SARS-CoV-2 Impairs Osteoblast Differentiation Through Spike Glycoprotein and Cytokine Dysregulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Expansion MSCs
2.2. Osteoblast and Adipocyte Differentiation from MSCs
2.3. Viral Infection of Osteoblasts, Adipocytes, and MSCs
2.4. Measurement of ACE2 Surface Expression in Osteoblasts and MSCs by Flow Cytometry
2.5. Detection and Quantification of SARS-CoV-2 Genomic RNA
2.6. Detection of Subgenomic (sg) SARS-CoV-2 RNA
2.7. Cell Death and Mitochondrial Reactive Oxygen Species (ROS) Production
2.8. UV-C Irradiation for SARS-CoV-2 Inactivation
2.9. Evaluation of SARS-CoV-2 Infectious Particles
2.10. Cellular mRNA Preparation and RT-qPCR
2.11. Spike (S) Protein Neutralization Assay
2.12. Evaluation of Osteogenic Differentiation
2.12.1. Alkaline Phosphatase (ALP) Activity
2.12.2. Assessment of Calcium Deposition by Alizarin Red S Staining
2.12.3. Assessment of Collagen Deposition by Sirius Red Staining
2.13. Evaluation of Adipocyte Differentiation Through Lipid Droplet Accumulation Analysis
2.14. Statistical Analysis
3. Results
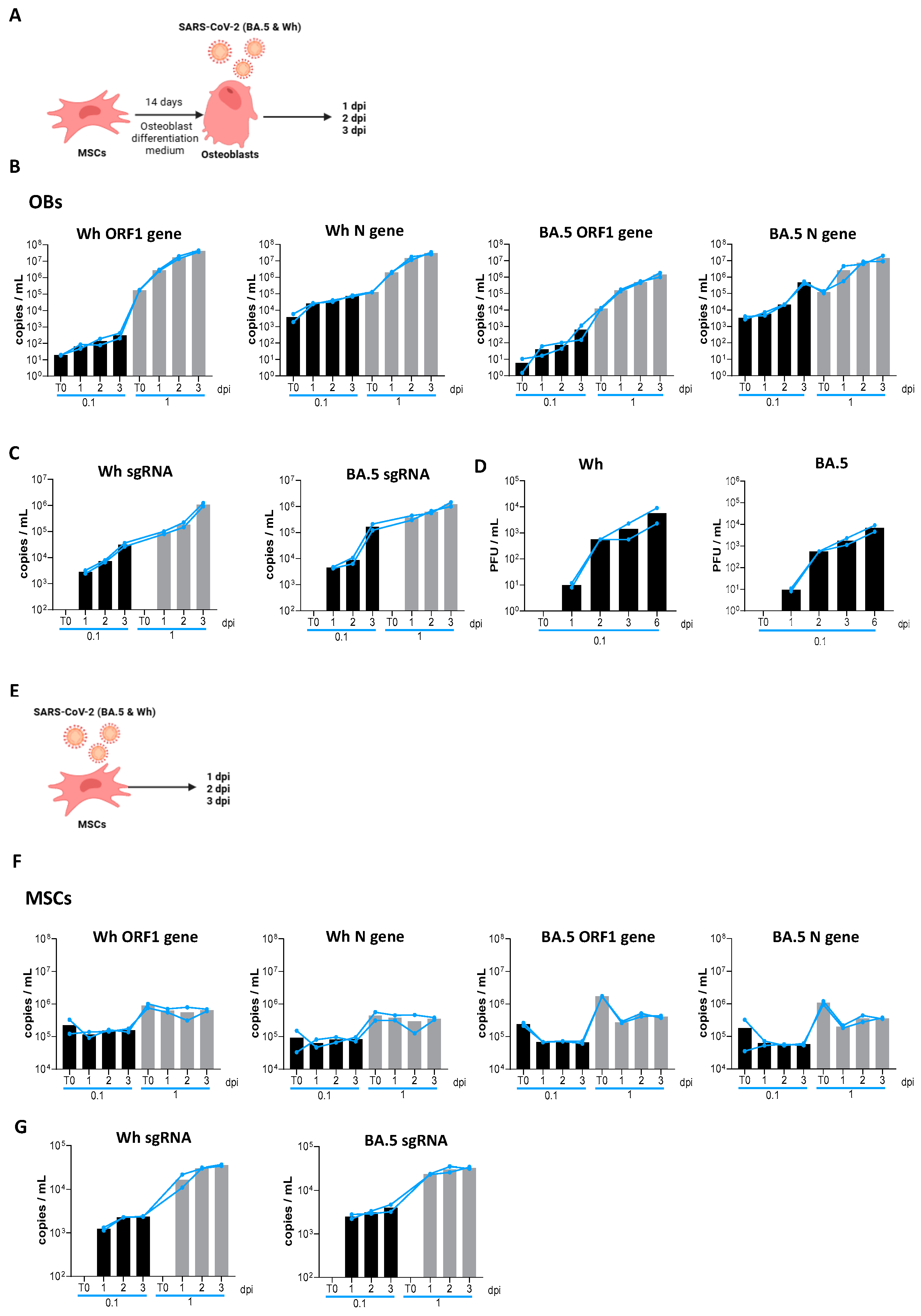
3.1. Osteoblasts Are Permissive for Productive SARS-CoV-2 Replication
3.2. SARS-CoV-2 Causes an Abortive Infection in MSCs
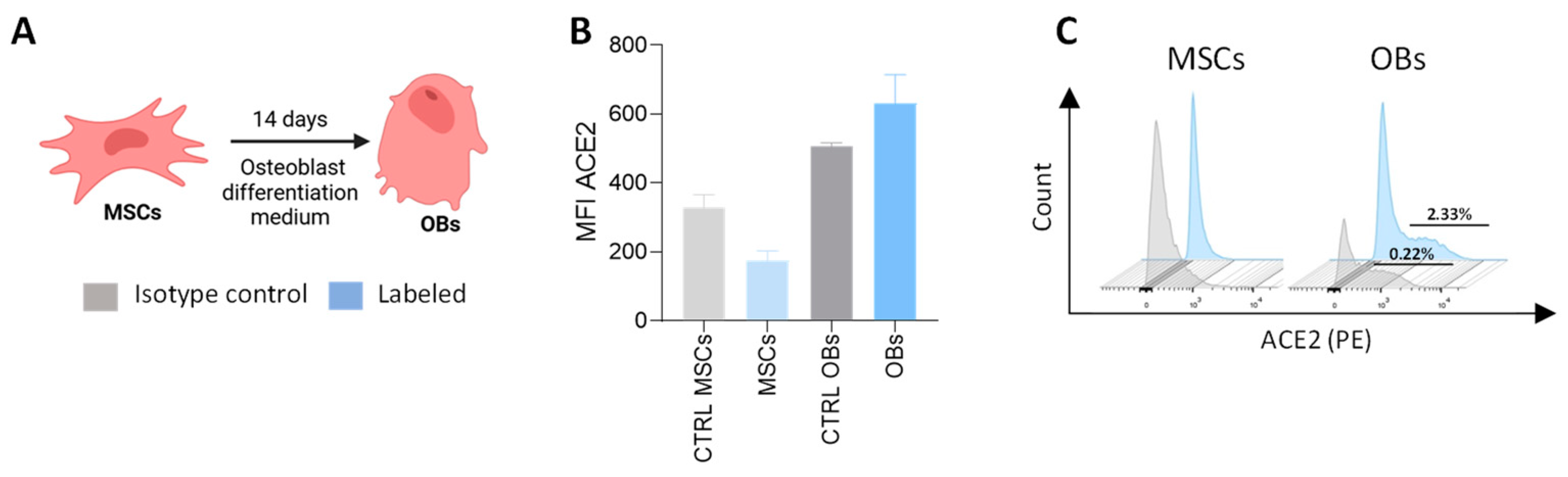
3.3. Expression of ACE2 in Precursor Cells and Differentiated Osteoblasts
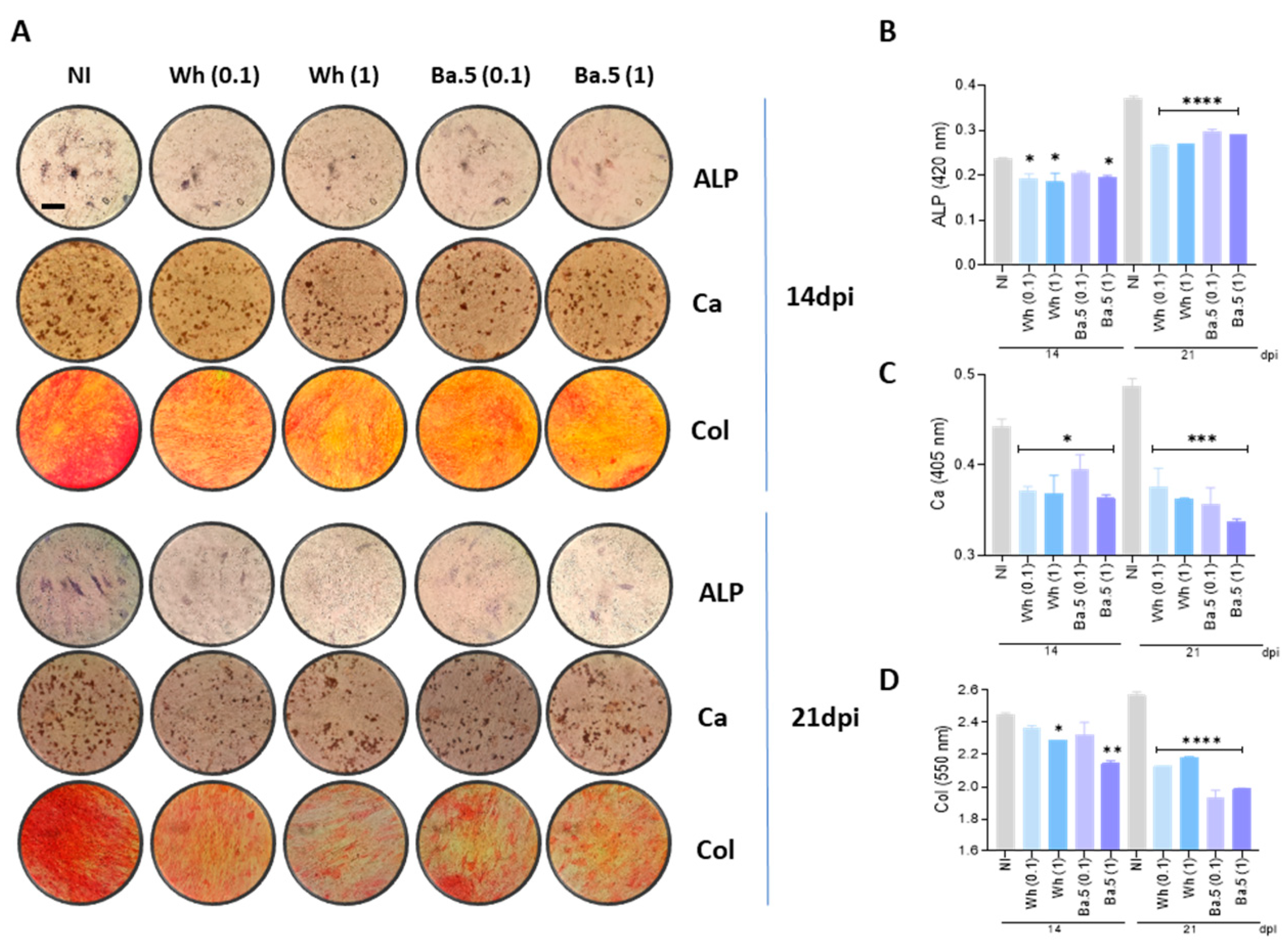
3.4. SARS-CoV-2 Inhibits Osteoblast Differentiation and Function
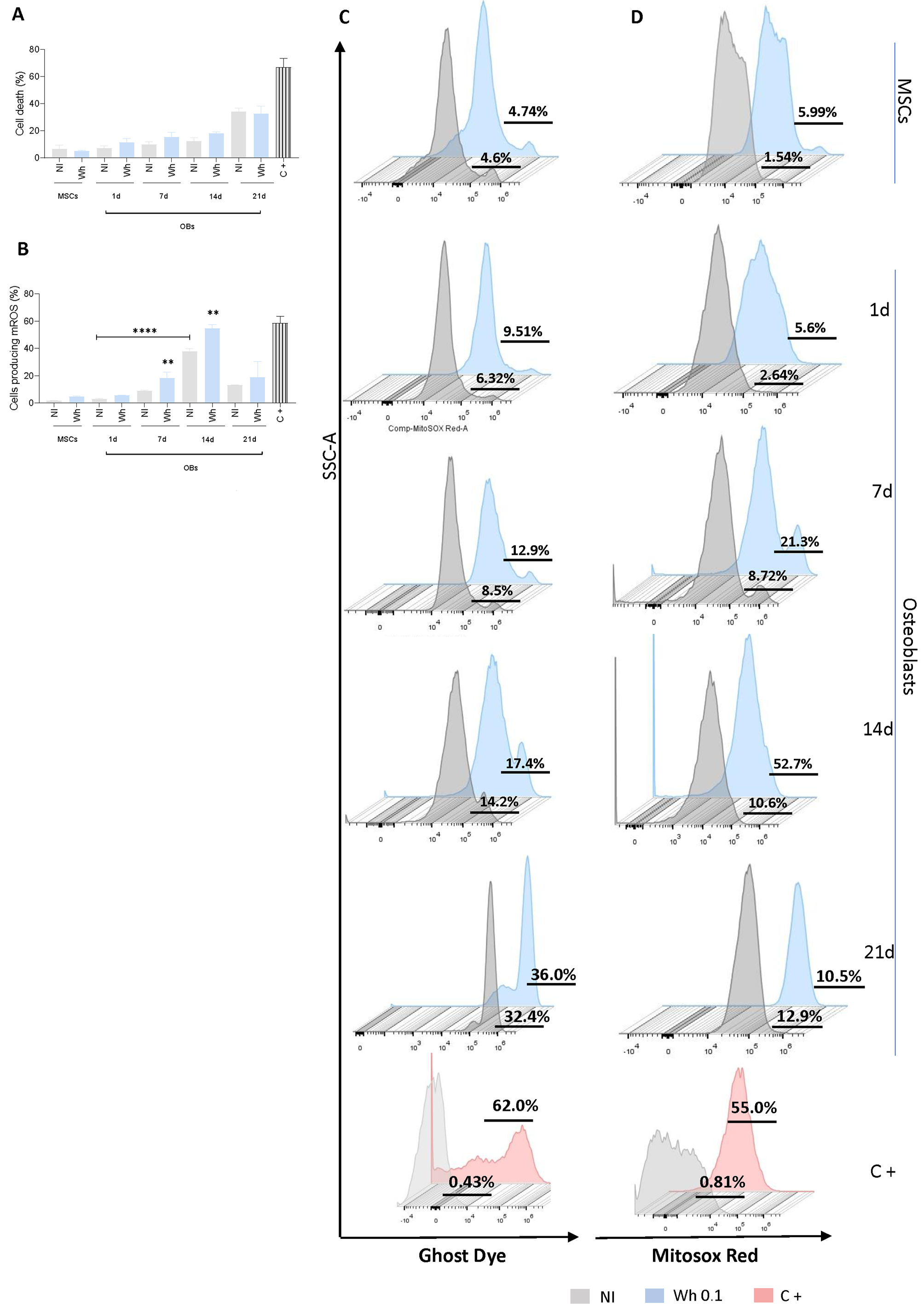
3.5. SARS-CoV-2-Abortively Infected MSCs Depict Redox Imbalance but Preserve Viability
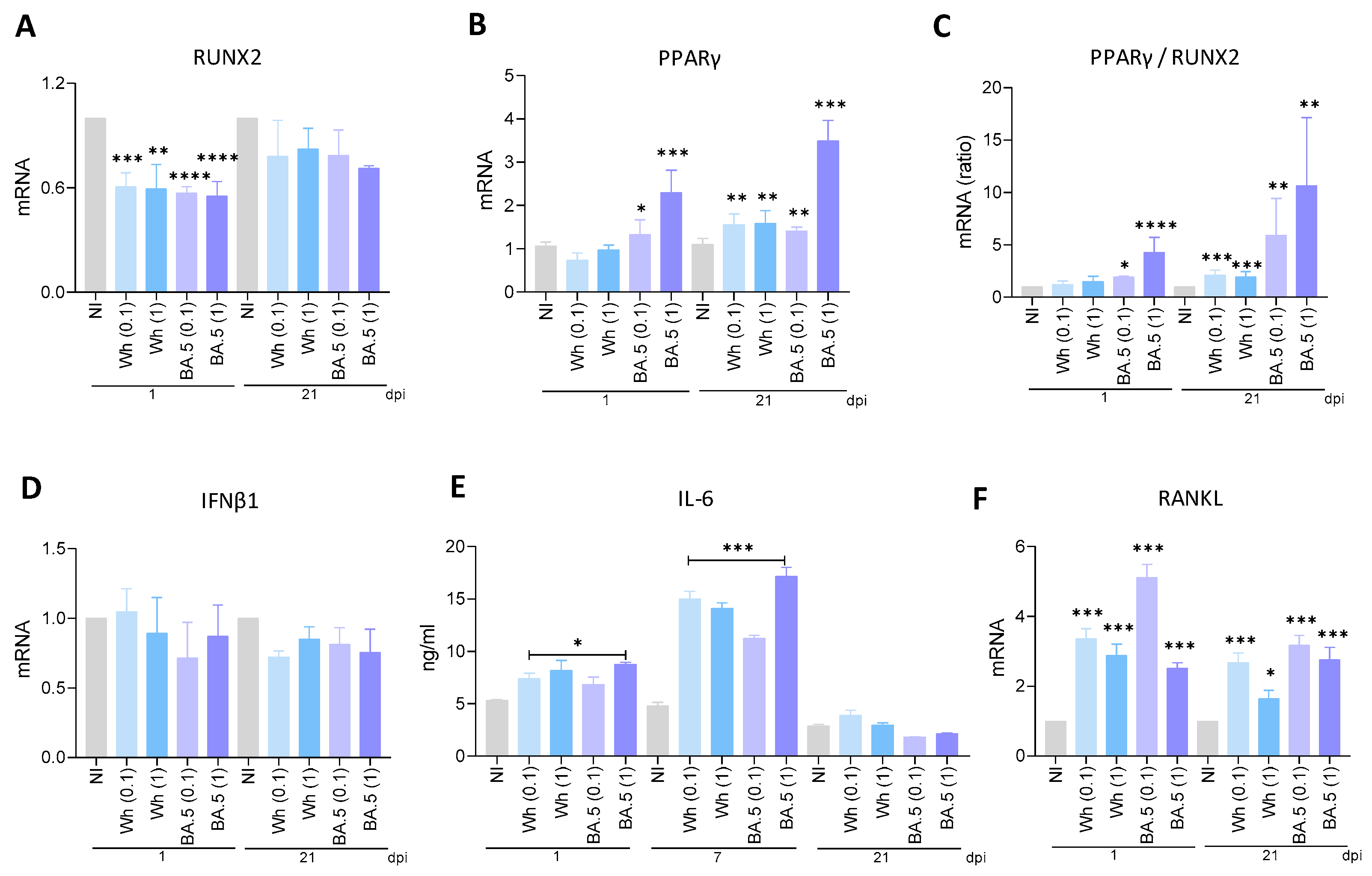
3.6. SARS-CoV-2 Modulates RUNX2 and PPARγ Transcription
3.7. SARS-CoV-2 Variants Were Unable to Promote IFNβ1 Transcription
3.8. Wh and BA.5 SARS-CoV-2 Strains Induce IL-6 Secretion During Osteoblast Differentiation
3.9. SARS-CoV-2 Induces RANKL Expression in Osteoblasts
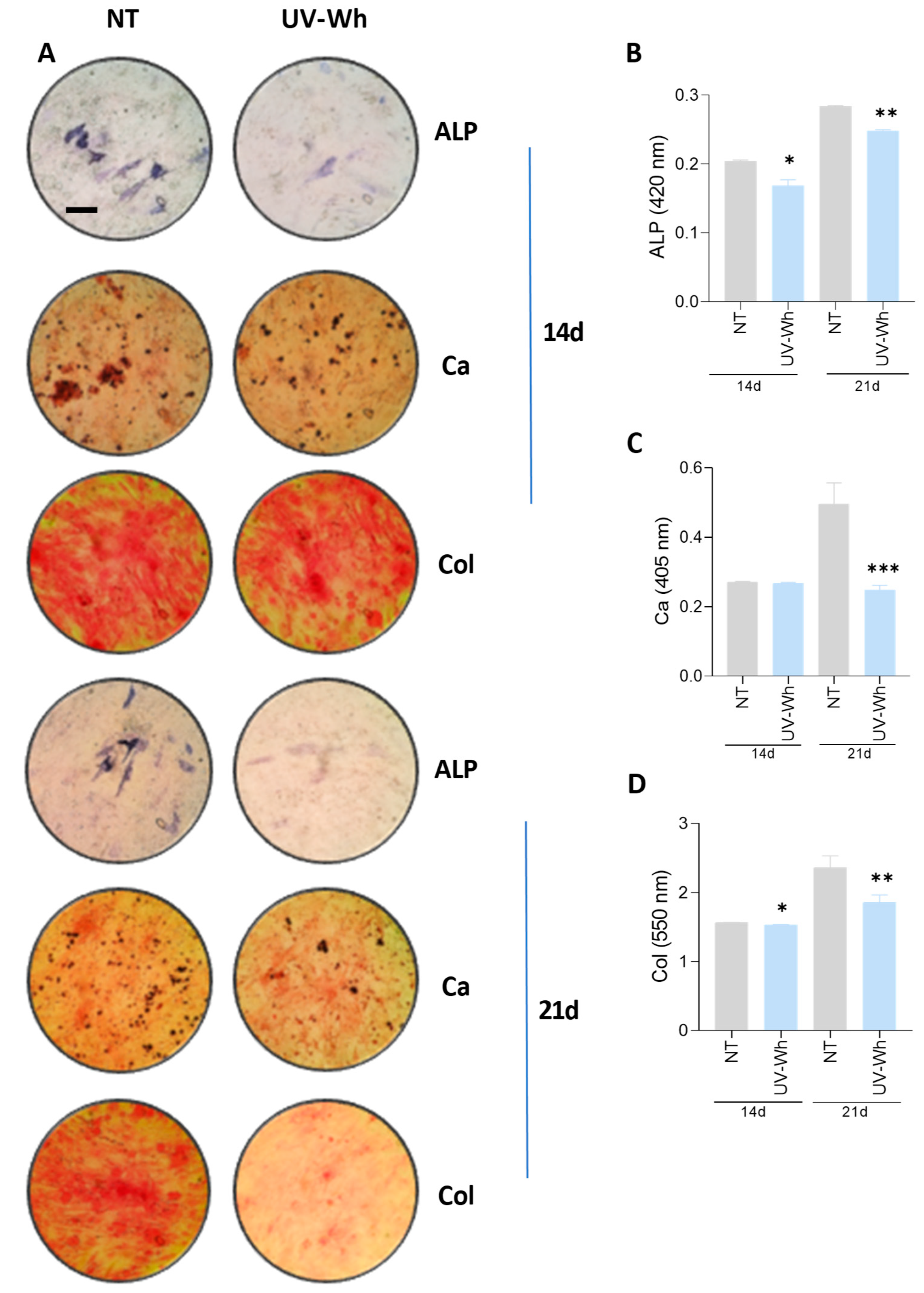
3.10. UV-Inactivated SARS-CoV-2 Was Able to Inhibit Osteoblast Differentiation
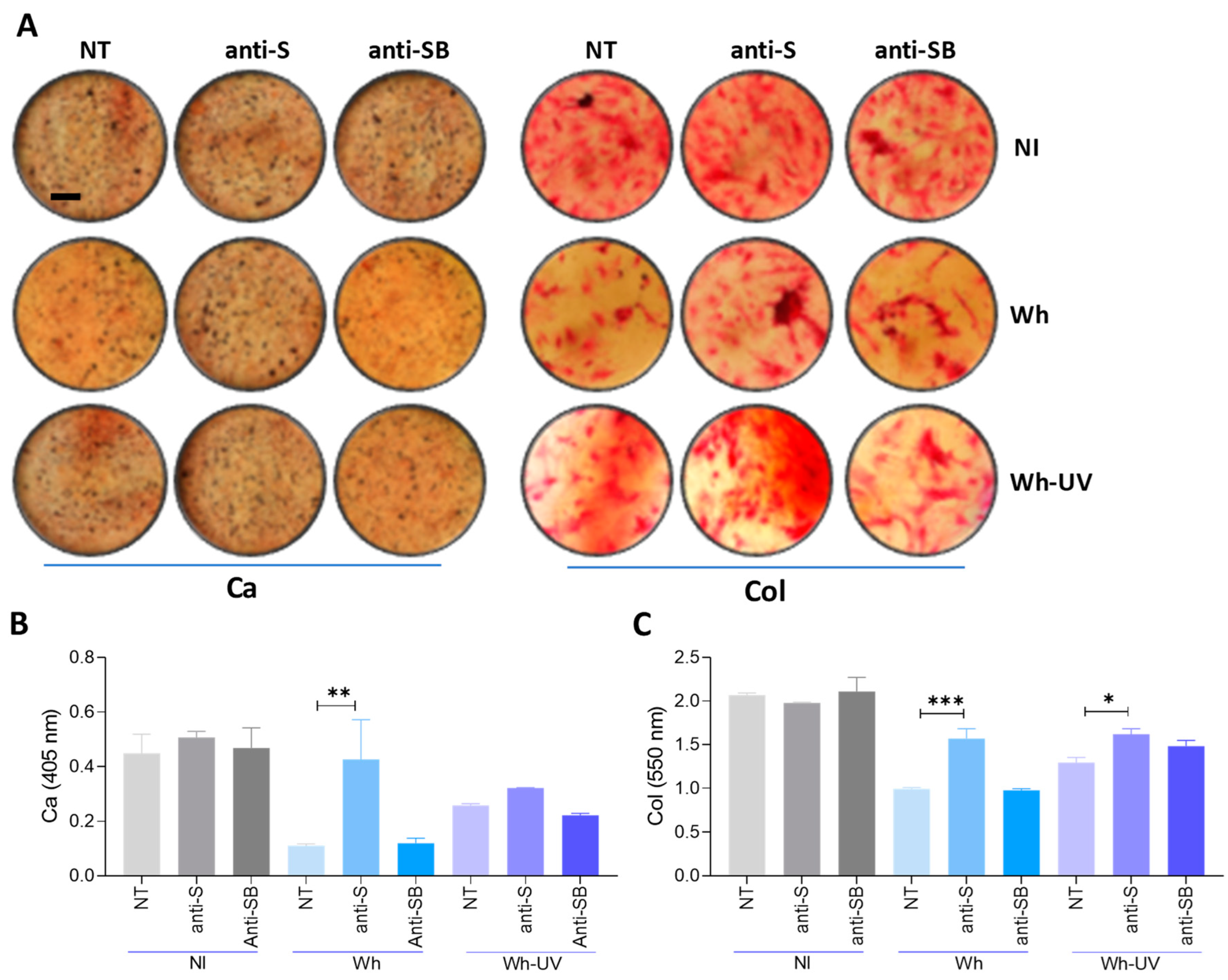
3.11. Spike–S-Protein Has a Role in the Inhibition of Osteoblast Differentiation
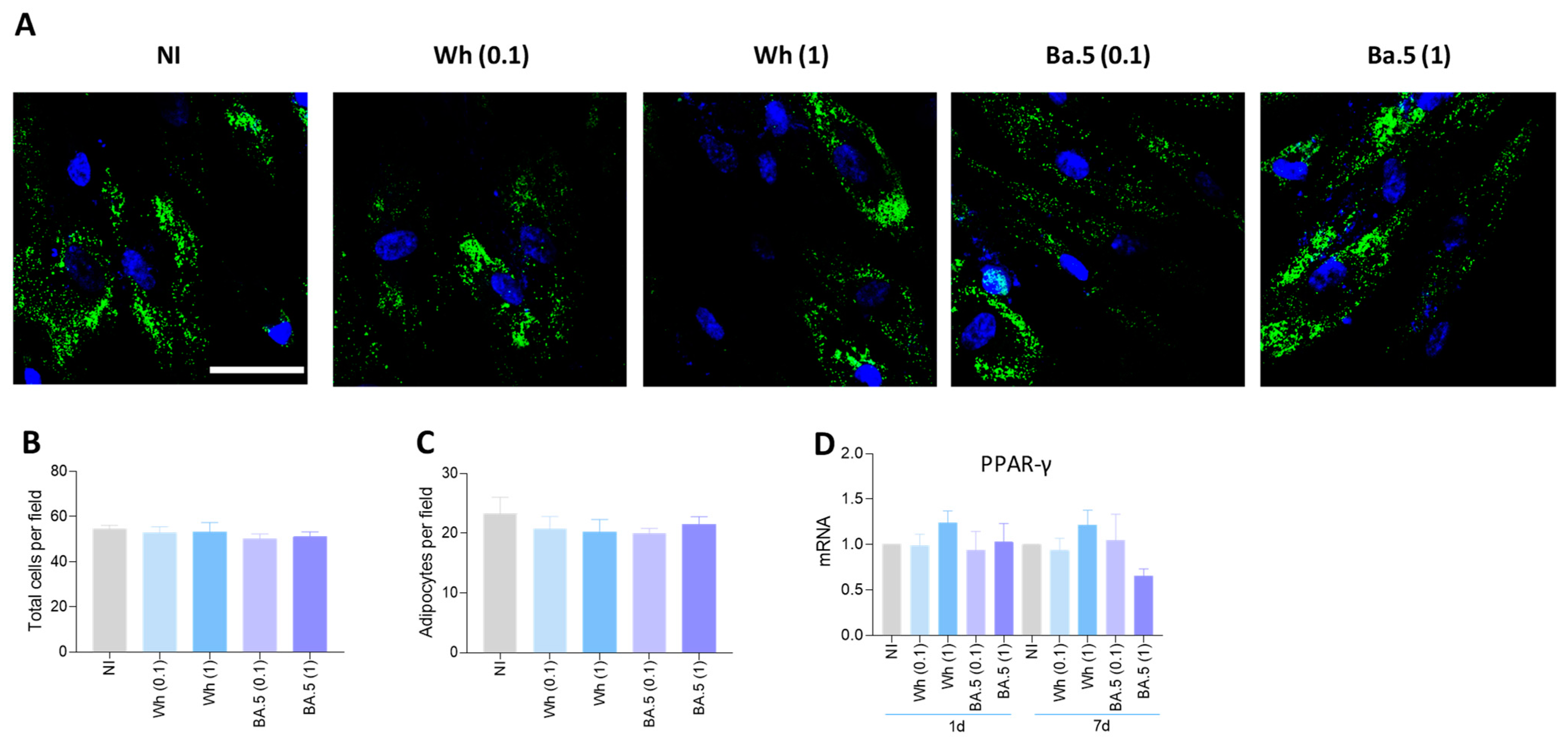
3.12. SARS-CoV-2 Could Not Modulate Adipocyte Differentiation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MSCs | mesenchymal stem cells |
| α-MEM | α-Minimal Essential Medium |
| DMEM | Dulbecco’s Modified Eagle Medium |
| FBS | fetal bovine serum |
| ATCC | American Type Culture Collection |
| PBS | phosphate-buffered saline |
| IBMX | 3-isobutyl-1-methylxanthine |
| mROS | mitochondrial reactive oxygen species |
| CPE | cytopathic effects |
| RUNX2 | runt-related transcription factor 2 |
| IL-6 | interleukin-6 |
| RANKL | receptor activator of nuclear factor kappa B ligand |
| RBD | receptor-binding domain |
| COVID-19 | Coronavirus Disease 2019 |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus |
| ALP | alkaline phosphatase |
| BMD | bone mineral density |
| α-MEM | α-Minimal Essential Medium |
| sg | subgenomic |
| mROS | mitochondrial reactive oxygen species |
| CPE | cytopathic effects |
| PFU | plaque-forming units |
| S | spike |
| RBD | spike protein receptor-binding domain |
| BCIP | 5-Bromo-4-chloro-3-indolylphosphate |
| NBT | nitroblue tetrazolium |
| pNPP | p-nitrophenylphosphate |
| OD | optical density |
| PPAR | peroxisome proliferator-activated receptor |
References
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Hill, P.A. Bone remodelling. Br. J. Orthod. 1998, 25, 101–107. [Google Scholar] [CrossRef]
- Buckwalter, J.A.; Cooper, R.R. Bone structure and function. Instr. Course Lect. 1987, 36, 27–48. [Google Scholar]
- Vimalraj, S. Alkaline phosphatase: Structure, expression and its function in bone mineralization. Gene 2020, 754, 144855. [Google Scholar] [CrossRef]
- Mohamed, A.M. An overview of bone cells and their regulating factors of differentiation. Malays. J. Med. Sci. 2008, 15, 4–12. [Google Scholar]
- Schlesinger, P.H.; Blair, H.C.; Beer Stolz, D.; Riazanski, V.; Ray, E.C.; Tourkova, I.L.; Nelson, D.J. Cellular and extracellular matrix of bone, with principles of synthesis and dependency of mineral deposition on cell membrane transport. Am. J. Physiol. Cell Physiol. 2020, 318, C111–C124. [Google Scholar] [CrossRef]
- Berktas, B.M.; Gokcek, A.; Hoca, N.T.; Koyuncu, A. COVID-19 illness and treatment decrease bone mineral density of surviving hospitalized patients. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 3046–3056. [Google Scholar]
- Redlich, K.; Smolen, J.S. Inflammatory bone loss: Pathogenesis and therapeutic intervention. Nat. Rev. Drug Discov. 2012, 11, 234–250. [Google Scholar] [CrossRef]
- di Filippo, L.; Frara, S.; Doga, M.; Giustina, A. The osteo-metabolic phenotype of COVID-19: An update. Endocrine 2022, 78, 247–254. [Google Scholar] [CrossRef]
- Awosanya, O.D.; Dadwal, U.C.; Imel, E.A.; Yu, Q.; Kacena, M.A. The Impacts of COVID-19 on Musculoskeletal Health. Curr. Osteoporos. Rep. 2022, 20, 213–225. [Google Scholar] [CrossRef]
- Sviercz, F.; Jarmoluk, P.; Godoy Coto, J.; Cevallos, C.; Freiberger, R.N.; Lopez, C.A.M.; Ennis, I.L.; Delpino, M.V.; Quarleri, J. The abortive SARS-CoV-2 infection of osteoclast precursors promotes their differentiation into osteoclasts. J. Med. Virol. 2024, 96, e29597. [Google Scholar] [CrossRef]
- Palma, M.B.; Luzzani, C.; Andrini, L.B.; Riccillo, F.; Buero, G.; Pelinski, P.; Inda, A.M.; Errecalde, A.L.; Miriuka, S.; Carosella, E.D.; et al. Wound Healing by Allogeneic Transplantation of Specific Subpopulation From Human Umbilical Cord Mesenchymal Stem Cells. Cell Transplant. 2021, 30, 963689721993774. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Gagne, M.; Flynn, B.J.; Honeycutt, C.C.; Flebbe, D.R.; Andrew, S.F.; Provost, S.J.; McCormick, L.; Van Ry, A.; McCarthy, E.; Todd, J.M.; et al. Variant-proof high affinity ACE2 antagonist limits SARS-CoV-2 replication in upper and lower airways. Nat. Commun. 2024, 15, 6894. [Google Scholar] [CrossRef]
- Jarmoluk, P.; Sviercz, F.A.; Cevallos, C.; Freiberger, R.N.; Lopez, C.A.; Poli, G.; Delpino, M.V.; Quarleri, J. SARS-CoV-2 Modulation of HIV Latency Reversal in a Myeloid Cell Line: Direct and Bystander Effects. Viruses 2024, 16, 1310. [Google Scholar] [CrossRef]
- Biasin, M.; Bianco, A.; Pareschi, G.; Cavalleri, A.; Cavatorta, C.; Fenizia, C.; Galli, P.; Lessio, L.; Lualdi, M.; Tombetti, E.; et al. UV-C irradiation is highly effective in inactivating SARS-CoV-2 replication. Sci. Rep. 2021, 11, 6260. [Google Scholar] [CrossRef]
- Tipnis, S.R.; Hooper, N.M.; Hyde, R.; Karran, E.; Christie, G.; Turner, A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000, 275, 33238–33243. [Google Scholar] [CrossRef]
- Yao, Y.; Wang, H.; Liu, Z. Expression of ACE2 in airways: Implication for COVID-19 risk and disease management in patients with chronic inflammatory respiratory diseases. Clin. Exp. Allergy 2020, 50, 1313–1324. [Google Scholar] [CrossRef]
- Lim, S.; Zhang, M.; Chang, T.L. ACE2-Independent Alternative Receptors for SARS-CoV-2. Viruses 2022, 14, 2535. [Google Scholar] [CrossRef]
- Sun, Y.; Shuang, F.; Chen, D.M.; Zhou, R.B. Treatment of hydrogen molecule abates oxidative stress and alleviates bone loss induced by modeled microgravity in rats. Osteoporos. Int. 2013, 24, 969–978. [Google Scholar] [CrossRef]
- Fatokun, A.A.; Stone, T.W.; Smith, R.A. Responses of differentiated MC3T3-E1 osteoblast-like cells to reactive oxygen species. Eur. J. Pharmacol. 2008, 587, 35–41. [Google Scholar] [CrossRef]
- Chen, Q.; Shou, P.; Zheng, C.; Jiang, M.; Cao, G.; Yang, Q.; Cao, J.; Xie, N.; Velletri, T.; Zhang, X.; et al. Fate decision of mesenchymal stem cells: Adipocytes or osteoblasts? Cell Death Differ. 2016, 23, 1128–1139. [Google Scholar] [CrossRef] [PubMed]
- Montenegro, A.F.L.; Clementino, M.A.F.; Yaochite, J.N.U. Type I interferon pathway genetic variants in severe COVID-19. Virus Res. 2024, 342, 199339. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Ng, C.; Inoue, K.; Chen, Z.; Xia, Y.; Hu, X.; Greenblatt, M.; Pernis, A.; Zhao, B. Def6 regulates endogenous type-I interferon responses in osteoblasts and suppresses osteogenesis. Elife 2020, 9, e59659. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Koga, T.; Isobe, M.; Kern, B.E.; Yokochi, T.; Chin, Y.E.; Karsenty, G.; Taniguchi, T.; Takayanagi, H. Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Genes. Dev. 2003, 17, 1979–1991. [Google Scholar] [CrossRef] [PubMed]
- Kaneshiro, S.; Ebina, K.; Shi, K.; Higuchi, C.; Hirao, M.; Okamoto, M.; Koizumi, K.; Morimoto, T.; Yoshikawa, H.; Hashimoto, J. IL-6 negatively regulates osteoblast differentiation through the SHP2/MEK2 and SHP2/Akt2 pathways in vitro. J. Bone Miner. Metab. 2014, 32, 378–392. [Google Scholar] [CrossRef] [PubMed]
- Harmer, D.; Falank, C.; Reagan, M.R. Interleukin-6 Interweaves the Bone Marrow Microenvironment, Bone Loss, and Multiple Myeloma. Front. Endocrinol. 2018, 9, 788. [Google Scholar] [CrossRef]
- Blanchard, F.; Duplomb, L.; Baud’huin, M.; Brounais, B. The dual role of IL-6-type cytokines on bone remodeling and bone tumors. Cytokine Growth Factor. Rev. 2009, 20, 19–28. [Google Scholar] [CrossRef]
- Franchimont, N.; Wertz, S.; Malaise, M. Interleukin-6: An osteotropic factor influencing bone formation? Bone 2005, 37, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.W.; Matsuura, R.; Iimura, K.; Wada, S.; Shinjo, A.; Benno, Y.; Nakagawa, M.; Takei, M.; Aida, Y. UVC disinfects SARS-CoV-2 by induction of viral genome damage without apparent effects on viral morphology and proteins. Sci. Rep. 2021, 11, 13804. [Google Scholar] [CrossRef] [PubMed]
- Gracheva, A.V.; Korchevaya, E.R.; Ammour, Y.I.; Smirnova, D.I.; Sokolova, O.S.; Glukhov, G.S.; Moiseenko, A.V.; Zubarev, I.V.; Samoilikov, R.V.; Leneva, I.A.; et al. Immunogenic properties of SARS-CoV-2 inactivated by ultraviolet light. Arch. Virol. 2022, 167, 2181–2191. [Google Scholar] [CrossRef]
- Verma, S.; Rajaratnam, J.H.; Denton, J.; Hoyland, J.A.; Byers, R.J. Adipocytic proportion of bone marrow is inversely related to bone formation in osteoporosis. J. Clin. Pathol. 2002, 55, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Gimble, J.M.; Zvonic, S.; Floyd, Z.E.; Kassem, M.; Nuttall, M.E. Playing with bone and fat. J. Cell Biochem. 2006, 98, 251–266. [Google Scholar] [CrossRef]
- Lowell, B.B. PPARgamma: An essential regulator of adipogenesis and modulator of fat cell function. Cell 1999, 99, 239–242. [Google Scholar] [CrossRef]
- Jeon, M.J.; Kim, J.A.; Kwon, S.H.; Kim, S.W.; Park, K.S.; Park, S.W.; Kim, S.Y.; Shin, C.S. Activation of peroxisome proliferator-activated receptor-gamma inhibits the Runx2-mediated transcription of osteocalcin in osteoblasts. J. Biol. Chem. 2003, 278, 23270–23277. [Google Scholar] [CrossRef]
- Hussain, M.; Khurram Syed, S.; Fatima, M.; Shaukat, S.; Saadullah, M.; Alqahtani, A.M.; Alqahtani, T.; Bin Emran, T.; Alamri, A.H.; Barkat, M.Q.; et al. Acute Respiratory Distress Syndrome and COVID-19: A Literature Review. J. Inflamm. Res. 2021, 14, 7225–7242. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Sun, N.N.; Gao, H.N.; Chen, Z.Y.; Yang, Y.; Ju, B.; Tang, L.L. Risk factors analysis of COVID-19 patients with ARDS and prediction based on machine learning. Sci. Rep. 2021, 11, 2933. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Kang, H.; Peng, R.; Song, K.; Guo, Q.; Guan, H.; Zhu, M.; Ye, D.; Li, F. Global, Regional, and National Burden of Low Bone Mineral Density From 1990 to 2019: Results From the Global Burden of Disease Study 2019. Front. Endocrinol. 2022, 13, 870905. [Google Scholar] [CrossRef]
- Liao, Z.; Wang, C.; Tang, X.; Yang, M.; Duan, Z.; Liu, L.; Lu, S.; Ma, L.; Cheng, R.; Wang, G.; et al. Human transferrin receptor can mediate SARS-CoV-2 infection. Proc. Natl. Acad. Sci. USA 2024, 121, e2317026121. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Qiu, Z.; Hou, Y.; Deng, X.; Xu, W.; Zheng, T.; Wu, P.; Xie, S.; Bian, W.; Zhang, C.; et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021, 31, 126–140. [Google Scholar] [CrossRef]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, C.Z.; Swaroop, M.; Xu, M.; Wang, L.; Lee, J.; Wang, A.Q.; Pradhan, M.; Hagen, N.; Chen, L.; et al. Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discov. 2020, 6, 80. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Dorival, I.; Cuesta-Geijo, M.A.; Barrado-Gil, L.; Galindo, I.; Garaigorta, U.; Urquiza, J.; Puerto, A.D.; Campillo, N.E.; Martinez, A.; Gastaminza, P.; et al. Identification of Niemann-Pick C1 protein as a potential novel SARS-CoV-2 intracellular target. Antiviral Res. 2021, 194, 105167. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Cao, J.; Zhang, X.; Gao, H.; Wang, Y.; Wang, J.; He, J.; Jiang, X.; Zhang, J.; Shen, G.; et al. Receptome profiling identifies KREMEN1 and ASGR1 as alternative functional receptors of SARS-CoV-2. Cell Res. 2022, 32, 24–37. [Google Scholar] [CrossRef]
- Sungnak, W.; Huang, N.; Becavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-Lopez, C.; Maatz, H.; Reichart, D.; Sampaziotis, F.; et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020, 26, 681–687. [Google Scholar] [CrossRef]
- Atashi, F.; Modarressi, A.; Pepper, M.S. The role of reactive oxygen species in mesenchymal stem cell adipogenic and osteogenic differentiation: A review. Stem Cells Dev. 2015, 24, 1150–1163. [Google Scholar] [CrossRef]
- Zhu, S.; Chen, W.; Masson, A.; Li, Y.P. Cell signaling and transcriptional regulation of osteoblast lineage commitment, differentiation, bone formation, and homeostasis. Cell Discov. 2024, 10, 71. [Google Scholar] [CrossRef]
- Arai, M.; Shibata, Y.; Pugdee, K.; Abiko, Y.; Ogata, Y. Effects of reactive oxygen species (ROS) on antioxidant system and osteoblastic differentiation in MC3T3-E1 cells. IUBMB Life 2007, 59, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Feng, Z.; Wang, X.; Zeng, M.; Liu, J.; Han, S.; Xu, J.; Chen, L.; Cao, K.; Long, J.; et al. SIRT3/SOD2 maintains osteoblast differentiation and bone formation by regulating mitochondrial stress. Cell Death Differ. 2018, 25, 229–240. [Google Scholar] [CrossRef]
- Hu, G.; Yu, Y.; Sharma, D.; Pruett-Miller, S.M.; Ren, Y.; Zhang, G.F.; Karner, C.M. Glutathione limits RUNX2 oxidation and degradation to regulate bone formation. JCI Insight 2023, 8, e166888. [Google Scholar] [CrossRef]
- Shockley, K.R.; Lazarenko, O.P.; Czernik, P.J.; Rosen, C.J.; Churchill, G.A.; Lecka-Czernik, B. PPARgamma2 nuclear receptor controls multiple regulatory pathways of osteoblast differentiation from marrow mesenchymal stem cells. J. Cell Biochem. 2009, 106, 232–246. [Google Scholar] [CrossRef]
- Rahman, S.; Czernik, P.J.; Lu, Y.; Lecka-Czernik, B. beta-catenin directly sequesters adipocytic and insulin sensitizing activities but not osteoblastic activity of PPARgamma2 in marrow mesenchymal stem cells. PLoS ONE 2012, 7, e51746. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bianco, P.; Robey, P.G. Skeletal stem cells. Development 2015, 142, 1023–1027. [Google Scholar] [CrossRef]
- Zheng, M.H.; Wood, D.J.; Papadimitriou, J.M. What’s new in the role of cytokines on osteoblast proliferation and differentiation? Pathol. Res. Pract. 1992, 188, 1104–1121. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Hu, W.; Ai, H.; Chen, Y.; Dong, S. The Dramatic Role of IFN Family in Aberrant Inflammatory Osteolysis. Curr. Gene Ther. 2021, 21, 112–129. [Google Scholar] [CrossRef]
- Nakamura, K.; Deyama, Y.; Yoshimura, Y.; Suzuki, K.; Morita, M. Toll-like receptor 3 ligand-induced antiviral response in mouse osteoblastic cells. Int. J. Mol. Med. 2007, 19, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, C.; Ito, J.; Nakayachi, M.; Okayasu, M.; Ohyama, Y.; Hakeda, Y.; Sato, T. Osteocytes produce interferon-beta as a negative regulator of osteoclastogenesis. J. Biol. Chem. 2014, 289, 11545–11555. [Google Scholar] [CrossRef]
- Lin, J.Y.; Huang, H.I. Respiratory viruses induce the expression of type I and III IFNs in MSCs through RLR/IRF3 signaling pathways. Microbes Infect. 2023, 25, 105171. [Google Scholar] [CrossRef]
- Kwan Tat, S.; Padrines, M.; Theoleyre, S.; Heymann, D.; Fortun, Y. IL-6, RANKL, TNF-alpha/IL-1: Interrelations in bone resorption pathophysiology. Cytokine Growth Factor. Rev. 2004, 15, 49–60. [Google Scholar]
- Grebennikov, D.; Kholodareva, E.; Sazonov, I.; Karsonova, A.; Meyerhans, A.; Bocharov, G. Intracellular Life Cycle Kinetics of SARS-CoV-2 Predicted Using Mathematical Modelling. Viruses 2021, 13, 1735. [Google Scholar] [CrossRef]
- Ponde, N.O.; Shoger, K.E.; Khatun, M.S.; Sarkar, M.K.; Dey, I.; Taylor, T.C.; Cisney, R.N.; Arunkumar, S.P.; Gudjonsson, J.E.; Kolls, J.K.; et al. SARS-CoV-2 ORF8 Mediates Signals in Macrophages and Monocytes through MyD88 Independently of the IL-17 Receptor. J. Immunol. 2023, 211, 252–260. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Freiberger, R.N.; López, C.A.M.; Jarmoluk, P.; Palma, M.B.; Cevallos, C.; Sviercz, F.A.; Grosso, T.M.; García, M.N.; Quarleri, J.; Delpino, M.V. SARS-CoV-2 Impairs Osteoblast Differentiation Through Spike Glycoprotein and Cytokine Dysregulation. Viruses 2025, 17, 143. https://doi.org/10.3390/v17020143
Freiberger RN, López CAM, Jarmoluk P, Palma MB, Cevallos C, Sviercz FA, Grosso TM, García MN, Quarleri J, Delpino MV. SARS-CoV-2 Impairs Osteoblast Differentiation Through Spike Glycoprotein and Cytokine Dysregulation. Viruses. 2025; 17(2):143. https://doi.org/10.3390/v17020143
Chicago/Turabian StyleFreiberger, Rosa Nicole, Cynthia Alicia Marcela López, Patricio Jarmoluk, María Belén Palma, Cintia Cevallos, Franco Agustin Sviercz, Tomás Martín Grosso, Marcela Nilda García, Jorge Quarleri, and M. Victoria Delpino. 2025. "SARS-CoV-2 Impairs Osteoblast Differentiation Through Spike Glycoprotein and Cytokine Dysregulation" Viruses 17, no. 2: 143. https://doi.org/10.3390/v17020143
APA StyleFreiberger, R. N., López, C. A. M., Jarmoluk, P., Palma, M. B., Cevallos, C., Sviercz, F. A., Grosso, T. M., García, M. N., Quarleri, J., & Delpino, M. V. (2025). SARS-CoV-2 Impairs Osteoblast Differentiation Through Spike Glycoprotein and Cytokine Dysregulation. Viruses, 17(2), 143. https://doi.org/10.3390/v17020143





