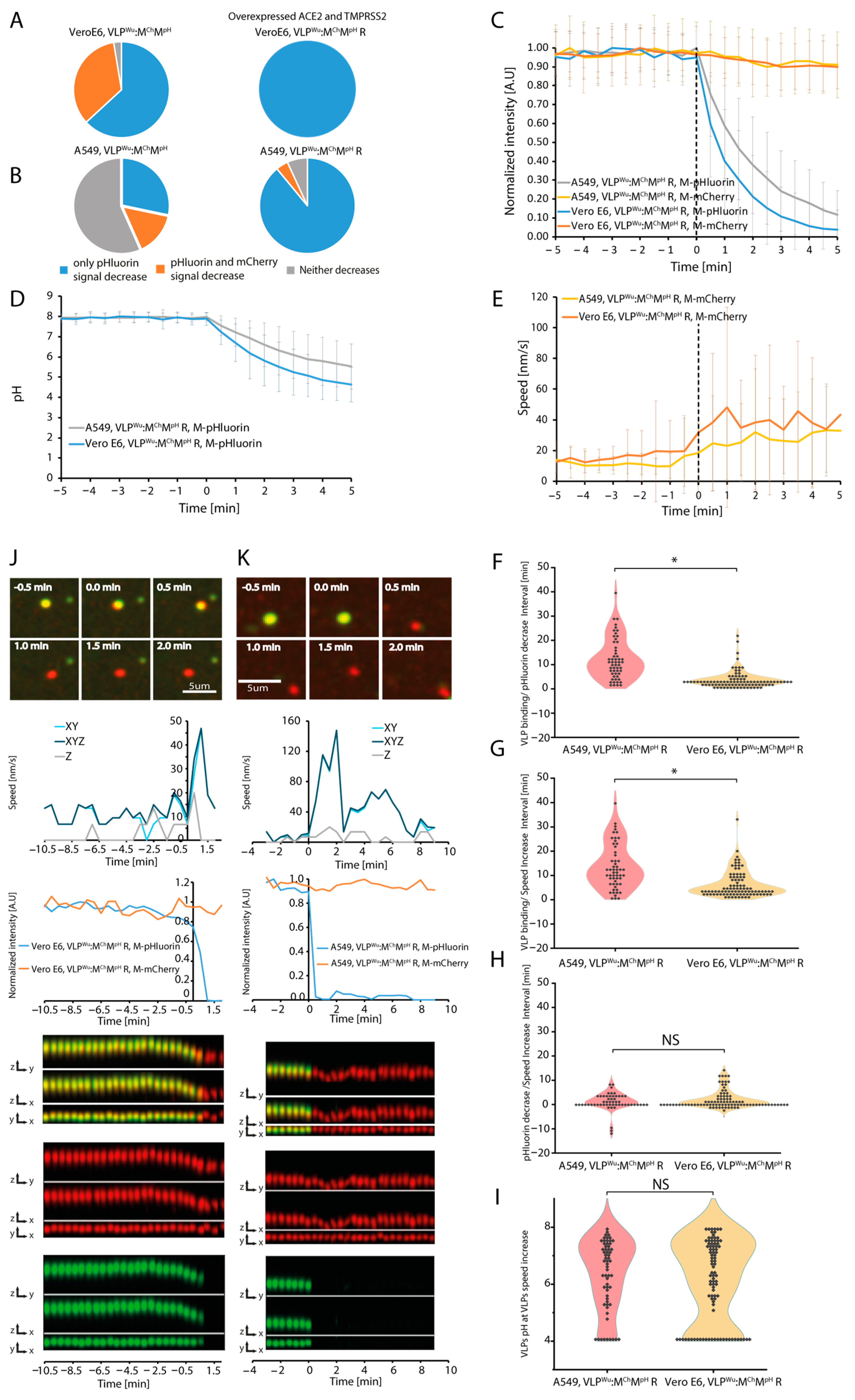
3.3. Dynamics of VLP Acidification
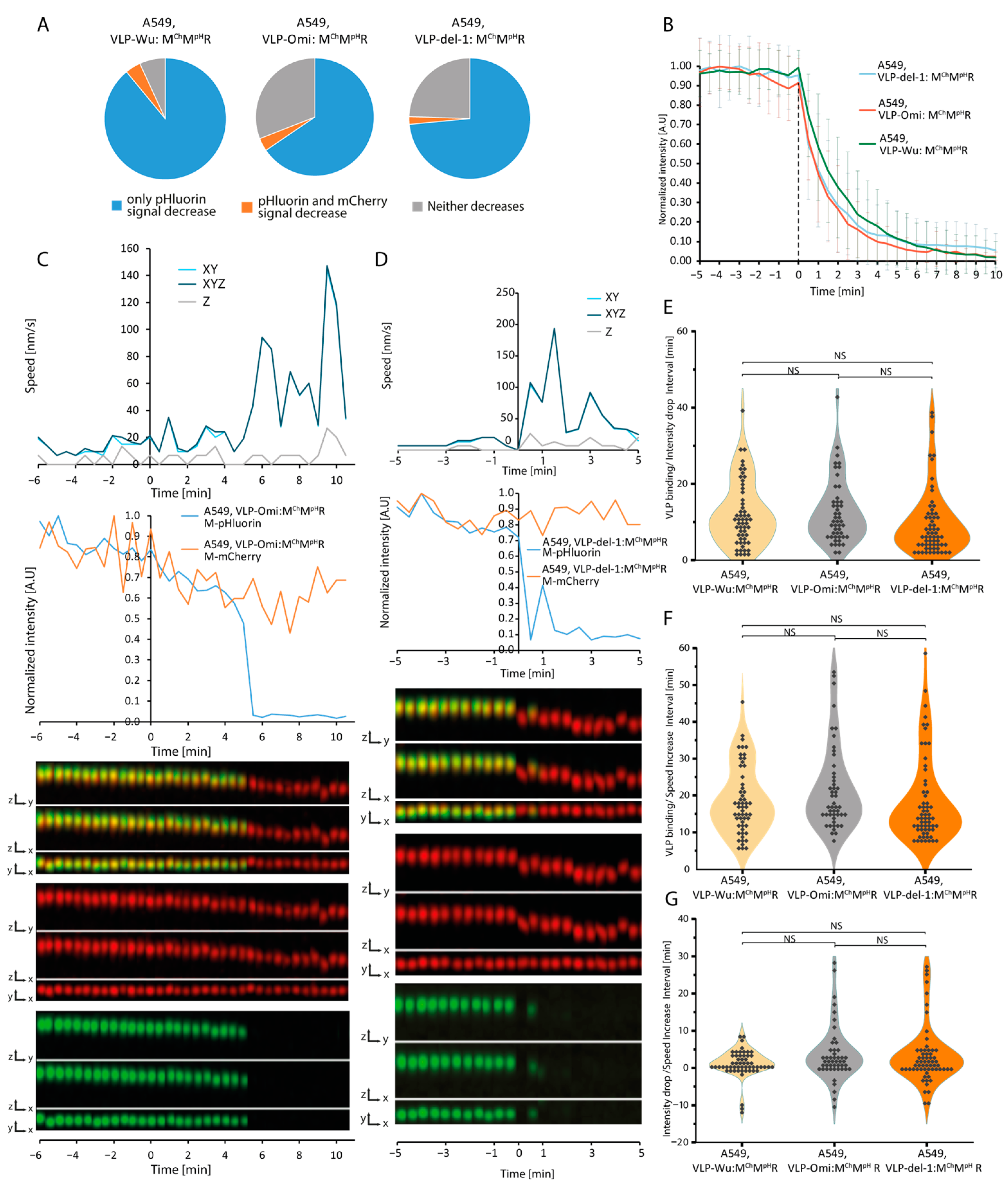
3.4. Kinetics of VLPs Lacking the Furin Cleavage Site
3.5. Kinetics of Omicron VLPs
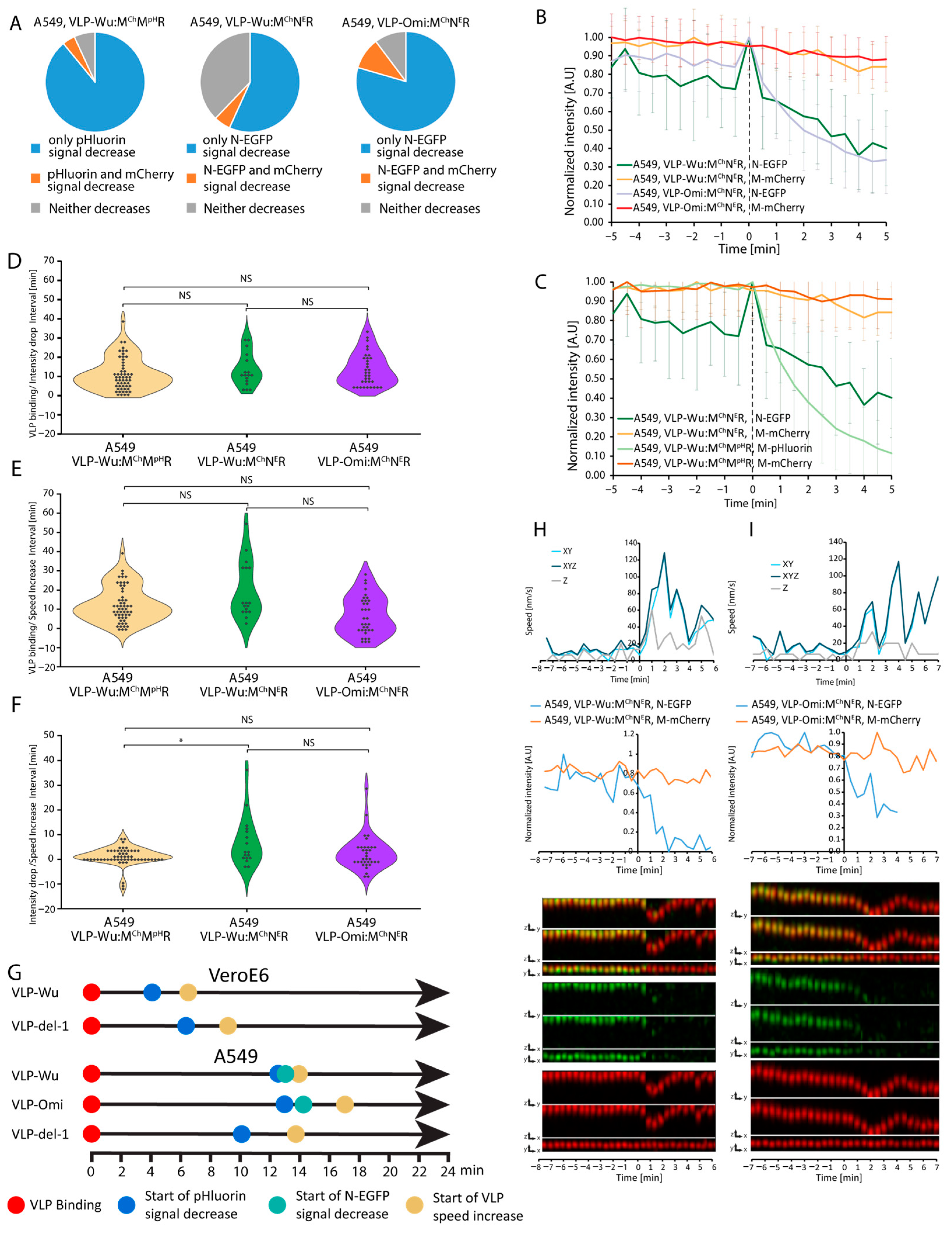
3.6. Rate of VLP Nucleocapsid Release
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, A.; Tiwari, S.; Deb, M.K.; Marty, J.L. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): A global pandemic and treatment strategies. Int. J. Antimicrob. Agents 2020, 56, 106054. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, I.; Maity, P. COVID-19 outbreak: Migration, effects on society, global environment and prevention. Sci. Total Environ. 2020, 728, 138882. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ho, W.; Huang, Y.; Jin, D.-Y.; Li, S.; Liu, S.-L.; Liu, X.; Qiu, J.; Sang, Y.; Wang, Q.; et al. SARS-CoV-2 is an appropriate name for the new coronavirus. Lancet 2020, 395, 949–950. [Google Scholar] [CrossRef]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef]
- Luytjes, W. Coronavirus Gene Expression. In The Coronaviridae; Siddell, S.G., Ed.; Springer US: Boston, MA, USA, 1995; pp. 33–54. ISBN 978-1-4899-1531-3. [Google Scholar]
- Satarker, S.; Nampoothiri, M. Structural Proteins in Severe Acute Respiratory Syndrome Coronavirus-2. Arch. Med. Res. 2020, 51, 482–491. [Google Scholar] [CrossRef]
- Syed, A.M.; Taha, T.Y.; Tabata, T.; Chen, I.P.; Ciling, A.; Khalid, M.M.; Sreekumar, B.; Chen, P.Y.; Hayashi, J.M.; Soczek, K.M.; et al. Rapid assessment of SARS-CoV-2–evolved variants using virus-like particles. Science 2021, 374, 1626–1632. [Google Scholar] [CrossRef]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like particles: Preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnol. 2021, 19, 59. [Google Scholar] [CrossRef]
- Xu, R.; Shi, M.; Li, J.; Song, P.; Li, N. Construction of SARS-CoV-2 Virus-Like Particles by Mammalian Expression System. Front. Bioeng. Biotechnol. 2020, 8, 862. [Google Scholar] [CrossRef]
- Swann, H.; Sharma, A.; Preece, B.; Peterson, A.; Eldridge, C.; Belnap, D.M.; Vershinin, M.; Saffarian, S. Minimal system for assembly of SARS-CoV-2 virus like particles. Sci. Rep. 2020, 10, 21877. [Google Scholar] [CrossRef]
- Gourdelier, M.; Swain, J.; Arone, C.; Mouttou, A.; Bracquemond, D.; Merida, P.; Saffarian, S.; Lyonnais, S.; Favard, C.; Muriaux, D. Optimized production and fluorescent labeling of SARS-CoV-2 virus-like particles. Sci. Rep. 2022, 12, 14651. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Preece, B.; Swann, H.; Fan, X.; McKenney, R.J.; Ori-McKenney, K.M.; Saffarian, S.; Vershinin, M.D. Structural stability of SARS-CoV-2 virus like particles degrades with temperature. Biochem. Biophys. Res. Commun. 2021, 534, 343–346. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Camargo, S.M.R.; Singer, D.; Makrides, V.; Huggel, K.; Pos, K.M.; Wagner, C.A.; Kuba, K.; Danilczyk, U.; Skovby, F.; Kleta, R.; et al. Tissue-specific amino acid transporter partners ACE2 and collectrin differentially interact with hartnup mutations. Gastroenterology 2009, 136, 872–882. [Google Scholar] [CrossRef]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020, 183, 1043–1057.e15. [Google Scholar] [CrossRef]
- Wei, C.; Wan, L.; Yan, Q.; Wang, X.; Zhang, J.; Yang, X.; Zhang, Y.; Fan, C.; Li, D.; Deng, Y.; et al. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat. Metab. 2020, 2, 1391–1400. [Google Scholar] [CrossRef]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.-E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef]
- Chen, Z.; Mi, L.; Xu, J.; Yu, J.; Wang, X.; Jiang, J.; Xing, J.; Shang, P.; Qian, A.; Li, Y.; et al. Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2005, 191, 755–760. [Google Scholar] [CrossRef]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.-Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.-X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef]
- Wang, S.; Qiu, Z.; Hou, Y.; Deng, X.; Xu, W.; Zheng, T.; Wu, P.; Xie, S.; Bian, W.; Zhang, C.; et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021, 31, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Fink, C.; Ichimura, T.; Sako, K.; Mori, M.; Lee, N.N.; Das, K.M.P.; Hong, S.; Song, M.; Padera, R.F., Jr.; et al. KIM-1/TIM-1 is a Receptor for SARS-CoV-2 in Lung and Kidney SARS-CoV-2 precipitates respiratory distress by infection of airway epithelial cells and is often domain 1 (KIM-1/TIM-1) is expressed in lung and kidney epithelial cells in COVID-19 patient. MedRxiv 2022, 2, 2020-09. [Google Scholar]
- Jeffers, S.A.; Tusell, S.M.; Gillim-Ross, L.; Hemmila, E.M.; Achenbach, J.E.; Babcock, G.J.; Thomas, W.D.J.; Thackray, L.B.; Young, M.D.; Mason, R.J.; et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 2004, 101, 15748–15753. [Google Scholar] [CrossRef] [PubMed]
- Amraei, R.; Yin, W.; Napoleon, M.A.; Suder, E.L.; Berrigan, J.; Zhao, Q.; Olejnik, J.; Chandler, K.B.; Xia, C.; Feldman, J.; et al. CD209L L-SIGN and CD209 DC-SIGN act as receptors for SARS-CoV-2 amraei. ACS Cent. Sci. 2021, 7, 1156–1165. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, M.; Hess, T.; Morbioli, G.G.; Sengar, A.; Kasson, P.M. The ACE2 receptor accelerates but is not biochemically required for SARS-CoV-2 membrane fusion. Chem. Sci. 2023, 14, 6997–7004. [Google Scholar] [CrossRef]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Bayati, A.; Kumar, R.; Francis, V.; McPherson, P.S. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021, 296, 100306. [Google Scholar] [CrossRef]
- Turk, B.; Dolenc, I.; Turk, V.; Bieth, J.G. Kinetics of the pH-induced inactivation of human cathepsin L. Biochemistry 1993, 32, 375–380. [Google Scholar] [CrossRef]
- Zhao, M.M.; Yang, W.L.; Yang, F.Y.; Zhang, L.; Huang, W.J.; Hou, W.; Fan, C.F.; Jin, R.H.; Feng, Y.M.; Wang, Y.C.; et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct. Target. Ther. 2021, 6, 134. [Google Scholar] [CrossRef]
- Luo, S.; Zhang, J.; Kreutzberger, A.J.B.; Eaton, A.; Edwards, R.J.; Jing, C.; Dai, H.Q.; Sempowski, G.D.; Cronin, K.; Parks, R.; et al. An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion. Sci. Immunol. 2022, 7, eadd5446. [Google Scholar] [CrossRef]
- Kreutzberger, A.J.B.; Sanyal, A.; Saminathan, A.; Bloyet, L.M.; Stumpf, S.; Liu, Z.; Ojha, R.; Patjas, M.T.; Geneid, A.; Scanavachi, G.; et al. SARS-CoV-2 requires acidic pH to infect cells. Proc. Natl. Acad. Sci. USA 2022, 119, e2209514119. [Google Scholar] [CrossRef]
- Sankaranarayanan, S.; De Angelis, D.; Rothman, J.E.; Ryan, T.A. The use of pHluorins for optical measurements of presynaptic activity. Biophys. J. 2000, 79, 2199–2208. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Li, Z.; Zhao, W.; Yang, H.; Zhao, S.; Tang, D.; Zhang, Q.; Li, Z.; Liu, H.; et al. Single particle tracking reveals SARS-CoV-2 regulating and utilizing dynamic filopodia for viral invasion. Sci. Bull. 2023, 68, 2210–2224. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Zhu, G.; Zhang, J.; Lai, Y.; Xu, Y.; He, J.; Xie, J. Betanodavirus-like particles enter host cells via clathrin-mediated endocytosis in a cholesterol-, pH- and cytoskeleton-dependent manner. Vet. Res. 2017, 48, 8. [Google Scholar] [CrossRef] [PubMed]
- Zepeda-Cervantes, J.; Ramírez-Jarquín, J.O.; Vaca, L. Interaction Between Virus-Like Particles (VLPs) and Pattern Recognition Receptors (PRRs) From Dendritic Cells (DCs): Toward Better Engineering of VLPs. Front. Immunol. 2020, 11, 1100. [Google Scholar] [CrossRef]
- Ogando, N.S.; Dalebout, T.J.; Zevenhoven-Dobbe, J.C.; Limpens, R.W.A.L.; van der Meer, Y.; Caly, L.; Druce, J.; de Vries, J.J.C.; Kikkert, M.; Barcena, M.; et al. SARS-coronavirus-2 replication in Vero E6 cells: Replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020, 101, 925–940. [Google Scholar] [CrossRef] [PubMed]
- Prichard, K.L.; O’Brien, N.S.; Murcia, S.R.; Baker, J.R.; McCluskey, A. Role of Clathrin and Dynamin in Clathrin Mediated Endocytosis/Synaptic Vesicle Recycling and Implications in Neurological Diseases. Front. Cell. Neurosci. 2022, 15, 754110. [Google Scholar] [CrossRef]
- Cocucci, E.; Gaudin, R.; Kirchhausen, T. Dynamin recruitment and membrane scission at the neck of a clathrin-coated pit. Mol. Biol. Cell 2014, 25, 3595–3609. [Google Scholar] [CrossRef]
- Antonny, B.; Burd, C.; De Camilli, P.; Chen, E.; Daumke, O.; Faelber, K.; Ford, M.; Frolov, V.A.; Frost, A.; Hinshaw, J.E.; et al. Membrane fission by dynamin: What we know and what we need to know. EMBO J. 2016, 35, 2270–2284. [Google Scholar] [CrossRef]
- Cheng, X.; Chen, K.; Dong, B.; Yang, M.; Filbrun, S.L.; Myoung, Y.; Huang, T.-X.; Gu, Y.; Wang, G.; Fang, N. Dynamin-dependent vesicle twist at the final stage of clathrin-mediated endocytosis. Nat. Cell Biol. 2021, 23, 859–869. [Google Scholar] [CrossRef]
- Hill, E.; Van Der Kaay, J.; Downes, C.P.; Smythe, E. The role of dynamin and its binding partners in coated pit invagination and scission. J. Cell Biol. 2001, 152, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Štimac, I.; Vučko, N.J.; Zagorac, G.B.; Marcelić, M.; Lučin, H.M.; Lučin, P. Dynamin inhibitors prevent the establishment of the cytomegalovirus assembly compartment in the early phase of infection. Life 2021, 11, 876. [Google Scholar] [CrossRef]
- Loerke, D.; Mettlen, M.; Yarar, D.; Jaqaman, K.; Jaqaman, H.; Danuser, G.; Schmid, S.L. Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 2009, 7, 0628–0639. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhao, T.; Li, Y.; Huang, G.; White, M.A.; Gao, J. Investigation of endosome and lysosome biology by ultra pH-sensitive nanoprobes. Adv. Drug Deliv. Rev. 2017, 113, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yu, M.; Zhang, S.; Ma, G.; Su, Z. Adsorption of virus-like particles on ion exchange surface: Conformational changes at different pH detected by dual polarization interferometry. J. Chromatogr. A 2015, 1408, 161–168. [Google Scholar] [CrossRef]
- Ausar, S.F.; Foubert, T.R.; Hudson, M.H.; Vedvick, T.S.; Middaugh, C.R. conformational stability and disassembly of norwalk virus-like particles: Effect of ph and temperature. J. Biol. Chem. 2006, 281, 19478–19488. [Google Scholar] [CrossRef]
- Maassen, S.J.; van der Schoot, P.; Cornelissen, J.J.L.M. Experimental and Theoretical Determination of the pH inside the Confinement of a Virus-Like Particle. Small 2018, 14, e1802081. [Google Scholar] [CrossRef] [PubMed]
- Martineau, M.; Somasundaram, A.; Grimm, J.B.; Gruber, T.D.; Choquet, D.; Taraska, J.W.; Lavis, L.D.; Perrais, D. Semisynthetic fluorescent pH sensors for imaging exocytosis and endocytosis. Nat. Commun. 2017, 8, 1412. [Google Scholar] [CrossRef]
- Emeny, J.M.; Morgan, M.J. Regulation of the interferon system: Evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 1979, 43, 247–252. [Google Scholar] [CrossRef]
- Wang, L.; Fan, X.; Bonenfant, G.; Cui, D.; Hossain, J.; Jiang, N.; Larson, G.; Currier, M.; Liddell, J.; Wilson, M.; et al. Susceptibility to SARS-CoV-2 of cell lines and substrates commonly used to diagnose and isolate influenza and other viruses. Emerg. Infect. Dis. 2021, 27, 1380–1392. [Google Scholar] [CrossRef]
- Cagno, V. SARS-CoV-2 cellular tropism. Lancet Microbe 2020, 1, e2–e3. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.A.; Sacco, O.; Mancino, E.; Cristiani, L.; Midulla, F. Differences and similarities between SARS-CoV and SARS-CoV-2: Spike receptor-binding domain recognition and host cell infection with support of cellular serine proteases. Infection 2020, 48, 665–669. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.Y.; Wang, P.; Mok, B.W.Y.; Zhang, A.J.; Chu, H.; Lee, A.C.Y.; Deng, S.; Chen, P.; Chan, K.H.; Song, W.; et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg. Microbes Infect. 2020, 9, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Uemura, K.; Sato, A.; Toba, S.; Sanaki, T.; Maenaka, K.; Hall, W.W.; Orba, Y.; Sawa, H. SARS-CoV-2 variants with mutations at the S1/ S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog. 2021, 17, e1009233. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.P.; Goldhill, D.H.; Zhou, J.; Baillon, L.; Frise, R.; Swann, O.C.; Kugathasan, R.; Penn, R.; Brown, J.C.; Sanchez-David, R.Y.; et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021, 6, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Lubinski, B.; Whittaker, G.R. The SARS-CoV-2 furin cleavage site: Natural selection or smoking gun? Lancet Microbe 2023, 4, e570. [Google Scholar] [CrossRef] [PubMed]
- Hui, K.P.Y.; Ho, J.C.W.; Cheung, M.-C.; Ng, K.-C.; Ching, R.H.H.; Lai, K.-L.; Kam, T.T.; Gu, H.; Sit, K.-Y.; Hsin, M.K.Y.; et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 2022, 603, 715–720. [Google Scholar] [CrossRef]
- Letoha, A.; Hudák, A.; Letoha, T. Exploring the Syndecan-Mediated Cellular Internalization of the SARS-CoV-2 Omicron Variant. Int. J. Mol. Sci. 2023, 24, 14140. [Google Scholar] [CrossRef]
- Hudák, A.; Letoha, A.; Szilák, L.; Letoha, T. Contribution of syndecans to the cellular entry of SARS-CoV-2. Int. J. Mol. Sci. 2021, 22, 5336. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef] [PubMed]


| VLP Variants/Cell Line | VLP Binding to pH Drop or N Release | pH Drop or N Release to Speed Increase | VLP Binding to Speed Increase |
|---|---|---|---|
| VLP-WT MM in Vero E6 | 4.1 ± 3.6 min | 2.4 ± 3.7 min | 6.5 ± 5.4 min |
| VLP-WT MM in A549 | 12.5 ± 8.4 min | 1.4 ± 3.7 min | 13.9 ± 9.2 min |
| VLP-WT NM in A549 | 13 ± 8.6 min | 7.0 ± 10.0 min | 20.1 ± 14.8 min |
| VLP-Omi NM in A549 | 14.2 ± 8.5 min | 2.9 ± 6.9 min | 17.1 ± 10.1 min |
| VLP-del1 MM in Vero E6 | 6.4 ± 5.4 min | 2.8 ± 8.2 min | 9.2 ± 8.9 min |
| VLP-del1 MM in A549 | 16.7 ± 12.7 min | 1.7 ± 1.6 min | 18.4 ± 12.9 min |
| VLP-del1 MM in A549 | 10.12 ± 8.72 min | 3.61 ± 7.79 min | 13.74 ± 11.57 min |
| VLP-OMI MM in A549 | 13.01 ± 8.4 min | 4.03 ± 7.44 min | 17.04 ± 11.48 min |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atemin, A.; Ivanova, A.; Peppel, W.; Stamatov, R.; Gallegos, R.; Durden, H.; Uzunova, S.; Vershinin, M.D.; Saffarian, S.; Stoynov, S.S. Kinetic Landscape of Single Virus-like Particles Highlights the Efficacy of SARS-CoV-2 Internalization. Viruses 2024, 16, 1341. https://doi.org/10.3390/v16081341
Atemin A, Ivanova A, Peppel W, Stamatov R, Gallegos R, Durden H, Uzunova S, Vershinin MD, Saffarian S, Stoynov SS. Kinetic Landscape of Single Virus-like Particles Highlights the Efficacy of SARS-CoV-2 Internalization. Viruses. 2024; 16(8):1341. https://doi.org/10.3390/v16081341
Chicago/Turabian StyleAtemin, Aleksandar, Aneliya Ivanova, Wiley Peppel, Rumen Stamatov, Rodrigo Gallegos, Haley Durden, Sonya Uzunova, Michael D. Vershinin, Saveez Saffarian, and Stoyno S. Stoynov. 2024. "Kinetic Landscape of Single Virus-like Particles Highlights the Efficacy of SARS-CoV-2 Internalization" Viruses 16, no. 8: 1341. https://doi.org/10.3390/v16081341
APA StyleAtemin, A., Ivanova, A., Peppel, W., Stamatov, R., Gallegos, R., Durden, H., Uzunova, S., Vershinin, M. D., Saffarian, S., & Stoynov, S. S. (2024). Kinetic Landscape of Single Virus-like Particles Highlights the Efficacy of SARS-CoV-2 Internalization. Viruses, 16(8), 1341. https://doi.org/10.3390/v16081341







