Investigations on the Potential Role of Free-Ranging Wildlife as a Reservoir of SARS-CoV-2 in Switzerland
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Set-Up
2.2. Sample Collection
2.2.1. Postmortem Examinations
2.2.2. Field Sampling of Culled Animals or Found Dead Animals
2.2.3. Field Sampling of Captured Felids
2.2.4. Overview of Animals and Samples Included in the Study
2.3. Serological Analyses
2.3.1. SARS-CoV-2 S1 and Omicron S1 Enzyme-Linked Immunosorbent Assay (ELISA)
2.3.2. SARS-CoV-2 RBD-ELISA
2.3.3. Indirect Immunofluorescence Test (iIFT)
2.3.4. Surrogate Virus Neutralization Test (sVNT)
2.3.5. Pseudotype-Based Virus Neutralization Assay (PVNA)
2.4. Immunohistology
2.5. Molecular Detection of SARS-CoV-2
2.5.1. Nucleic Acid Extraction
2.5.2. SARS-CoV-2 RT-qPCRs
2.6. Submission to ProMed and OIE
2.7. Statistical Analysis and Software
3. Results
3.1. Serological Analyses
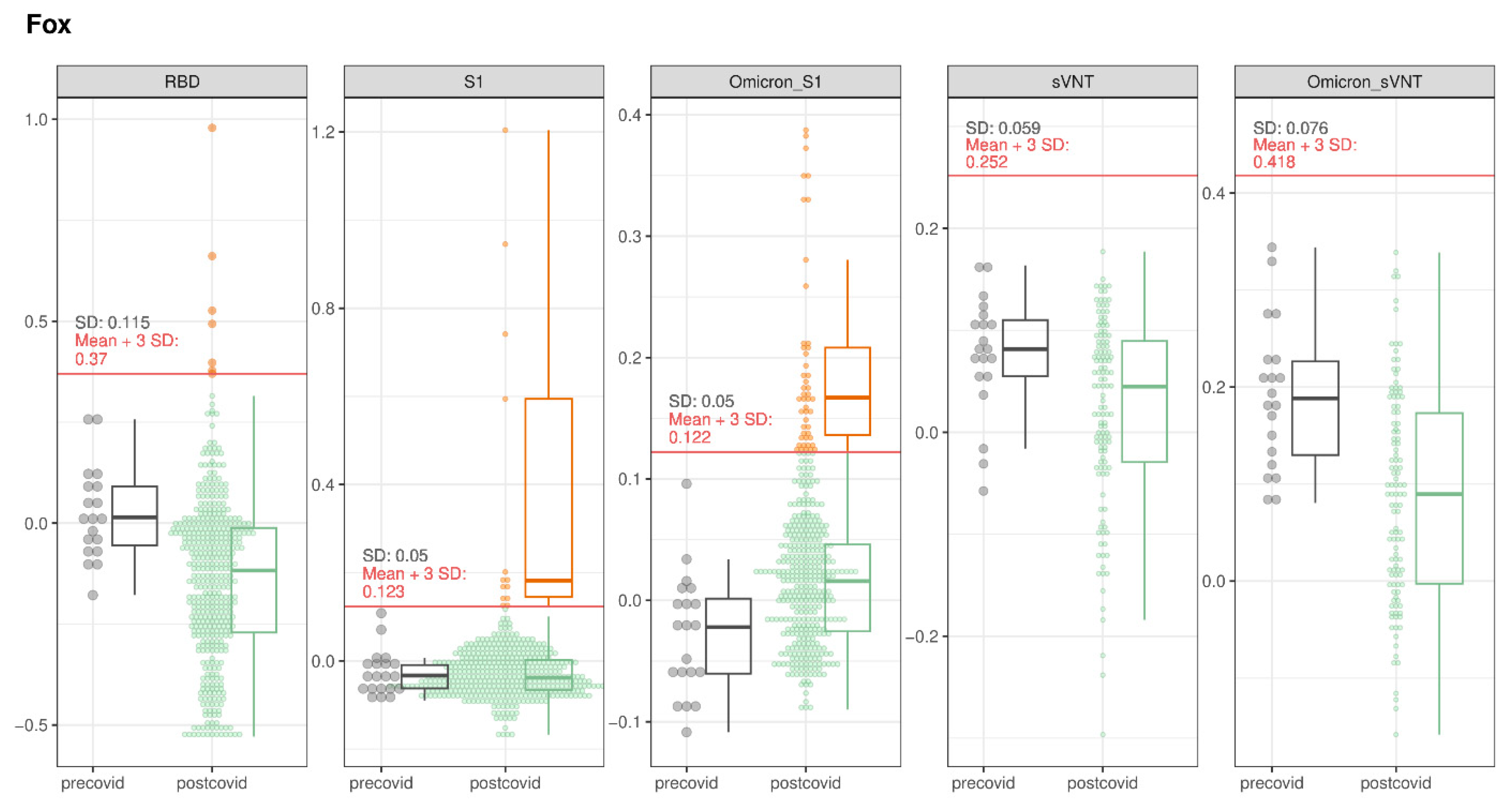
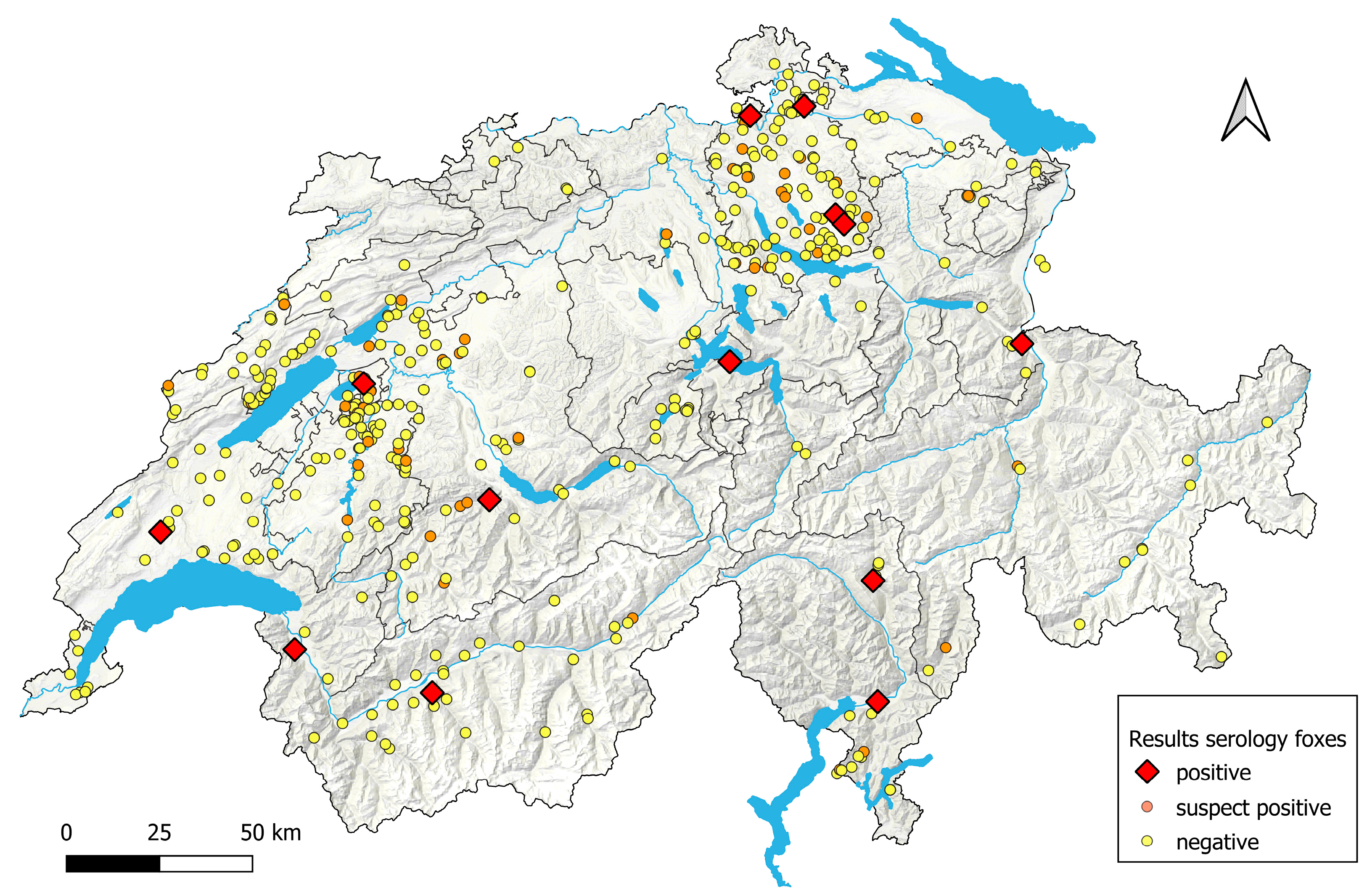
3.1.1. Foxes
3.1.2. Lynx
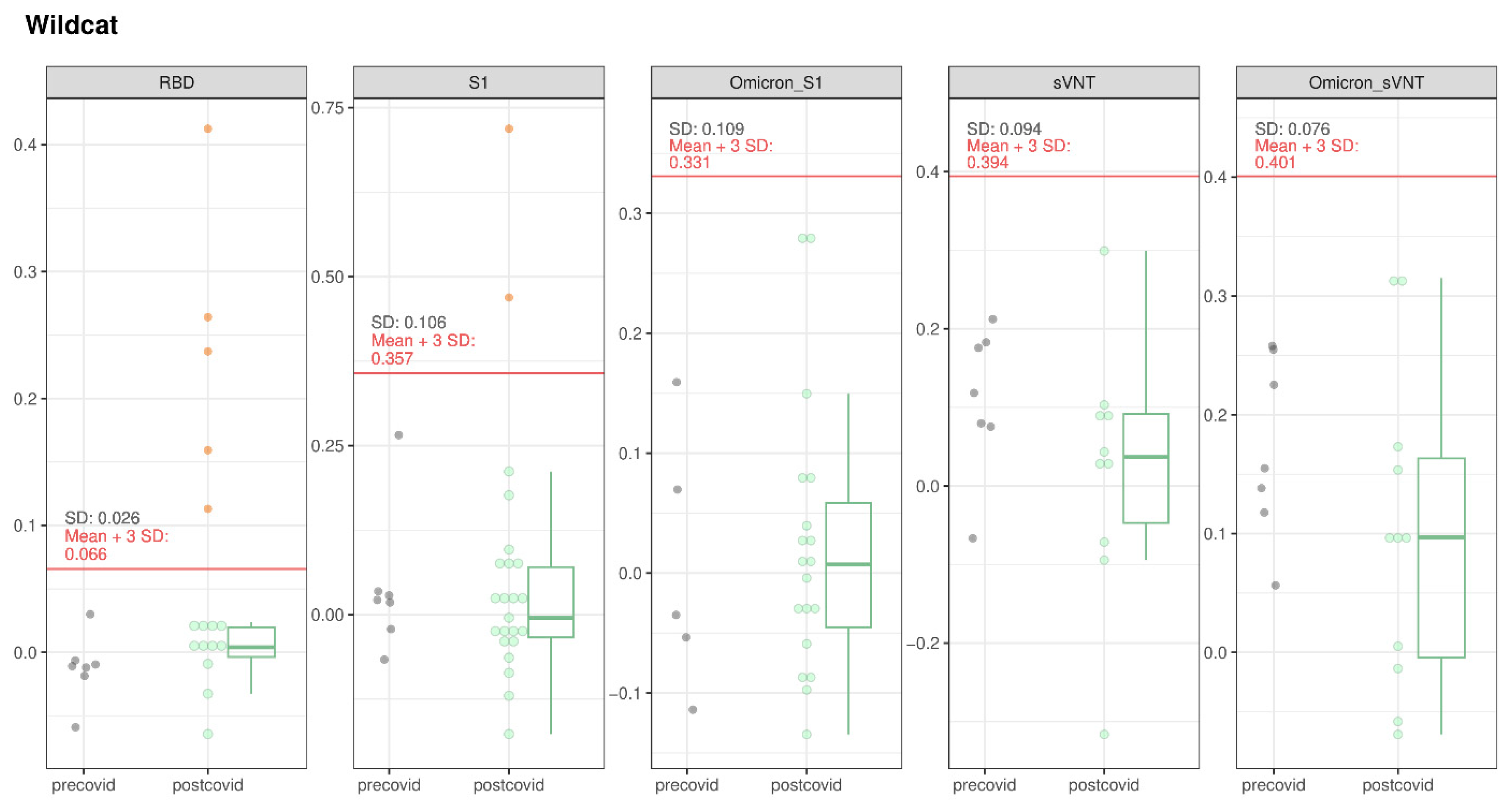
3.1.3. Wildcats
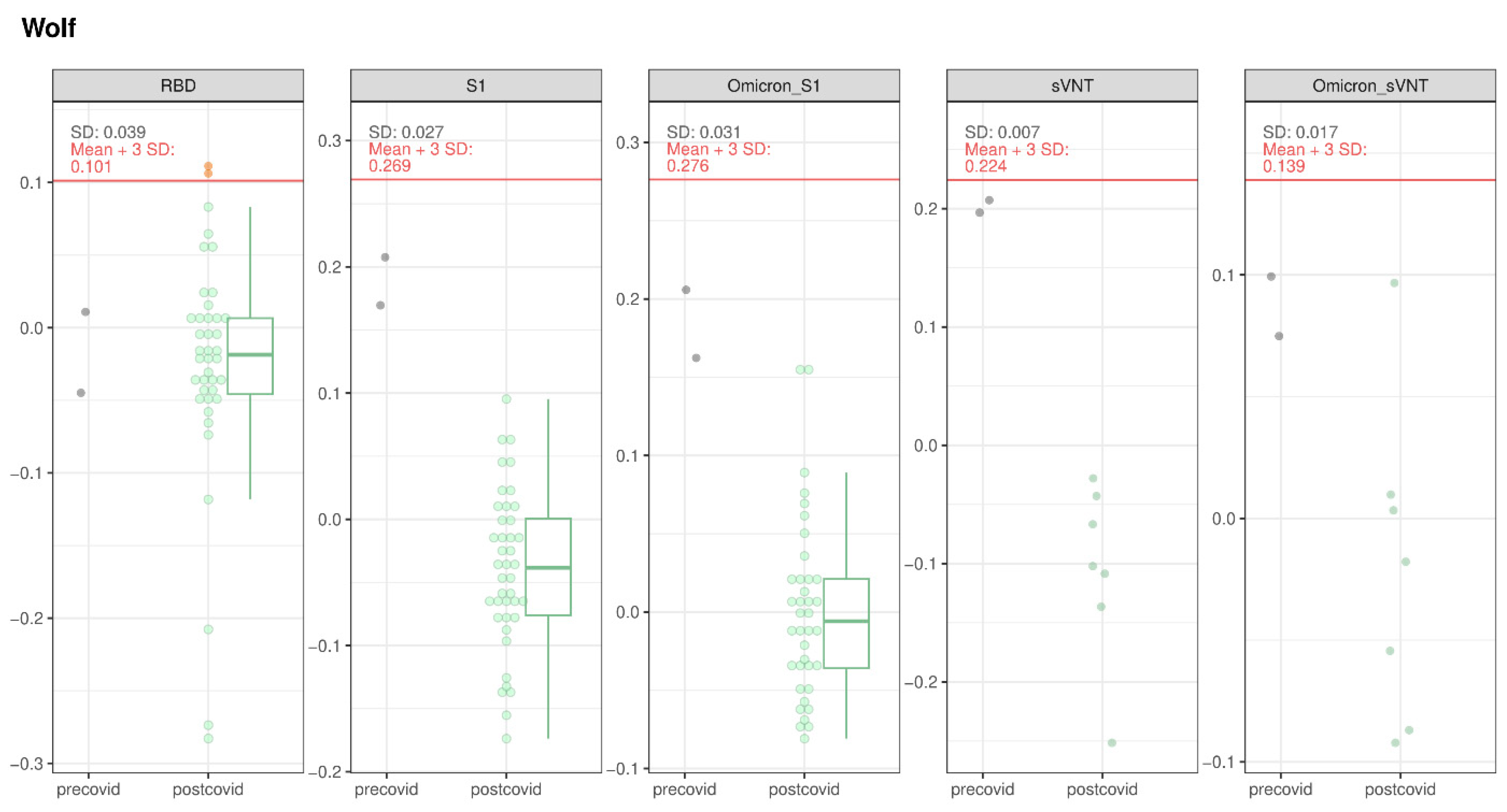
3.1.4. Wolves and Golden Jackal
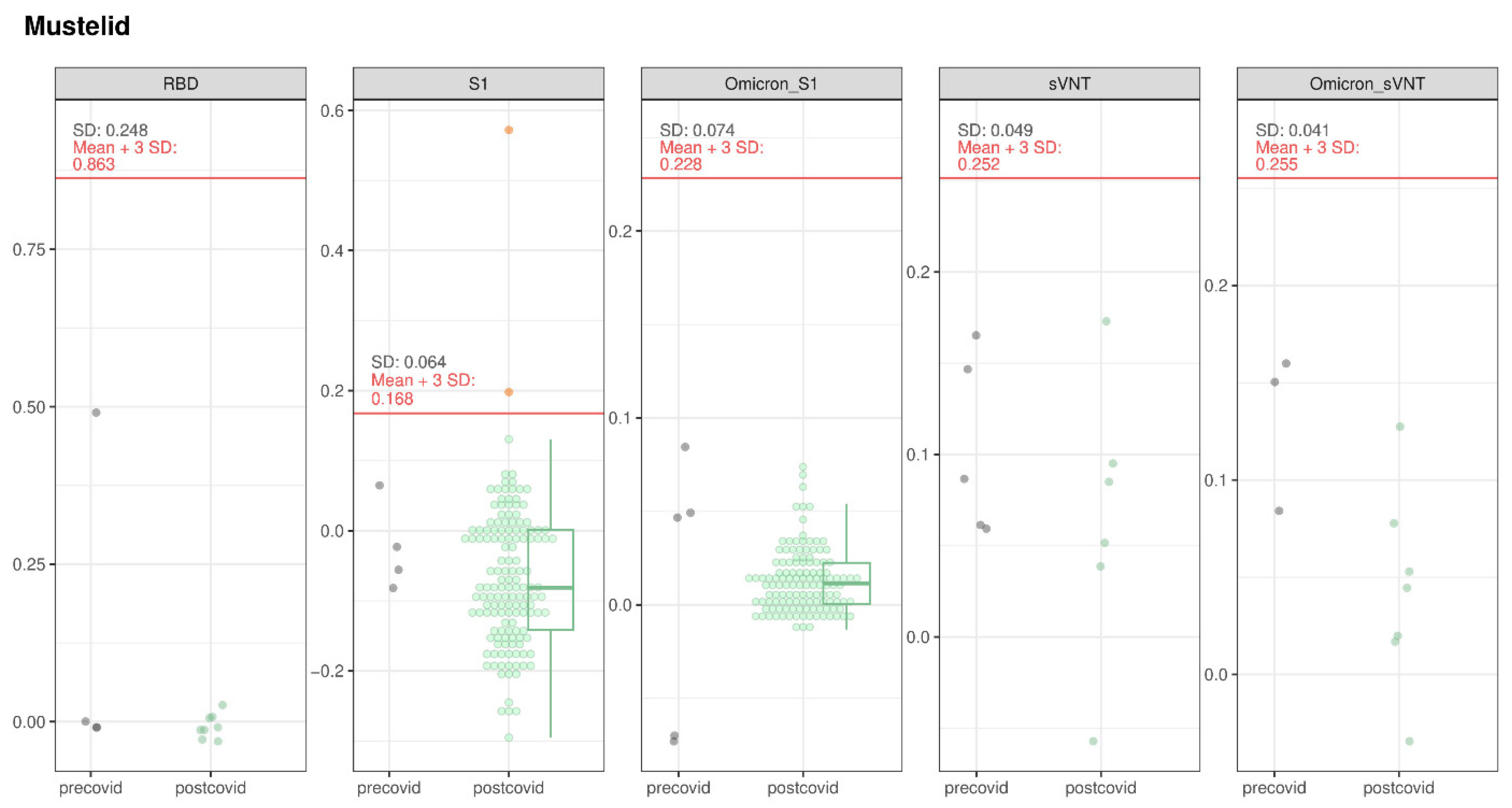
3.1.5. Mustelids
3.2. Detection of Viral RNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Temmam, S.; Vongphayloth, K.; Salazar, E.B.; Munier, S.; Bonomi, M.; Régnault, B.; Douangboubpha, B.; Karami, Y.; Chretien, D.; Sanamxay, D.; et al. Coronaviruses with a SARS-CoV-2-like Receptor-Binding Domain Allowing ACE2-Mediated Entry into Human Cells Isolated from Bats of Indochinese Peninsula. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Schlottau, K.; Rissmann, M.; Graaf, A.; Schön, J.; Sehl, J.; Wylezich, C.; Höper, D.; Mettenleiter, T.C.; Balkema-Buschmann, A.; Harder, T.; et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: An experimental transmission study. Lancet Microbe 2020, 1, e218–e225. [Google Scholar] [CrossRef]
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 18 December 2023).
- WOAH—World Organisation for Animal Health. SARS-CoV-2 in Animals—Situation Report 22—WOAH—World Organisation for Animal Health. Available online: https://www.woah.org/en/document/sars-cov-2-in-animals-situation-report-22/ (accessed on 7 September 2023).
- Cui, S.; Liu, Y.; Zhao, J.; Peng, X.; Lu, G.; Shi, W.; Pan, Y.; Zhang, D.; Yang, P.; Wang, Q. An Updated Review on SARS-CoV-2 Infection in Animals. Viruses 2022, 14, 1527. [Google Scholar] [CrossRef] [PubMed]
- SARS-ANI VIS. Available online: https://vis.csh.ac.at/sars-ani/#infections (accessed on 16 May 2024).
- Sharun, K.; Tiwari, R.; Natesan, S.; Dhama, K. SARS-CoV-2 Infection in Farmed Minks, Associated Zoonotic Concerns, and Importance of the One Health Approach during the Ongoing COVID-19 Pandemic; Taylor and Francis Ltd.: Abingdon, UK, 2021; Volume 41. [Google Scholar]
- Roundy, C.M.; Nunez, C.M.; Thomas, L.F.; Auckland, L.D.; Tang, W.; Richison, J.J.; Green, B.R.; Hilton, C.D.; Cherry, M.J.; Pauvolid-Correa, A.; et al. High seroprevalence of SARS-CoV-2 in white-tailed deer (Odocoileus virginianus) at one of three captive cervid facilities in Texas. Microbiol. Spectr. 2022, 10, e00576-22. [Google Scholar] [CrossRef]
- Hale, V.L.; Dennis, P.M.; McBride, D.S.; Nolting, J.M.; Madden, C.; Huey, D.; Ehrlich, M.; Grieser, J.; Winston, J.; Lombardi, D.; et al. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature 2022, 602, 481–486. [Google Scholar] [CrossRef]
- Porter, S.M.; Hartwig, A.E.; Bielefeldt-Ohmann, H.; Bosco-Lauth, A.M.; Root, J.J. Susceptibility of Wild Canids to SARS-CoV-2. Emerg. Infect. Dis. 2022, 28, 1852–1855. [Google Scholar] [CrossRef]
- Jemeršić, L.; Lojkić, I.; Krešić, N.; Keros, T.; Zelenika, T.A.; Jurinović, L.; Skok, D.; Bata, I.; Boras, J.; Habrun, B.; et al. Investigating the Presence of SARS CoV-2 in Free-Living and Captive Animals. Pathogens 2021, 10, 635. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Jin, Y.; Liu, Y.; Sun, J.; Hao, L.; Bai, J.; Huang, T.; Lin, D.; Jin, Y.; Tian, K. Serological survey of SARS-CoV-2 for experimental, domestic, companion and wild animals excludes intermediate hosts of 35 different species of animals. Transbound. Emerg. Dis. 2020, 67, 1745–1749. [Google Scholar] [CrossRef]
- ProMED-Mail. Promed Post—ProMED-Mail. Available online: https://promedmail.org/promed-post/?place=8705766,105#promedmailmap (accessed on 31 October 2023).
- Chan, T.; Ginders, J.; Kuhlmeier, E.; Meli, M.L.; Bönzli, E.; Meili, T.; Hüttl, J.; Hatt, J.-M.; Hindenlang Clerc, K.; Kipar, A.; et al. Detection of SARS-CoV-2 RNA in a Zoo-Kept Red Fox (Vulpes vulpes). Viruses 2024, 16, 521. [Google Scholar] [CrossRef]
- Tizzani, P. SARS-CoV-2 in animals—Situation Report 17; WOAH: Paris, France, 2022. [Google Scholar]
- WOAH. Situation Report n.14; WOAH: Paris, France, 2022. [Google Scholar]
- USDA APHIS|Confirmation of COVID-19 in a Canada Lynx at a Pennsylvania Zoo. Available online: https://www.aphis.usda.gov/aphis/newsroom/stakeholder-info/sa_by_date/sa-2021/sa-12/covid-lynx-pa (accessed on 18 December 2023).
- Molenaar, R.J.; Vreman, S.; Hakze-van der Honing, R.W.; Zwart, R.; De Rond, J.; Weesendorp, E.; Smit, L.A.M.; Koopmans, M.; Bouwstra, R.; Stegeman, A.; et al. Clinical and Pathological Findings in SARS-CoV-2 Disease Outbreaks in Farmed Mink (Neovison vison). Vet. Pathol. 2020, 57, 653–657. [Google Scholar] [CrossRef]
- Chu, H.; Chan, J.F.-W.; Yuen, K.-Y. Animal models in SARS-CoV-2 research. Nat. Methods 2022, 19, 392–394. [Google Scholar] [CrossRef]
- Klaus, J.; Meli, M.L.; Willi, B.; Nadeau, S.; Beisel, C.; Stadler, T.; Eth, S.-C.-S.T.; Egberink, H.; Zhao, S.; Lutz, H.; et al. Detection and Genome Sequencing of SARS-CoV-2 in a Domestic Cat with Respiratory Signs in Switzerland. Viruses 2021, 13, 496. [Google Scholar] [CrossRef] [PubMed]
- Klaus, J.; Palizzotto, C.; Zini, E.; Meli, M.L.; Leo, C.; Egberink, H.; Zhao, S.; Hofmann-Lehmann, R. SARS-CoV-2 Infection and Antibody Response in a Symptomatic Cat from Italy with Intestinal B-Cell Lymphoma. Viruses 2021, 13, 527. [Google Scholar] [CrossRef] [PubMed]
- Koeppel, K.N.; Mendes, A.; Strydom, A.; Rotherham, L.; Mulumba, M.; Venter, M. SARS-CoV-2 Reverse Zoonoses to Pumas and Lions, South Africa. Viruses 2022, 14, 120. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Smits, C.; Schuurman, N.; Barnum, S.; Pusterla, N.; van Kuppeveld, F.; Bosch, B.-J.; van Maanen, K.; Egberink, H. Development and Validation of a S1 Protein-Based ELISA for the Specific Detection of Antibodies against Equine Coronavirus. Viruses 2019, 11, 1109. [Google Scholar] [CrossRef]
- Kuhlmeier, E.; Chan, T.; Agüí, C.V.; Willi, B.; Wolfensberger, A.; Beisel, C.; Topolsky, I.; Beerenwinkel, N.; Stadler, T.; Swiss, S.-C.-S.C.; et al. Detection and Molecular Characterization of the SARS-CoV-2 Delta Variant and the Specific Immune Response in Companion Animals in Switzerland. Viruses 2023, 15, 245. [Google Scholar] [CrossRef]
- Wernike, K.; Aebischer, A.; Michelitsch, A.; Hoffmann, D.; Freuling, C.; Balkema-Buschmann, A.; Graaf, A.; Müller, T.; Osterrieder, N.; Rissmann, M.; et al. Multi-species ELISA for the detection of antibodies against SARS-CoV-2 in animals. Transbound. Emerg. Dis. 2021, 68, 1779–1785. [Google Scholar] [CrossRef]
- Voller, A.; Bidwell, D.E.; Bartlett, A. Enzyme immunoassays in diagnostic medicine. Theory and practice. Bull. World Health Organ. 1976, 53, 55–65. [Google Scholar]
- Berguido, F.J.; Burbelo, P.D.; Bortolami, A.; Bonfante, F.; Wernike, K.; Hoffmann, D.; Balkema-Buschmann, A.; Beer, M.; Dundon, W.G.; Lamien, C.E.; et al. Serological Detection of SARS-CoV-2 Antibodies in Naturally-Infected Mink and Other Experimentally-Infected Animals. Viruses 2021, 13, 1649. [Google Scholar] [CrossRef]
- Boenzli, E.; Hadorn, M.; Hartnack, S.; Huder, J.; Hofmann-Lehmann, R.; Lutz, H. Detection of antibodies to the feline leukemia Virus (FeLV) transmembrane protein p15E: An alternative approach for serological FeLV detection based on antibodies to p15E. J. Clin. Microbiol. 2014, 52, 2046–2052. [Google Scholar] [CrossRef]
- Michelitsch, A.; Hoffmann, D.; Wernike, K.; Beer, M. Occurrence of Antibodies against SARS-CoV-2 in the Domestic Cat Population of Germany. Vaccines 2020, 8, 772. [Google Scholar] [CrossRef]
- Klaus, J.; Zini, E.; Hartmann, K.; Egberink, H.; Kipar, A.; Bergmann, M.; Palizzotto, C.; Zhao, S.; Rossi, F.; Franco, V.; et al. SARS-CoV-2 Infection in Dogs and Cats from Southern Germany and Northern Italy during the First Wave of the COVID-19 Pandemic. Viruses 2021, 13, 1453. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.B.; Jones, S.; Logan, N.; McDonald, M.; Marshall, L.; Murcia, P.R.; Willett, B.J.; Weir, W.; Hosie, M.J. SARS-CoV-2 Seroprevalence and Cross-Variant Antibody Neutralization in Cats, United Kingdom. Emerg. Infect. Dis. 2023, 29, 1223–1227. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.B.; Jones, S.; Montreuil-Spencer, C.; Logan, N.; Scott, S.; Sasvari, H.; McDonald, M.; Marshall, L.; Murcia, P.R.; Willett, B.J.; et al. Increase in SARS-CoV-2 Seroprevalence in UK Domestic Felids Despite Weak Immunogenicity of Post-Omicron Variants. Viruses 2023, 15, 1661. [Google Scholar] [CrossRef]
- Tyson, G.B.; Jones, S.; Logan, N.; McDonald, M.; Murcia, P.R.; Willett, B.J.; Weir, W.; Hosie, M.J. Rising SARS-CoV-2 Seroprevalence and Patterns of Cross-Variant Antibody Neutralization in UK Domestic Cats. BioRxiv 2022. [Google Scholar] [CrossRef]
- Davis, C.; Logan, N.; Tyson, G.; Orton, R.; Harvey, W.T.; Perkins, J.S.; Mollett, G.; Blacow, R.M.; Peacock, T.P.; Barclay, W.S.; et al. Reduced neutralisation of the Delta (B.1.617.2) SARS-CoV-2 variant of concern following vaccination. PLoS Pathog. 2021, 17, e1010022. [Google Scholar] [CrossRef]
- Kant, R.; Kareinen, L.; Smura, T.; Freitag, T.L.; Jha, S.K.; Alitalo, K.; Meri, S.; Sironen, T.; Saksela, K.; Strandin, T.; et al. Common Laboratory Mice Are Susceptible to Infection with the SARS-CoV-2 Beta Variant. Viruses 2021, 13, 2263. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. Bull. Eur. Mal. Transm.=Eur. Commun. Dis. Bull. 2020, 25, 2000045. [Google Scholar] [CrossRef]
- COVID-19 Schweiz|Coronavirus|Dashboard. Available online: https://www.covid19.admin.ch/de/epidemiologic/virus-variants?variantsHosp=VariantBA1,VariantBA2,VariantBA275,VariantBA4,VariantBA5,VariantBQ1,VariantOtherWgs,VariantXBB (accessed on 31 May 2024).
- Goldberg, A.R.; Langwig, K.E.; Brown, K.L.; Marano, J.M.; Rai, P.; King, K.M.; Sharp, A.K.; Ceci, A.; Kailing, C.D.; Kailing, M.J.; et al. Widespread exposure to SARS-CoV-2 in wildlife communities. Nat. Commun. 2024, 15, 6210. [Google Scholar] [CrossRef]
- Bordes, L.; Vreman, S.; Heutink, R.; Roose, M.; Venema, S.; Pritz-Verschuren, S.B.E.; Rijks, J.M.; Gonzales, J.L.; Germeraad, E.A.; Engelsma, M.; et al. Highly Pathogenic Avian Influenza H5N1 Virus Infections in Wild Red Foxes (Vulpes vulpes) Show Neurotropism and Adaptive Virus Mutations. Microbiol. Spectr. 2023, 11, e0286722. [Google Scholar] [CrossRef]
- Jung, S.; Yeo, D.; Wang, Z.; Choi, C. Survival of severe acute respiratory syndrome coronavirus 2 in foods and its inactivation by different methods. Curr. Opin. Food Sci. 2024, 55, 101106. [Google Scholar] [CrossRef]
- Helldin, J.O.; Liberg, O.; Glöersen, G. Lynx (Lynx lynx) killing red foxes (Vulpes vulpes) in boreal Sweden—Frequency and population effects. J. Zool. 2006, 270, 657–663. [Google Scholar] [CrossRef]
- Nájera, F.; Sánchez-Cuerda, S.; López, G.; Del Rey-Wamba, T.; Rueda, C.; Vallverdú-Coll, N.; Panadero, J.; Palacios, M.J.; López-Bao, J.V.; Jiménez, J. Lynx eats cat: Disease risk assessment during an Iberian lynx intraguild predation. Eur. J. Wildl. Res. 2019, 65, 39. [Google Scholar] [CrossRef]
- van Aart, A.E.; Velkers, F.C.; Fischer, E.A.; Broens, E.M.; Egberink, H.; Zhao, S.; Engelsma, M.; der Honing, R.W.H.; Harders, F.; de Rooij, M.M.; et al. SARS-CoV-2 infection in cats and dogs in infected mink farms. Transbound. Emerg. Dis. 2021, 69, 3001–3007. [Google Scholar] [CrossRef]
- Padilla-Blanco, M.; Aguiló-Gisbert, J.; Rubio, V.; Lizana, V.; Chillida-Martínez, E.; Cardells, J.; Maiques, E.; Rubio-Guerri, C. The Finding of the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) in a Wild Eurasian River Otter (Lutra lutra) Highlights the Need for Viral Surveillance in Wild Mustelids. Front. Vet. Sci. 2022, 9, 826991. [Google Scholar] [CrossRef] [PubMed]
- Aguiló-Gisbert, J.; Padilla-Blanco, M.; Lizana, V.; Maiques, E.; Muñoz-Baquero, M.; Chillida-Martínez, E.; Cardells, J.; Rubio-Guerri, C. First Description of SARS-CoV-2 Infection in Two Feral American Mink (Neovison vison) Caught in the Wild. Animals 2021, 11, 1422. [Google Scholar] [CrossRef]
- Felten, S.; Klein-Richers, U.; Hofmann-Lehmann, R.; Bergmann, M.; Unterer, S.; Leutenegger, C.M.; Hartmann, K. Correlation of Feline Coronavirus Shedding in Feces with Coronavirus Antibody Titer. Pathogens 2020, 9, 598. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Liu, Y.; Lyu, G.; Shang, S.; Xia, H.; Zhang, J.; Irwin, D.M.; Wang, Z.; Zhang, S. Adaptive Evolution of the Fox Coronavirus Based on Genome-Wide Sequence Analysis. Biomed Res. Int. 2022, 2022, 9627961. [Google Scholar] [CrossRef]
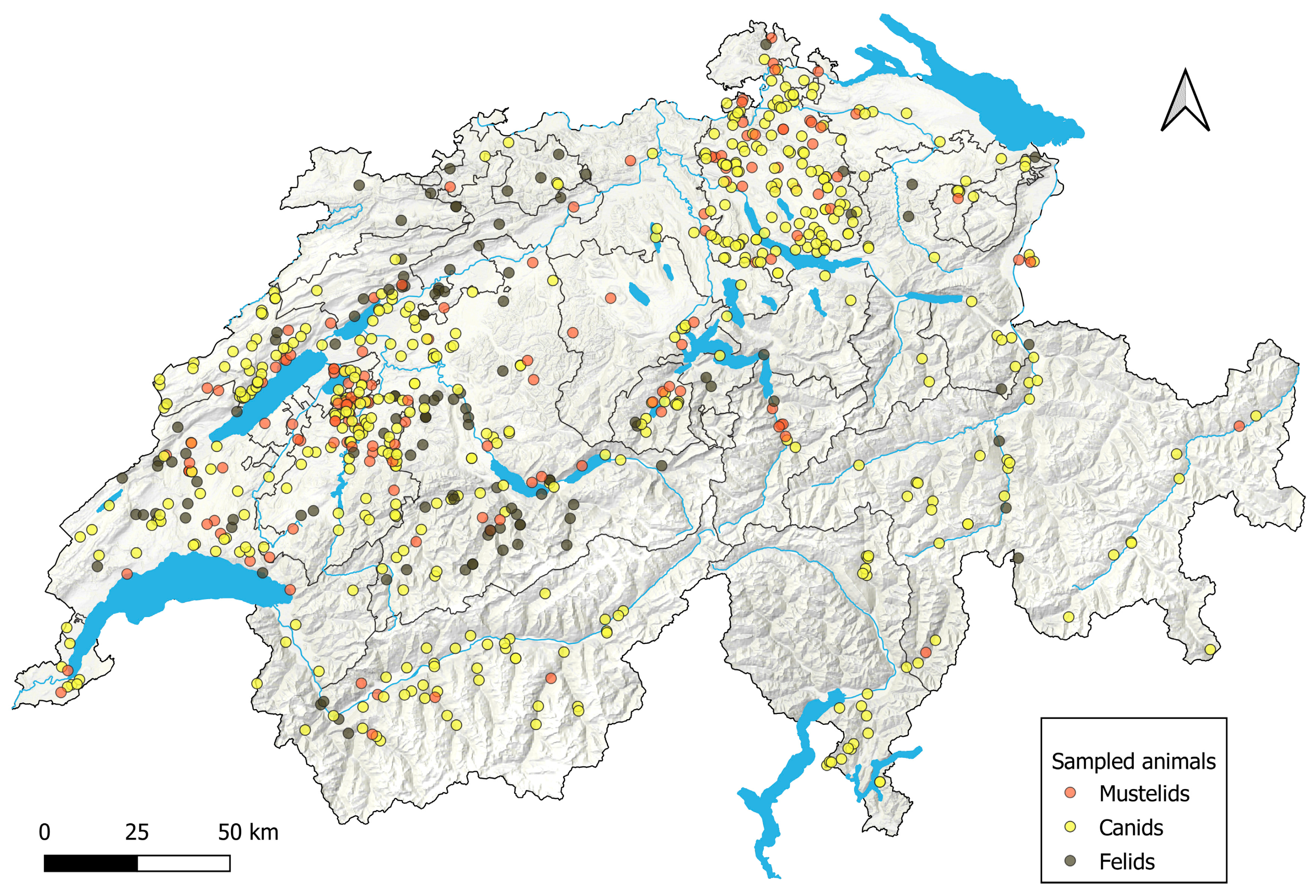



| Taxonomy | Species | Behavior | Protected | Necropsies 1 | Field Sampling | Immobilization | Total/Estimated Sample Sizes |
|---|---|---|---|---|---|---|---|
| Canids | Red fox | Social | No | 30 | 419 | 449/600 | |
| Gray wolf | Social | Yes | 41 | 41/20 | |||
| Golden jackal | Variable | Yes | 1 | 1/<5 | |||
| Felids | Eurasian lynx | Solitary | Yes | 56 | 39 | 95/60 | |
| European wildcat | Solitary | Yes | 18 | 6 | 24/10 | ||
| Mustelids | Stone and pine marten | Solitary | No | 20 | 44 | 64/210 | |
| European badger | Social | No | 8 | 65 | 73/210 | ||
| European polecat | Solitary | Yes | 2 | 8 | 10/10 | ||
| Total | 8 | 176 | 536 | 45 | 757/1115 |
| Taxonomy | Species | Serum | Oronasal/ Oropharyngeal Swabs | Rectal Swabs | Lung Tissue |
|---|---|---|---|---|---|
| Canids | Red fox | 446 | 29 | 27 | 29 |
| Gray wolf | 41 | 39 | 38 | 36 | |
| Golden jackal | 1 | 1 | 1 | 1 | |
| Felids | Eurasian lynx | 92 * | 91 | 91 | 52 |
| European wildcat | 23 | 22 | 22 | 16 | |
| Mustelids | European marten | 62 | 18 | 18 | 18 |
| European badger | 72 | 8 | 8 | 8 | |
| European polecat | 10 | 2 | 2 | 2 | |
| Total | 8 | 747 | 210 | 207 | 162 |
| Species | No of Samples | Suspect Positive Binding Antibodies | IFT Confirmed Binding Antibodies ° | sVNT Positive ° | PVNA Positive ° |
|---|---|---|---|---|---|
| Red fox | 446 | 64/446 | 12/64 | 0/116 | 2/120 |
| Grey wolf | 41 | 2/41 | 0/7 | 0/7 | 0/7 |
| Golden jackal | 1 | 0/1 | nt* | nt* | nt* |
| Eurasian lynx | 92 * | 4 */92 | 3 */11 | 1/16 * | 0 */10 |
| European wildcat | 24 | 6/24 | 0 °/7 | 0 °/11 | 1 °/11 |
| Stone marten | 48 | 1/48 | 0/1 | 0/4 | 0/1 |
| Pine marten | 13 | 0/13 | nt* | nt* | nt* |
| European badger | 72 | 0/72 | nt* | nt* | nt* |
| European polecat | 10 | 1/10 | 0/4 | 0/2 | 0/2 |
| Fox ID | Sampling Date | Sex | Age | Clinical Signs | Localization | Binding/Neutralizing |
|---|---|---|---|---|---|---|
| 80 | 3 December 2021 | Female | Over a year | Sarcoptic mange | Canton Nidwalden, near dogs | Binding |
| 811 | 27 January 2022 | Male | Over a year | Wounded right foreleg | Canton Bern, near cattle | Binding |
| 943 | 31 January 2022 | Unknown | Over a year | Apparently healthy | Canton Valais | Binding |
| W22_6633 | 26 April 2022 | Male | Over a year | Signs of canine distemper (CDV) * | Canton Zurich | Binding |
| 1091 | 11 November 2022 | Female | Over a year | Apparently healthy | Canton Zurich | Binding |
| 1434 | 12 November 2022 | Male | Over a year | Unknown | Canton Zurich | Binding |
| 1787 | 8 December 2022 | Unknown | Over a year | Apparently healthy | Canton St. Gallen | Binding |
| 866 | 5 January 2023 | Female | Under a year | Apparently healthy | Canton Ticino | Binding |
| 1655 | 16 January 2023 | Male | Over a year | Hit by car, apparently healthy | Canton Fribourg | Binding |
| 1827 | 23 February 2023 | Female | Over a year | Apparently healthy | Canton Ticino | Binding |
| 1501 | 14 March 2023 | Male | Over a year | Apparently healthy | Canton Vaud | Binding |
| 1761 | 29 March 2023 | Male | Over a year | Apparently healthy | Canton Valais | Binding |
| 1577 | 9 November 2022 | Male | Over a year | Apparently healthy | Canton Zurich, near sheep | Neutralizing |
| 371 | 28 February 2023 | Male | Over a year | Suspected signs of distemper (CDV) | Canton St. Gallen | Neutralizing |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuhn, J.; Marti, I.; Ryser-Degiorgis, M.-P.; Wernike, K.; Jones, S.; Tyson, G.; Delalay, G.; Scherrer, P.; Borel, S.; Hosie, M.J.; et al. Investigations on the Potential Role of Free-Ranging Wildlife as a Reservoir of SARS-CoV-2 in Switzerland. Viruses 2024, 16, 1407. https://doi.org/10.3390/v16091407
Kuhn J, Marti I, Ryser-Degiorgis M-P, Wernike K, Jones S, Tyson G, Delalay G, Scherrer P, Borel S, Hosie MJ, et al. Investigations on the Potential Role of Free-Ranging Wildlife as a Reservoir of SARS-CoV-2 in Switzerland. Viruses. 2024; 16(9):1407. https://doi.org/10.3390/v16091407
Chicago/Turabian StyleKuhn, Juliette, Iris Marti, Marie-Pierre Ryser-Degiorgis, Kerstin Wernike, Sarah Jones, Grace Tyson, Gary Delalay, Patrick Scherrer, Stéphanie Borel, Margaret J. Hosie, and et al. 2024. "Investigations on the Potential Role of Free-Ranging Wildlife as a Reservoir of SARS-CoV-2 in Switzerland" Viruses 16, no. 9: 1407. https://doi.org/10.3390/v16091407







