Baculovirus Genetic Diversity and Population Structure
Abstract
1. Introduction
1.1. Variation in the Virus Genome
1.2. Variation in the Virus Population
1.3. Variant Interactions Affect Phenotype
2. Mechanisms and Processes Affecting Diversity
2.1. In the Environment
2.2. In the Host Organism
2.3. In the Cell
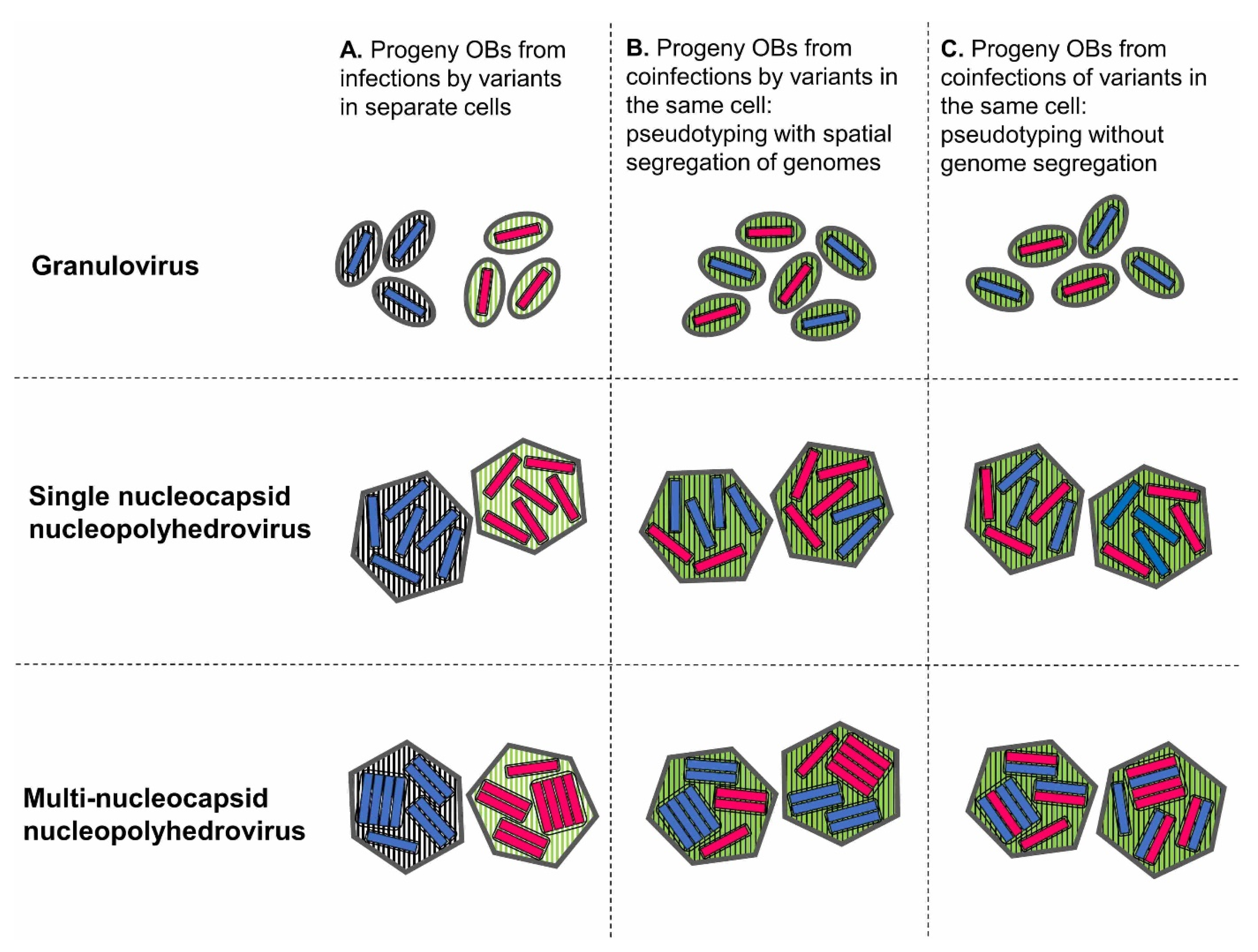
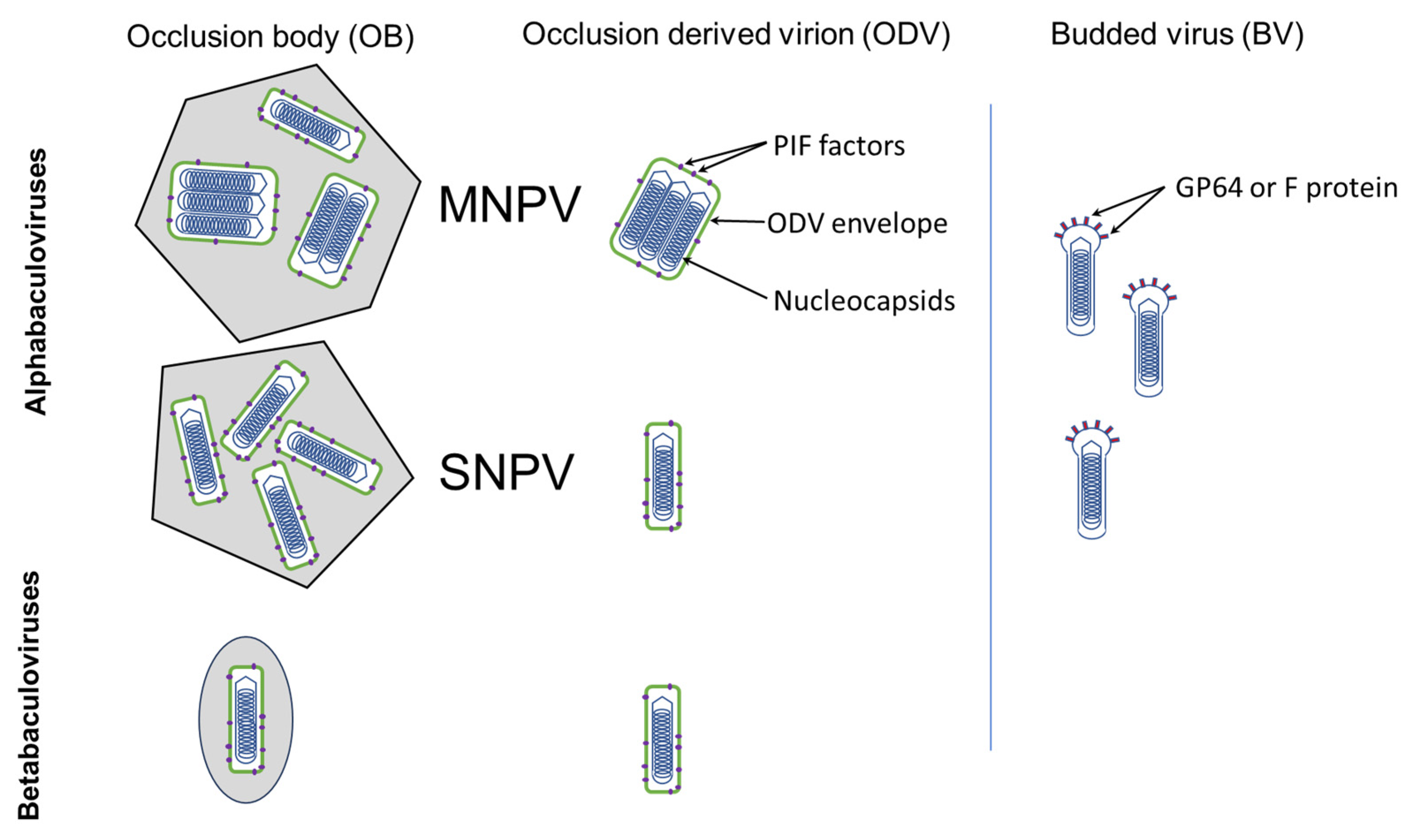
2.4. In the Occlusion Body
2.5. In the Occlusion-Derived Virion
3. Processes That Favor Genotypic Diversity
3.1. Trade-Offs Between Components of Virus Fitness
3.2. Interactions Between Virus Genotypes
3.3. Differential Selection for Genotypes
3.4. Genotype × Environment Interactions
4. Genetic Diversity in Biological Insecticides
Risks of Resistance
5. Future Issues
5.1. Physical Segregation of Variants
5.2. Transmission of Diversity
5.3. Host Resistance
5.4. Independent Action of Virions
Author Contributions
Funding
Conflicts of Interest
References
- Harrison, R.L.; Herniou, E.A.; Jehle, J.A.; Theilmann, D.A.; Burand, J.P.; Becnel, J.J.; Krell, P.J.; van Oers, M.M.; Mowery, J.D.; Bauchan, G.R. ICTV virus taxonomy profile: Baculoviridae. J. Gen. Virol. 2018, 99, 1185–1986. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.; Jukes, M. Advances in microbial control in IPM: Entomopathogenic viruses. In Integrated Management of Insect Pests; Kogan, M., Higley, L., Eds.; Burleigh Dodds Science Publish: Cambridge, UK, 2019; pp. 593–648. [Google Scholar]
- van Oers, M.M.; Pijlman, G.P.; Vlak, J.M. Thirty years of baculovirus–insect cell protein expression: From dark horse to mainstream technology. J. Gen. Virol. 2015, 96, 6–23. [Google Scholar] [CrossRef]
- Williams, T.; Bergoin, M.; van Oers, M.M. Diversity of large DNA viruses of invertebrates. J. Invertebr. Pathol. 2017, 147, 4–22. [Google Scholar] [CrossRef]
- Erlandson, M.A.; Toprak, U.; Hegedus, D.D. Role of the peritrophic matrix in insect-pathogen interactions. J. Insect Physiol. 2019, 117, 103894. [Google Scholar] [CrossRef] [PubMed]
- Blissard, G.W.; Theilmann, D.A. Baculovirus entry and egress from insect cells. Annu. Rev. Virol. 2018, 5, 113–139. [Google Scholar] [CrossRef]
- Rohrmann, G.F. Baculovirus Molecular Biology, 4th ed.; US National Center for Biotechnology Information: Bethesda, MD, USA, 2019.
- Fuxa, J.R. Ecology of insect nucleopolyhedroviruses. Agric. Ecosyst. Environ. 2004, 103, 27–43. [Google Scholar] [CrossRef]
- Williams, T. Soil as an environmental reservoir for baculoviruses: Persistence, dispersal and role in pest control. Soil Syst. 2023, 7, 29. [Google Scholar] [CrossRef]
- Craveiro, S.R.; Melo, F.L.; Ribeiro, Z.M.A.; Ribeiro, B.M.; Báo, S.N.; Inglis, P.W.; Castro, M.E.B. Pseudoplusia includens single nucleopolyhedrovirus: Genetic diversity, phylogeny and hypervariability of the pif-2 gene. J. Invertebr. Pathol. 2013, 114, 258–267. [Google Scholar] [CrossRef]
- D’Amico, V.; Slavicek, J.; Podgwaite, J.D.; Webb, R.; Fuester, R.; Peiffer, R.A. Deletion of v-chiA from a baculovirus reduces horizontal transmission in the field. Appl. Environ. Microbiol. 2013, 79, 4056–4064. [Google Scholar] [CrossRef]
- de Brito, A.F.D.; Braconi, C.T.; Weidmann, M.; Dilcher, M.; Alves, J.M.P.; Gruber, A.; Zanotto, P.M.D.A. The pangenome of the Anticarsia gemmatalis multiple nucleopolyhedrovirus (AgMNPV). Genome Biol. Evol. 2016, 8, 94–108. [Google Scholar] [CrossRef]
- Harrison, R.L. Concentration-and time-response characteristics of plaque isolates of Agrotis ipsilon multiple nucleopolyhedrovirus derived from a field isolate. J. Invertebr. Pathol. 2013, 112, 159–161. [Google Scholar] [CrossRef]
- Kitchin, D.; Bouwer, G. Significant differences in the intra-host genetic diversity of Helicoverpa armigera nucleopolyhedrovirus dnapol after serial in vivo passages in the same insect population. Arch. Virol. 2018, 163, 713–718. [Google Scholar] [CrossRef]
- Martemyanov, V.V.; Kabilov, M.R.; Tupikin, A.E.; Baturin, O.A.; Belousova, I.A.; Podgwaite, J.D.; Ilynykh, A.V.; Vlassov, V.V. The enhancin gene: One of the genetic determinants of population variation in baculoviral virulence. Dokl. Biochem. Biophys. 2015, 465, 351–353. [Google Scholar] [CrossRef] [PubMed]
- Masson, T.; Fabre, M.L.; Pidre, M.L.; Niz, J.M.; Berretta, M.F.; Romanowski, V.; Ferrelli, M.L. Genomic diversity in a population of Spodoptera frugiperda nucleopolyhedrovirus. Infect. Genet. Evol. 2021, 90, 104749. [Google Scholar] [CrossRef] [PubMed]
- Niz, J.M.; Salvador, R.; Ferrelli, M.L.; Sciocco de Cap, A.; Romanowski, V.; Berretta, M.F. Genetic variants in Argentinean isolates of Spodoptera frugiperda multiple nucleopolyhedrovirus. Virus Genes 2020, 56, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Thézé, J.; Cabodevilla, O.; Palma, L.; Williams, T.; Caballero, P.; Herniou, E.A. Genomic diversity in european Spodoptera exigua multiple nucleopolyhedrovirus isolates. J. Gen. Virol. 2014, 95, 2297–2309. [Google Scholar] [CrossRef]
- Xu, Y.P.; Cheng, R.L.; Xi, Y.; Zhang, C.X. Genomic diversity of Bombyx mori nucleopolyhedrovirus strains. Genomics 2013, 102, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Milks, M.L.; Myers, J.H. The development of larval resistance to a nucleopolyhedrovirus is not accompanied by an increased virulence in the virus. Evol. Ecol. 2000, 14, 645–664. [Google Scholar] [CrossRef]
- Boezen, D.; Ali, G.; Wang, M.; Wang, X.; van der Werf, W.; Vlak, J.M.; Zwart, M.P. Empirical estimates of the mutation rate for n alphabaculovirus. PLoS Genet. 2021, 18, e1009806. [Google Scholar] [CrossRef]
- Chateigner, A.; Bézier, A.; Labrousse, C.; Jiolle, D.; Barbe, V.; Herniou, E.A. Ultra deep sequencing of a baculovirus population reveals widespread genomic variations. Viruses 2015, 7, 3625–3646. [Google Scholar] [CrossRef]
- Wennmann, J.T.; Radtke, P.; Eberle, K.E.; Gueli-Alletti, G.; Jehle, J.A. Deciphering single nucleotide polymorphisms and evolutionary trends in isolates of the Cydia pomonella granulovirus. Viruses 2017, 9, 227. [Google Scholar] [CrossRef]
- McDougal, V.V.; Guarino, L.A. Autographa californica nuclear polyhedrosis virus DNA polymerase: Measurements of processivity and strand displacement. J. Virol. 1999, 73, 4908–4918. [Google Scholar] [CrossRef]
- Fan, J.; Wennmann, J.T.; Wang, D.; Jehle, J.A. Novel diversity and virulence patterns found in new isolates of Cydia pomonella granulovirus from China. Appl. Environ. Microbiol. 2020, 86, e02000-19. [Google Scholar] [CrossRef] [PubMed]
- López-Ferber, M.; Argaud, O.; Croizier, L.; Croizier, G. Diversity, distribution and mobility of bro gene sequences in Bombyx mori nucleopolyhedrovirus. Virus Genes 2001, 22, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Barrera, G.P.; Belaich, M.N.; Patarroyo, M.A.; Villamizar, L.F.; Ghiringhelli, P.D. Evidence of recent interspecies horizontal gene transfer regarding nucleopolyhedrovirus infection of Spodoptera frugiperda. BMC Genom. 2015, 16, 1008. [Google Scholar] [CrossRef] [PubMed]
- Croizier, G.; Ribeiro, H.C.T. Recombination as a possible major cause of genetic heterogeneity in Anticarsia gemmatalis nuclear polyhedrosis virus wild populations. Virus Res. 1992, 26, 183–196. [Google Scholar] [CrossRef]
- Simón, O.; Williams, T.; Possee, R.D.; López-Ferber, M.; Caballero, P. Stability of a Spodoptera frugiperda nucleopolyhedrovirus deletion recombinant during serial passage in insects. Appl. Environ. Microbiol. 2010, 76, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, C.; Chateigner, A.; Ernenwein, L.; Barbe, V.; Bézier, A.; Herniou, E.A.; Cordaux, R. Population genomics supports baculoviruses as vectors of horizontal transfer of insect transposons. Nat. Commun. 2014, 5, 3348. [Google Scholar] [CrossRef]
- Loiseau, V.; Peccoud, J.; Bouzar, C.; Guillier, S.; Fan, J.; Gueli Alletti, G.; Meignin, C.; Herniou, E.A.; Federici, B.A.; Wennmann, J.T.; et al. Monitoring insect transposable elements in large double-stranded DNA viruses reveals host-to-virus and virus-to-virus transposition. Mol. Biol. Evol. 2021, 38, 3512–3530. [Google Scholar] [CrossRef] [PubMed]
- Yanase, Y.; Hashimoto, Y.; Kawarabata, T. Identification of insertion and deletion genes in Autographa californica nucleopolyhedrovirus variants isolated from Galleria mellonella, Spodoptera exigua, Spodoptera litura and Xestia c-nigrum. Virus Genes 2000, 21, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Serrano, A.; Williams, T.; Simón, O.; López-Ferber, M.; Caballero, P.; Muñoz, D. Analogous population structures in two alphabaculoviruses highlight functional role for deletion mutants Appl. Environ. Microbiol. 2013, 79, 1118–1125. [Google Scholar] [CrossRef]
- Erlandson, M.A. Genetic variation in field populations of baculoviruses: Mechanisms for generating variation and its potential role in baculovirus epizootiology. Virol. Sin. 2009, 24, 458–469. [Google Scholar] [CrossRef]
- Pijlman, G.P.; van den Born, E.; Martens, D.E.; Vlak, J.M. Autographa californica baculoviruses with large genomic deletions are rapidly generated in infected insect cells. Virology 2001, 283, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Cory, J.S.; Green, B.M.; Paul, R.K.; Hunter-Fujita, F. Genotypic and phenotypic diversity of a baculovirus population within an individual insect host. J. Invertebr. Pathol. 2005, 89, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Barrera, G.; Williams, T.; Villamizar, L.; Caballero, P.; Simón, O. Deletion genotypes reduce occlusion body potency but increase occlusion body production in a Colombian Spodoptera frugiperda nucleopolyhedrovirus population. PLoS ONE 2013, 8, e77271. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, D.; Castillejo, J.I.; Caballero, P. Naturally occurring deletion mutants are parasitic genotypes in a wild-type nucleopolyhedrovirus population of Spodoptera exigua. Appl. Environ. Microbiol. 1998, 64, 4372–4377. [Google Scholar] [CrossRef] [PubMed]
- Simón, O.; Williams, T.; López-Ferber, M.; Caballero, P. Genetic structure of a Spodoptera frugiperda nucleopolyhedrovirus population: High prevalence of deletion genotypes. Appl. Environ. Microbiol. 2004, 70, 5579–5588. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, B.C.; Melo, F.L.; Silva, A.M.R.; Sanches, M.M.; Moscardi, F.; Ribeiro, B.M.; Souza, M.L. Biological and molecular characterization of two Anticarsia gemmatalis multiple nucleopolyhedrovirus clones exhibiting contrasting virulence. J. Invertebr. Pathol. 2019, 164, 23–31. [Google Scholar] [CrossRef]
- Aguirre, E.; Beperet, I.; Williams, T.; Caballero, P. Generation of variability in Chrysodeixis includens nucleopolyhedrovirus (ChinNPV): The role of a single variant. Viruses 2021, 13, 1895. [Google Scholar] [CrossRef] [PubMed]
- Baillie, V.L.; Bouwer, G. High levels of genetic variation within Helicoverpa armigera nucleopolyhedrovirus populations in individual host insects. Arch. Virol. 2012, 157, 2281–2289. [Google Scholar] [CrossRef]
- Redman, E.M.; Wilson, K.; Grzywacz, D.; Cory, J.S. High levels of genetic diversity in Spodoptera exempta NPV from Tanzania. J. Invertebr. Pathol. 2010, 105, 190–193. [Google Scholar] [CrossRef] [PubMed]
- Vickers, J.M.; Cory, J.S.; Entwisle, P.F. DNA characterization of eight geographic isolates of granulosis virus from the potato tuber moth (Phthorimaea operculella) (Lepidoptera, Gelechiidae). J. Invertebr. Pathol. 1991, 57, 334–342. [Google Scholar] [CrossRef]
- Fan, J.; Jehle, J.A.; Wennmann, J.T. Population structure of Cydia pomonella granulovirus isolates revealed by quantitative analysis of genetic variation. Virus Evol. 2021, 7, veaa073. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, E.; Beperet, I.; Williams, T.; Caballero, P. Genetic variability of Chrysodeixis includens nucleopolyhedrovirus (ChinNPV) and the insecticidal characteristics of selected genotypic variants. Viruses 2019, 11, 581. [Google Scholar] [CrossRef] [PubMed]
- Briese, D.T.; Mende, H.A. Differences in susceptibility to a granulosis virus between field populations of the potato tuber moth, Phthorimaea operculella (Zeller) (Lepidoptera: Gelechiidae). Bull. Entomol. Res. 1981, 71, 11–18. [Google Scholar] [CrossRef]
- Espinel-Correal, C.; López-Ferber, M.; Zeddam, J.L.; Villamizar, L.; Gomez, J.; Cotes, A.M.; Léry, X. Experimental mixtures of Phthorimaea operculella granulovirus isolates provide high biological efficacy on both Phthorimaea operculella and Tecia solanivora (Lepidoptera: Gelechiidae). J. Invertebr. Pathol. 2012, 110, 375–381. [Google Scholar] [CrossRef]
- Cabodevilla, O.; Ibañez, I.; Simón, O.; Murillo, R.; Caballero, P.; Williams, T. Occlusion body pathogenicity, virulence and productivity traits vary with transmission strategy in a nucleopolyhedrovirus. Biol. Control 2011, 56, 184–192. [Google Scholar] [CrossRef]
- Berling, M.; Blachère-Lopez, C.; Soubabère, O.; Léry, X.; Bonhomme, A.; Sauphanor, B.; López-Ferber, M. Cydia pomonella granulovirus (CpGV) genotypes overcome virus resistance in the codling moth and improve virus efficiency by selection against resistant hosts. App. Environ. Microbiol. 2009, 75, 925–930. [Google Scholar] [CrossRef] [PubMed]
- Eberle, K.E.; Asser-Kaiser, S.; Sayed, S.M.; Nguyen, H.T.; Jehle, J.A. Overcoming the resistance of codling moth against conventional Cydia pomonella granulovirus (CpGV-M) by a new isolate CpGV-I12. J. Invertebr. Pathol. 2008, 98, 293–298. [Google Scholar] [CrossRef]
- Simon, O.; Williams, T.; López-Ferber, M.; Taulemesse, J.M.; Caballero, P. Population genetic structure determines speed of kill and occlusion body production in Spodoptera frugiperda multiple nucleopolyhedrovirus. Biol. Control 2008, 44, 321–330. [Google Scholar] [CrossRef]
- Williams, T.; Melo-Molina, G.D.C.; Jiménez-Fernández, J.A.; Weissenberger, H.; Gómez-Díaz, J.S.; Navarro-de-la-Fuente, L.; Richards, A.R. Presence of Spodoptera frugiperda multiple nucleopolyhedrovirus (SfMNPV) occlusion bodies in maize field soils of Mesoamerica. Insects 2023, 14, 80. [Google Scholar] [CrossRef] [PubMed]
- Clavijo, G.; Williams, T.; Muñoz, D.; Caballero, P.; López-Ferber, M. Mixed genotype transmission bodies and virions contribute to the maintenance of diversity in an insect virus. Proc. R. Soc. B Biol. Sci. 2010, 277, 943–945. [Google Scholar] [CrossRef]
- Graham, R.I.; Tyne, W.I.; Possee, R.D.; Sait, S.M.; Hails, R.S. Genetically variable nucleopolyhedroviruses isolated from spatially separate populations of the winter moth Operophtera brumata (Lepidoptera: Geometridae) in Orkney. J. Invertebr. Pathol. 2004, 87, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Hinsberger, A.; Blachère-López, C.; López-Ferber, M. Promoting mixed genotype infections in CpGV: Analysis on field and laboratory sprayed apple leaves. Biocontrol Sci. Technol. 2020, 30, 975–982. [Google Scholar] [CrossRef]
- Rezapanah, M.; Shojai-Estabragh, S.; Huber, J.; Jehle, J.A. Molecular and biological characterization of new isolates of Cydia pomonella granulovirus from Iran. J. Pest Sci. 2008, 81, 187–191. [Google Scholar] [CrossRef]
- Ferrelli, M.L.; Salvador, R. Effects of mixed baculovirus infections in biological control: A comprehensive historical and technical analysis. Viruses 2023, 15, 1838. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.W.; Carner, G.R.; Lange, M.; Jehle, J.A.; Arif, B.M. Biological and molecular characterization of a multicapsid nucleopolyhedrovirus from Thysanoplusia orichalcea (L.) (Lepidoptera: Noctuidae). J. Invertebr. Pathol. 2005, 88, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Rincon Castro, M.C.D.; Ibarra, J.E. Caracterizacion de cepas silvestres de virus de poliedrosis nuclear aisladas de Trichoplusia ni (Lepidoptera: Noctuidae) en el centro de México. Vedalia 1995, 2, 7–15. [Google Scholar]
- Lauzon, H.A.; Jamieson, P.B.; Krell, P.J.; Arif, B.M. Gene organization and sequencing of the Choristoneura fumiferana defective nucleopolyhedrovirus genome. J. Gen. Virol. 2005, 86, 945–961. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, V.A.; Belaich, M.N.; Quintana, G.; Cap, A.S.; Ghiringhelli, P.D. Isolation and characterization of a nucleopolyhedrovirus from Rachiplusia nu (Guenée) (Lepidoptera: Noctuidae). Int. J. Virol. Mol. Biol. 2012, 1, 28–34. [Google Scholar]
- Jakubowicz, V.; Taibo, C.B.; Sciocco-Cap, A.; Arneodo, J.D. Biological and molecular characterization of Rachiplusia nu single nucleopolyhedrovirus, a promising biocontrol agent against the South American soybean pest Rachiplusia nu. J. Invertebr. Pathol. 2019, 166, 107211. [Google Scholar] [CrossRef] [PubMed]
- Decker-Franco, C.; Taibo, C.B.; Di Rienzo, J.A.; Alfonso, V.; Arneodo, J.D. Comparative pathogenesis of generalist AcMNPV and specific RanuNPV in larvae of Rachiplusia nu (Lepidoptera: Noctuidae) following single and mixed inoculations. J. Econ. Entomol. 2021, 114, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Clavijo, G.; Williams, T.; Muñoz, D.; López-Ferber, M.; Caballero, P. Entry into midgut epithelial cells is a key step in the selection of genotypes in a nucleopolyhedrovirus. Virol. Sin. 2009, 24, 350–358. [Google Scholar] [CrossRef]
- Zwart, M.P.; van Oers, M.M.; Cory, J.S.; van Lent, J.W.; van der Werf, W.; Vlak, J.M. Development of a quantitative real-time PCR for determination of genotype frequencies for studies in baculovirus population biology. J. Virol. Meth. 2008, 148, 146–154. [Google Scholar] [CrossRef]
- Zwart, M.P.; Elena, S.F. Matters of size: Genetic bottlenecks in virus infection and their potential impact on evolution. Annu. Rev. Virol. 2015, 2, 161–179. [Google Scholar] [CrossRef] [PubMed]
- Washburn, J.O.; Kirkpatrick, B.A.; Haas-Stapleton, E.; Volkman, L.E. M2R enhances Autographa californica M nucleopolyhedrovirus infection of Trichoplusia ni and Heliothis virescens by preventing sloughing of infected midgut epithelial cells. Biol. Control 1998, 11, 58–69. [Google Scholar] [CrossRef]
- Burden, J.P.; Possee, R.D.; Sait, S.M.; King, L.A.; Hails, R.S. Phenotypic and genotypic characterisation of persistent baculovirus infections in populations of the cabbage moth (Mamestra brassicae) within the British Isles. Arch. Virol. 2006, 151, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Vilaplana, L.; Wilson, K.; Redman, E.M.; Cory, J.S. Pathogen persistence in migratory insects: High levels of vertically-transmitted virus infection in field populations of the African armyworm. Evol. Ecol. 2010, 24, 147–160. [Google Scholar] [CrossRef]
- Wilson, K.; Grzywacz, D.; Cory, J.S.; Donkersley, P.; Graham, R.I. Trans-generational viral transmission and immune priming are dose-dependent. J. Anim. Ecol. 2021, 90, 1560–1569. [Google Scholar] [CrossRef]
- Qin, F.; Xu, C.; Hu, J.; Lei, C.; Zheng, Z.; Peng, K.; Wang, H.; Sun, X. Dissecting the cell entry pathway of baculovirus by single-particle tracking and quantitative electron microscopic analysis. J. Virol. 2019, 93, e00033-1. [Google Scholar] [CrossRef] [PubMed]
- Beperet, I.; Irons, S.; Simón, O.; King, L.A.; Williams, T.; Possee, R.D.; López-Ferber, M.; Caballero, P. Superinfection exclusion in alphabaculovirus infections is concomitant with actin reorganization. J. Virol. 2014, 88, 3548–3556. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.C.; Godfray, H.C.J.; O’Reilly, D.R. Persistence of an occlusion-negative recombinant nucleopolyhedrovirus in Trichoplusia ni indicates high multiplicity of cellular infection. Appl. Environ. Microbiol. 2001, 67, 5204–5209. [Google Scholar] [CrossRef]
- Fu, Q.-M.; Fang, Z.; Ren, L.; Wu, Q.-S.; Zhang, J.-B.; Liu, Q.-P.; Tan, L.-T.; Weng, Q.-B. Partial Alleviation of Homologous Superinfection Exclusion of SeMNPV Latently Infected Cells by G1 Phase Infection and G2/M Phase Arrest. Viruses 2024, 16, 736. [Google Scholar] [CrossRef] [PubMed]
- Sanjuán, R. Collective infectious units in viruses. Trends Microbiol. 2017, 25, 402–412. [Google Scholar] [CrossRef]
- Adams, J.R.; McClintock, J.T. Baculoviridae. part II. Nuclear polyhedrosis viruses of insects. In Atlas of Invertebrate Viruses; Adams, J.R., Bonami, J.R., Eds.; CRC Press: Boca Raton, FL, USA, 1991; pp. 87–204. [Google Scholar]
- Sanjuán, R. The social life of viruses. Annu. Rev. Virol. 2021, 8, 183–199. [Google Scholar] [CrossRef] [PubMed]
- Falcon, L.A.; Hess, R.T. Electron microscope observations of multiple occluded virions in the granulosis virus of the codling moth, Cydia pomonella. J. Invertebr. Pathol. 1985, 45, 356–359. [Google Scholar] [CrossRef]
- Sciocco de Cap, A.; Parola, A.D.; Goldberg, A.V.; Ghiringhelli, P.D.; Romanowski, V. Characterization of a granulovirus isolated from Epinotia aporema Wals. (Lepidoptera: Tortricidae) larvae. Appl. Environ. Microbiol. 2001, 67, 3702–3706. [Google Scholar] [CrossRef]
- Hamblin, M.; van Beek, N.A.M.; Hughes, P.R.; Wood, H.A. Co-occlusion and persistence of a baculovirus mutant lacking the polyhedrin gene. Appl. Environ. Microbiol. 1990, 56, 3057–3062. [Google Scholar] [CrossRef] [PubMed]
- van Beek, N.A.M.; Wood, H.A.; Hughes, P.R. Quantitative aspects of nuclear polyhedrosis virus infections in lepidopterous larvae: The dose-survival time relationship. J. Invertebr. Pathol. 1988, 51, 58–63. [Google Scholar] [CrossRef]
- Arrizubieta, M.; Simón, O.; Ricarte-Bermejo, A.; López-Ferber, M.; Williams, T.; Caballero, P. Coocclusion of Helicoverpa armigera single nucleopolyhedrovirus (HearSNPV) and Helicoverpa armigera multiple nucleopolyhedrovirus (HearMNPV): Pathogenicity and stability in homologous and heterologous hosts. Viruses 2022, 14, 687. [Google Scholar] [CrossRef] [PubMed]
- Munsamy, T.; Bouwer, G. Determination of the virulence of single nucleopolyhedrovirus occlusion bodies using a novel laser capture microdissection method. J. Gen. Virol. 2020, 101, 1300–1304. [Google Scholar] [CrossRef] [PubMed]
- Hinsberger, A.; Graillot, B.; Blachère-López, C.; Juliant, S.; Cerutti, M.; King, L.A.; Possee, R.D.; Gallardo, F.; López-Ferber, M. Tracing baculovirus AcMNPV infection using a real-time method based on ANCHOR™ DNA labeling technology. Viruses 2020, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, S.; Kokusho, R.; Kakemizu, H.; Izaku, T.; Katsuma, S.; Iwashita, Y.; Kawasaki, H.; Iwanaga, M. Characterization of a Bombyx mori nucleopolyhedrovirus variant isolated in Laos. J. Insect Biotechnol. Sericol. 2017, 86, 3085–3094. [Google Scholar]
- Beperet, I.; Barrera, G.; Simón, O.; Williams, T.; López-Ferber, M.; Caballero, P. The sf32 unique gene of Spodoptera frugiperda multiple nucleopolyhedrovirus (SfMNPV) is a non-essential gene that could be involved in nucleocapsid organization in occlusion-derived virions. PLoS ONE 2013, 8, e77683. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.-L.; Xu, Y.-P.; Zhang, C.-X. Genome sequence of a Bombyx mori nucleopolyhedrovirus strain with cubic occlusion bodies. J. Virol. 2012, 86, 10245. [Google Scholar] [CrossRef]
- Li, S.N.; Wang, J.Y.; Yuan, M.J.; Yang, K. Disruption of the baculovirus core gene ac78 results in decreased production of multiple nucleocapsid-enveloped occlusion-derived virions and the failure of primary infection in vivo. Virus Res. 2014, 191, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Yu, I.L.; Bray, D.; Lin, Y.C.; Lung, O. Autographa californica multiple nucleopolyhedrovirus ORF 23 null mutant produces occlusion-derived virions with fewer nucleocapsids. J. Gen. Virol. 2009, 90, 1499–1504. [Google Scholar] [CrossRef]
- Rohrmann, G.F. Baculovirus nucleocapsid aggregation (MNPV vs SNPV): An evolutionary strategy, or a product of replication conditions? Virus Genes 2014, 49, 351–357. [Google Scholar] [CrossRef]
- Freedman, A.S.; Huang, A.Y.; Dixon, K.P.; Polivka, C.; Dwyer, G. Effects of host-tree foliage on polymorphism in an insect pathogen. bioRxiv 2024. preprint. [Google Scholar]
- Granados, R.R.; Lawler, K.A. In vivo pathway of Autographa californica baculovirus invasion and infection. Virology 1981, 108, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Washburn, J.O.; Lyons, E.H.; Haas-Stapleton, E.J.; Volkman, L.E. Multiple nucleocapsid packaging of Autographa californica nucleopolyhedrovirus accelerates the onset of systemic infection in Trichoplusia ni. J. Virol. 1999, 73, 411–416. [Google Scholar] [CrossRef]
- Ikeda, M.; Yamada, H.; Hamajima, R.; Kobayashi, M. Baculovirus genes modulating intracellular innate antiviral immunity of lepidopteran insect cells. Virology 2013, 435, 1–13. [Google Scholar] [CrossRef]
- Nagamine, T. Apoptotic arms races in insect-baculovirus coevolution. Physiol. Entomol. 2022, 47, 1–10. [Google Scholar] [CrossRef]
- Beperet, I.; Simón, O.; López-Ferber, M.; Van Lent, J.; Williams, T.; Caballero, P. Mixtures of insect pathogenic viruses in a single virion: Towards the development of custom designed insecticides. Appl. Environ.Microbiol. 2021, 87, e02180-20. [Google Scholar] [CrossRef] [PubMed]
- Pazmiño-Ibarra, V.; Herrero, S.; Sanjuán, R. Spatially segregated transmission of co-occluded baculoviruses limits virus–virus interactions mediated by cellular coinfection during primary infection. Viruses 2022, 14, 1697. [Google Scholar] [CrossRef]
- Xia, J.; Fei, S.; Huang, Y.; Lai, W.; Yu, Y.; Liang, L.; Wu, H.; Swevers, L.; Sun, J.; Feng, M. Single-nucleus sequencing of silkworm larval midgut reveals the immune escape strategy of BmNPV in the midgut during the late stage of infection. Insect Biochem. Mol. Biol. 2024, 164, 104043. [Google Scholar] [CrossRef]
- Nagamine, T.; Kawasaki, Y.; Abe, A.; Matsumoto, S. Nuclear marginalization of host cell chromatin associated with expansion of two discrete virus-induced subnuclear compartments during baculovirus infection. J. Virol. 2008, 82, 6409–6418. [Google Scholar] [CrossRef]
- Cory, J.S.; Clarke, E.E.; Brown, M.L.; Hails, R.S.; O’Reilly, D.R. Microparasite manipulation of an insect: The influence of the egt gene on the interaction between a baculovirus and its lepidopteran host. Funct. Ecol. 2004, 18, 443–450. [Google Scholar] [CrossRef]
- Fleming-Davies, A.E.; Dukic, V.; Andreasen, V.; Dwyer, G. Effects of host heterogeneity on pathogen diversity and evolution. Ecol. Lett. 2015, 18, 1252–1261. [Google Scholar] [CrossRef] [PubMed]
- Fleming-Davies, A.E.; Dwyer, G. Phenotypic variation in overwinter environmental transmission of a baculovirus and the cost of virulence. Am. Nat. 2015, 186, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Páez, D.J.; Fleming-Davies, A.E. Understanding the evolutionary ecology of host–pathogen interactions provides insights into the outcomes of insect pest biocontrol. Viruses 2020, 12, 141. [Google Scholar] [CrossRef] [PubMed]
- Lepore, L.S.; Roelvink, P.R.; Granados, R.R. Enhancin, the granulosis virus protein that facilitates nucleopolyhedrovirus (NPV) infections, is a metalloprotease. J. Invertebr. Pathol. 1996, 68, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, D.J.; Hitchman, R.B.; Vanbergen, A.J.; Hails, R.S.; Possee, R.D.; Cory, J.S. Host ecology determines the relative fitness of virus genotypes in mixed-genotype nucleopolyhedrovirus infections. J. Evol. Biol. 2004, 17, 1018–1025. [Google Scholar] [CrossRef] [PubMed]
- Boogaard, B.; Van Oers, M.M.; Van Lent, J.W.M. An advanced view on baculovirus per os infectivity factors. Insects 2018, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Arrizubieta, M.; Simón, O.; Williams, T.; Caballero, P. A novel binary mixture of Helicoverpa armigera single nucleopolyhedrovirus genotypic variants has improved insecticidal characteristics for control of cotton bollworms. Appl. Environ. Microbiol. 2015, 81, 3984–3993. [Google Scholar] [CrossRef] [PubMed]
- del Angel, C.; Lasa, R.; Rodríguez del Bosque, L.A.; Mercado, G.; Beperet, I.; Caballero, P.; Williams, T. Anticarsia gemmatalis nucleopolyhedrovirus from soybean crops in Tamaulipas, Mexico: Diversity and insecticidal characteristics of individual variants and their co-occluded mixtures. Fla. Entomol. 2018, 101, 404–410. [Google Scholar] [CrossRef]
- López-Ferber, M.; Simón, O.; Williams, T.; Caballero, P. Defective or effective? Mutualistic interactions between virus genotypes. Proc. R. Soc. Lond. B. Biol. Sci. 2003, 270, 2249–2255. [Google Scholar] [CrossRef] [PubMed]
- Clavijo, G.; Williams, T.; Simón, O.; Muñoz, D.; Cerutti, M.; López-Ferber, M.; Caballero, P. Mixtures of complete and pif1- and pif2-deficient genotypes are required for increased potency of an insect nucleopolyhedrovirus. J. Virol. 2009, 83, 5127–5136. [Google Scholar] [CrossRef] [PubMed]
- Simón, O.; Williams, T.; Cerutti, M.; Caballero, P.; López-Ferber, M. Expression of a peroral infection factor determines pathogenicity and population structure in an insect virus. PLoS ONE 2013, 8, e78834. [Google Scholar] [CrossRef]
- Hinsberger, A.; Blachère-López, C.; Knox, C.; Moore, S.; Marsberg, T.; López-Ferber, M. CpGV-M replication in type I resistant insects: Helper virus and order of ingestion are important. Viruses 2021, 13, 1695. [Google Scholar] [CrossRef]
- Erez, Z.; Steinberger-Levy, I.; Shamir, M.; Doron, S.; Stokar-Avihail, A.; Peleg, Y.; Melamed, S.; Leavitt, A.; Savidor, A.; Albeck, S.; et al. Communication between viruses guides lysis-lysogeny decisions. Nature 2017, 541, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Graillot, B.; Bayle, S.; Blachère-López, C.; Besse, S.; Siegwart, M.; López-Ferber, M. Biological characteristics of experimental genotype mixtures of Cydia pomonella granulovirus (CpGV): Ability to control susceptible and resistant pest populations. Viruses 2016, 8, 147. [Google Scholar] [CrossRef] [PubMed]
- Gueli-Alletti, G.; Sauer, A.J.; Weihrauch, B.; Fritsch, E.; Undorf-Spahn, K.; Wennmann, J.T.; Jehle, J.A. Using next generation sequencing to identify and quantify the genetic composition of resistance-breaking commercial isolates of Cydia pomonella granulovirus. Viruses 2017, 9, 250. [Google Scholar] [CrossRef]
- Belda, I.M.; Beperet, I.; Williams, T.; Caballero, P. Genetic variation and biological activity of two closely related alphabaculoviruses during serial passage in permissive and semi-permissive heterologous hosts. Viruses 2019, 11, 660. [Google Scholar] [CrossRef]
- Graillot, B.; Blachère-López, C.; Besse, S.; Siegwart, M.; López-Ferber, M. Host range extension of Cydia pomonella granulovirus: Adaptation to oriental fruit moth, Grapholita molesta. Biocontrol 2016, 62, 19–27. [Google Scholar] [CrossRef]
- Hitchman, R.B.; Hodgson, D.J.; King, L.A.; Hails, R.S.; Cory, J.S.; Possee, R.D. Host mediated selection of pathogen genotypes as a mechanism for the maintenance of baculovirus diversity in the field. J. Invertebr. Pathol. 2007, 94, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Kolodny-Hirsch, D.M.; Van Beek, N.A.M. Selection of a morphological variant of Autographa californica nuclear polyhedrosis virus with increased virulence following serial passage in Plutella xylostella. J. Invertebr. Pathol. 1997, 69, 205–211. [Google Scholar] [CrossRef]
- Molina-Ruiz, C.S.; Zamora-Briseño, J.A.; Simón, O.; Lasa, R.; Williams, T. A qPCR assay for the quantification of selected genotypic variants of Spodoptera frugiperda multiple nucleopolyhedrovirus (Baculoviridae). Viruses 2024, 16, 881. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.A.; Dwyer, G. Effects of multiple sources of genetic drift on pathogen variation within hosts. PLoS Biol. 2018, 16, e2004444. [Google Scholar] [CrossRef] [PubMed]
- van der Werf, W.; Hemerik, L.; Vlak, J.M.; Zwart, M.P. Heterogeneous host susceptibility enhances prevalence of mixed-genotype micro-parasite infections. PLoS Comput. Biol. 2011, 7, e1002097. [Google Scholar] [CrossRef]
- Hudson, A.I.; Fleming-Davies, A.E.; Páez, D.J.; Dwyer, G. Genotype-by-genotype interactions between an insect and its pathogen. J. Evol. Biol. 2016, 29, 2480–2490. [Google Scholar] [CrossRef]
- Siegwart, M.; Maugin, S.; Besse, S.; López-Ferber, M.; Hinsberger, A.; Gauffre, B. Le carpocapse des pommes résiste au virus dela granulose. Phytoma 2020, 738, 45–50. [Google Scholar]
- Murillo, R.; Muñoz, D.; Ruíz-Portero, M.C.; Alcázar, M.D.; Belda, J.E.; Williams, T.; Caballero, P. Abundance and genetic structure of nucleopolyhedrovirus populations in greenhouse substrate reservoirs. Biol. Control 2007, 42, 216–225. [Google Scholar] [CrossRef]
- Martínez-Solís, M.; Collado, M.C.; Herrero, S. Influence of diet, sex, and viral infections on the gut microbiota composition of Spodoptera exigua caterpillars. Front. Microbiol. 2020, 11, 753. [Google Scholar]
- Shikano, I. Evolutionary ecology of multitrophic interactions between plants, insect herbivores and entomopathogens. J. Chem. Ecol. 2017, 43, 586–598. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, D.J.; Vanbergen, A.J.; Hartley, S.E.; Hails, R.S.; Cory, J.S. Differential selection of baculovirus genotypes mediated by different species of host food plant. Ecol. Lett. 2002, 5, 512–518. [Google Scholar] [CrossRef]
- Raymond, B.; Vanbergen, A.; Pearce, I.; Hartley, S.; Cory, J.; Hails, R. Host plant species can influence the fitness of herbivore pathogens: The winter moth and its nucleopolyhedrovirus. Oecologia 2002, 131, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Cory, J.S.; Myers, J.H. Within and between population variation in disease resistance in cyclic populations of western tent caterpillars: A test of the disease defence hypothesis. J. Anim. Ecol. 2009, 78, 646–655. [Google Scholar] [CrossRef]
- Lee, H.H.; Miller, L.K. Isolation of genotypic variants of Autographa californica nuclear polyhedrosis virus. J. Virol. 1978, 27, 754–767. [Google Scholar] [CrossRef]
- Lynn, D.E.; Shapiro, M.; Dougherty, E.M. Selection and screening of clonal isolates of the Abington strain of gypsy moth nuclear polyhedrosis virus. J. Invertebr. Pathol. 1993, 62, 191–195. [Google Scholar] [CrossRef]
- Anonymous. AgBiTech launches lepidopteran biocontrol options. Outlooks Pest Manag. 2019, 30, 277–281. [Google Scholar] [CrossRef]
- Popham, H.J.R.; Nusawardani, T.; Bonning, B.C. Introduction to the use of baculoviruses as biological insecticides. In Baculovirus and Insect Cell Expression Protocols; Methods in Molecular Biology; Murhammer, D., Ed.; Humana Press: New York, NY, USA, 2016; Volume 1350, pp. 383–392. [Google Scholar]
- Kroemer, J.A.; Bonning, B.C.; Harrison, R.L. Expression, delivery and function of insecticidal proteins expressed by recombinant baculoviruses. Viruses 2015, 7, 422–455. [Google Scholar] [CrossRef]
- Williams, T.; López-Ferber, M.; Caballero, P. Nucleopolyhedrovirus coocclusion technology: A new concept in the development of biological insecticides. Front. Microbiol. 2022, 12, 810026. [Google Scholar] [CrossRef]
- Caballero, P.; Williams, T.; Muñoz-Labiano, D.; Murillo-Perez, R.; Lasa-Covarrubias, R. Nuevos Genotipos del Nucleopoliedrovirus de Spodoptera exigua y Uso de los Mismos en el Control de las Plagas Producidas por Este Insecto. ES P200601065A. Oficina Española de Patentes ES2301352A1, 1 May 2006. [Google Scholar]
- Carballo, A.; Murillo, R.; Jakubowska, A.; Herrero, S.; Williams, T.; Caballero, P. Co-infection with iflaviruses influences the insecticidal properties of Spodoptera exigua multiple nucleopolyhedrovirus occlusion bodies: Implications for the production and biosecurity of baculovirus insecticides. PLoS ONE 2017, 12, e0177301. [Google Scholar] [CrossRef]
- Briese, D.T. Genetic basis for resistance to a granulosis virus in the potato tuber moth, Phthorimaea operculella. J. Invertebr. Pathol. 1982, 39, 215–218. [Google Scholar] [CrossRef]
- Abot, A.R.; Moscardi, F.; Fuxa, J.R.; Sosa-Gomez, D.R.; Richter, A.R. Development of resistance by Anticarsia gemmatalis from Brazil and the United States to a nuclear polyhedrosis virus under laboratory selection pressure. Biol. Control 1996, 7, 126–130. [Google Scholar] [CrossRef]
- Fuxa, J.R.; Richter, A.R. Reversion of resistance by Spodoptera frugiperda to nuclear polyhedrosis virus. J. Invertebr. Pathol. 1989, 53, 52–56. [Google Scholar] [CrossRef]
- Martínez, A.M.; Caballero, P.; Villanueva, M.; Miralles, N.; San Martín, I.; López, E.; Williams, T. Formulation with an optical brightener does not increase probability of developing resistance to Spodoptera frugiperda nucleopolyhedrovirus in the laboratory. J. Econ. Entomol. 2004, 97, 1202–1208. [Google Scholar] [CrossRef][Green Version]
- Nakai, M.; Takahashi, K.; Iwata, K.; Tanaka, K.; Koyanagi, J.; Ookuma, A.; Takatsuka, J.; Okuno, S.; Kunimi, Y. Acquired resistance to a nucleopolyhedrovirus in the smaller tea tortrix Adoxophyes honmai (Lepidoptera: Tortricidae) after selection by serial viral administration. J. Invertebr. Pathol. 2017, 145, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Moscardi, F. Assessment of the application of baculoviruses for control of Lepidoptera. Annu. Rev. Entomol. 1999, 44, 257–289. [Google Scholar] [CrossRef]
- Tanada, Y. A granulosis virus of the codling moth, Carpocapsa pomonella (Linnaeus) (Olethreutidae, Lepidoptera). J. Insect Pathol. 1964, 6, 378–380. [Google Scholar]
- Fritsch, E.; Undorf-Spahn, K.; Kienzle, J.; Zebitz, C.; Huber, J. Apfelwickler-Granulovirus: Erste Hinweise auf Unterschiede in der Empfindlichkeit lokaler Apfelwicklerpopulationen. Nachr Dt Pflanzenschutzdienstes 2005, 57, 29–34. [Google Scholar]
- Sauphanor, B.; Berling, M.; Toubon, J.F.; Reyes, M.; Delnatte, J.; Allemoz, P. Cases of resistance to granulosis virus in the codling moth. Phytoma 2006, 590, 24–27. [Google Scholar]
- Eberle, K.E.; Sayed, S.; Rezapanah, M.; Shojai-Estabragh, S.; Jehle, J.A. Diversity and evolution of the Cydia pomonella granulovirus. J. Gen. Virol. 2009, 90, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Sauer, A.J.; Fritsch, E.; Undorf-Spahn, K.; Nguyen, P.; Marec, F.; Heckel, D.G.; Jehle, J.A. Novel resistance to Cydia pomonella granulovirus (CpGV) in codling moth shows autosomal and dominant inheritance and confers cross-resistance to different CpGV genome groups. PLoS ONE 2017, 12, e0179157. [Google Scholar] [CrossRef]
- Feng, G.; Thumbi, D.K.; de Jong, J.; Hodgson, J.J.; Arif, B.; Doucet, D.; Krell, P.J. Selection and characterization of Autographa californica multiple nucleopolyhedrovirus DNA polymerase mutations. J. Virol. 2012, 86, 13576–13588. [Google Scholar] [CrossRef] [PubMed]
- Baillie, V.L.; Bouwer, G. The effect of inoculum dose on the genetic diversity detected within Helicoverpa armigera nucleopolyhedrovirus populations. J. Gen. Virol. 2013, 94, 2524–2529. [Google Scholar] [CrossRef]
- Díaz-Muñoz, S.L.; Sanjuán, R.; West, S. Sociovirology: Conflict, cooperation, and communication among viruses. Cell Host Microbe 2017, 22, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Leeks, A.; Bono, L.M.; Ampolini, E.A.; Souza, L.S.; Höfler, T.; Mattson, C.L.; Dye, A.E.; Díaz-Muñoz, S.L. Open questions in the social lives of viruses. J. Evol. Biol. 2023, 36, 1551–1567. [Google Scholar] [CrossRef] [PubMed]
- Cory, J.S. Evolution of host resistance to insect pathogens. Curr. Opin. Insect Sci. 2017, 21, 54–59. [Google Scholar] [CrossRef]
- Zheng-Li, L.Y. The Role of Pathogen Diversity on the Evolution of Resistance. Master’s Thesis, Department of Biological Sciences, Simon Fraser University, Burnaby, BC, Canada, 2017. Available online: https://summit.sfu.ca/item/17543 (accessed on 2 December 2024).
- Visher, E.; Uricchio, L.; Bartlett, L.; de Namur, N.; Yarcan, A.; Alhassani, D.; Boots, M. The evolution of host specialization in an insect pathogen. Evolution 2022, 76, 2375–2388. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, L.J.; Wilfert, L.; Boots, M. XA genotypic trade-off between constitutive resistance to viral infection and host growth rate. Evolution 2022, 72, 2749–2757. [Google Scholar] [CrossRef] [PubMed]
- Boots, M. The evolution of resistance to a parasite is determined by resources. Am. Nat. 2011, 178, 214–220. [Google Scholar] [CrossRef]
- Roberts, K.E.; Meaden, S.; Sharpe, S.; Kay, S.; Doyle, T.; Wilson, D.; Bartlett, L.J.; Paterson, S.; Boots, M. Resource quality determines the evolution of resistance and its genetic basis. Mol. Ecol. 2020, 29, 4128–4142. [Google Scholar] [CrossRef]
- Karlsson-Green, K. The effects of host plant species and larval density on immune function in the polyphagous moth Spodoptera littoralis. Ecol. Evol. 2021, 11, 10090–10097. [Google Scholar] [CrossRef]
- Undorf-Spahn, K.; Fritsch, E.; Huber, J.; Kienzle, J.; Zebitz, C.P.W.; Jehle, J.A. High stability and no fitness costs of the resistance of codling moth to Cydia pomonella granulovirus (CpGV-M). J. Invertebr. Pathol. 2012, 111, 136–142. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Ferber, M.; Caballero, P.; Williams, T. Baculovirus Genetic Diversity and Population Structure. Viruses 2025, 17, 142. https://doi.org/10.3390/v17020142
López-Ferber M, Caballero P, Williams T. Baculovirus Genetic Diversity and Population Structure. Viruses. 2025; 17(2):142. https://doi.org/10.3390/v17020142
Chicago/Turabian StyleLópez-Ferber, Miguel, Primitivo Caballero, and Trevor Williams. 2025. "Baculovirus Genetic Diversity and Population Structure" Viruses 17, no. 2: 142. https://doi.org/10.3390/v17020142
APA StyleLópez-Ferber, M., Caballero, P., & Williams, T. (2025). Baculovirus Genetic Diversity and Population Structure. Viruses, 17(2), 142. https://doi.org/10.3390/v17020142







