The Genome Sequences of Baculoviruses from the Tufted Apple Bud Moth, Platynota idaeusalis, Reveal Recombination Between an Alphabaculovirus and a Betabaculovirus from the Same Host
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus Samples
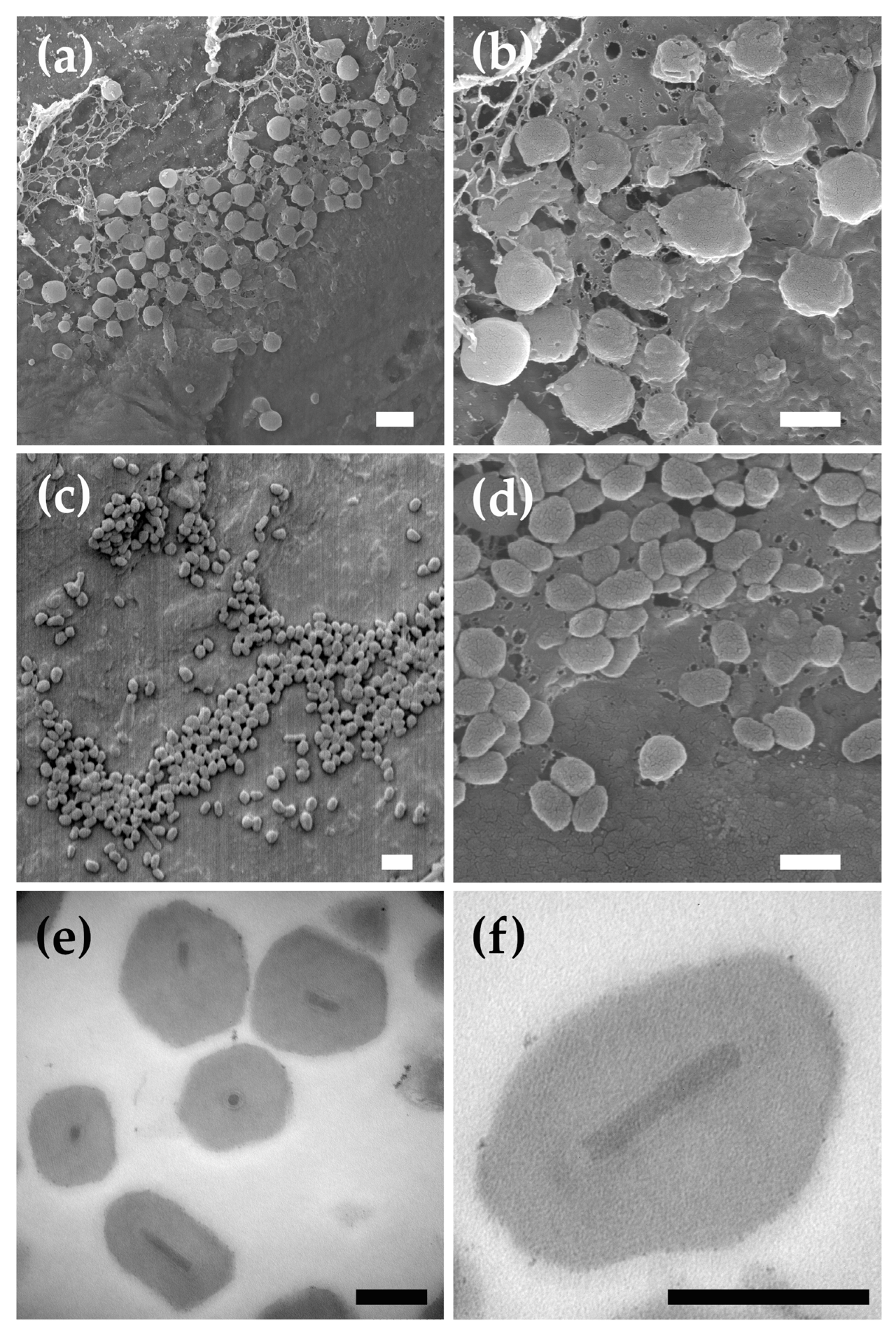
2.2. Electron Microscopy
2.2.1. LT-SEM
2.2.2. TEM
2.3. DNA Isolation and Sequencing
2.4. Genome Annotation
2.5. Phylogeny
2.6. Gene Synteny and Pairwise Distance Estimation
3. Results
3.1. Ultrastructure Features of P. idaeusalis Baculovirus Occlusion Bodies
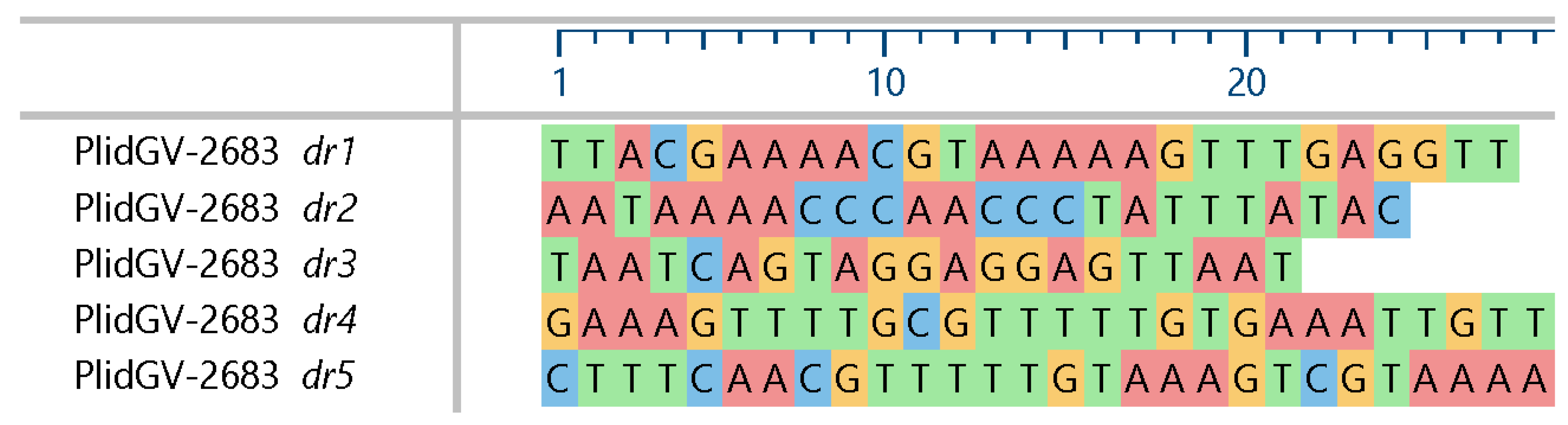
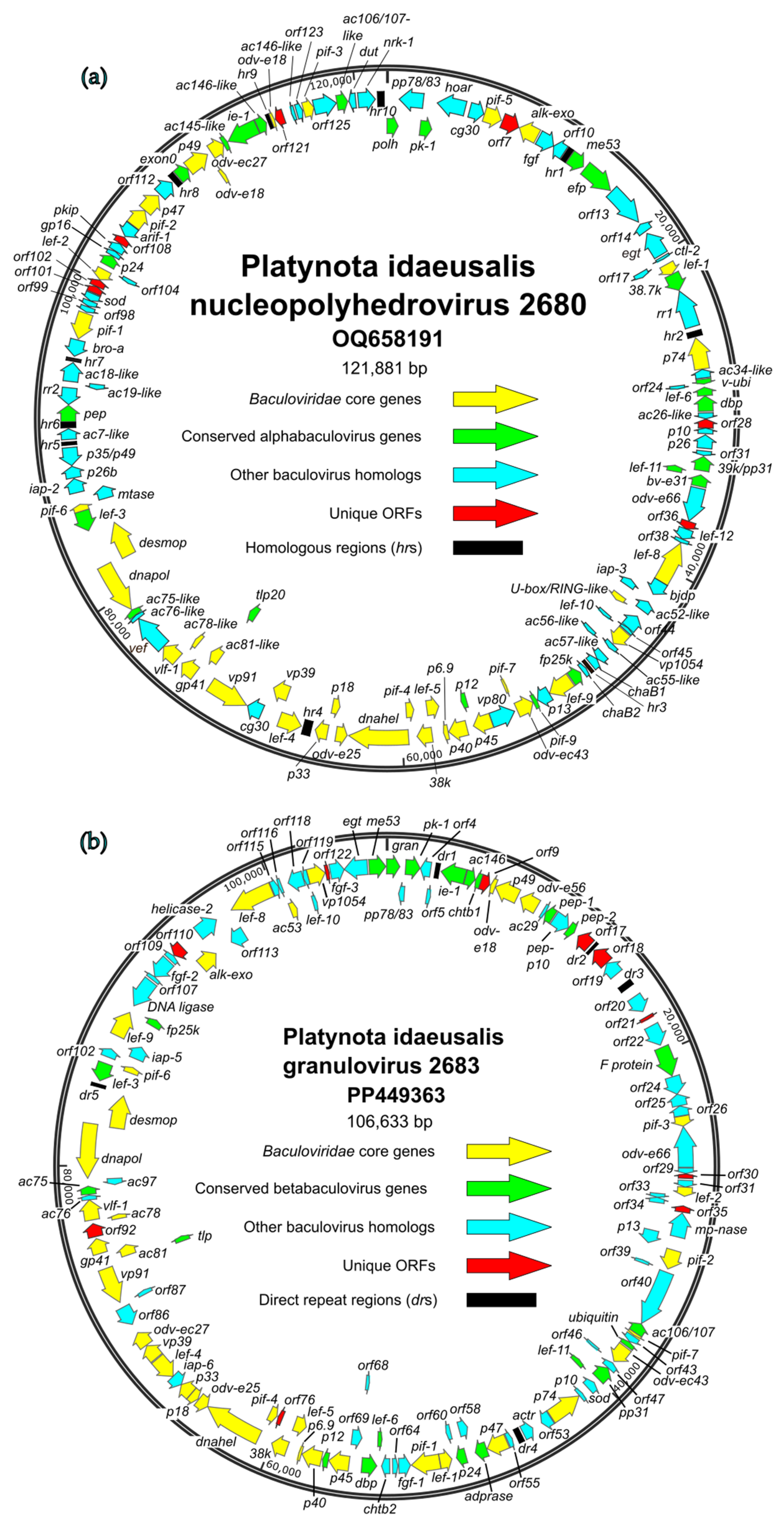
3.2. Properties of P. idaeusalis Alphabaculovirus and Betabaculovirus Genomes
3.3. ORF Content
3.3.1. Conserved Baculovirus Genes
3.3.2. Unique Genes
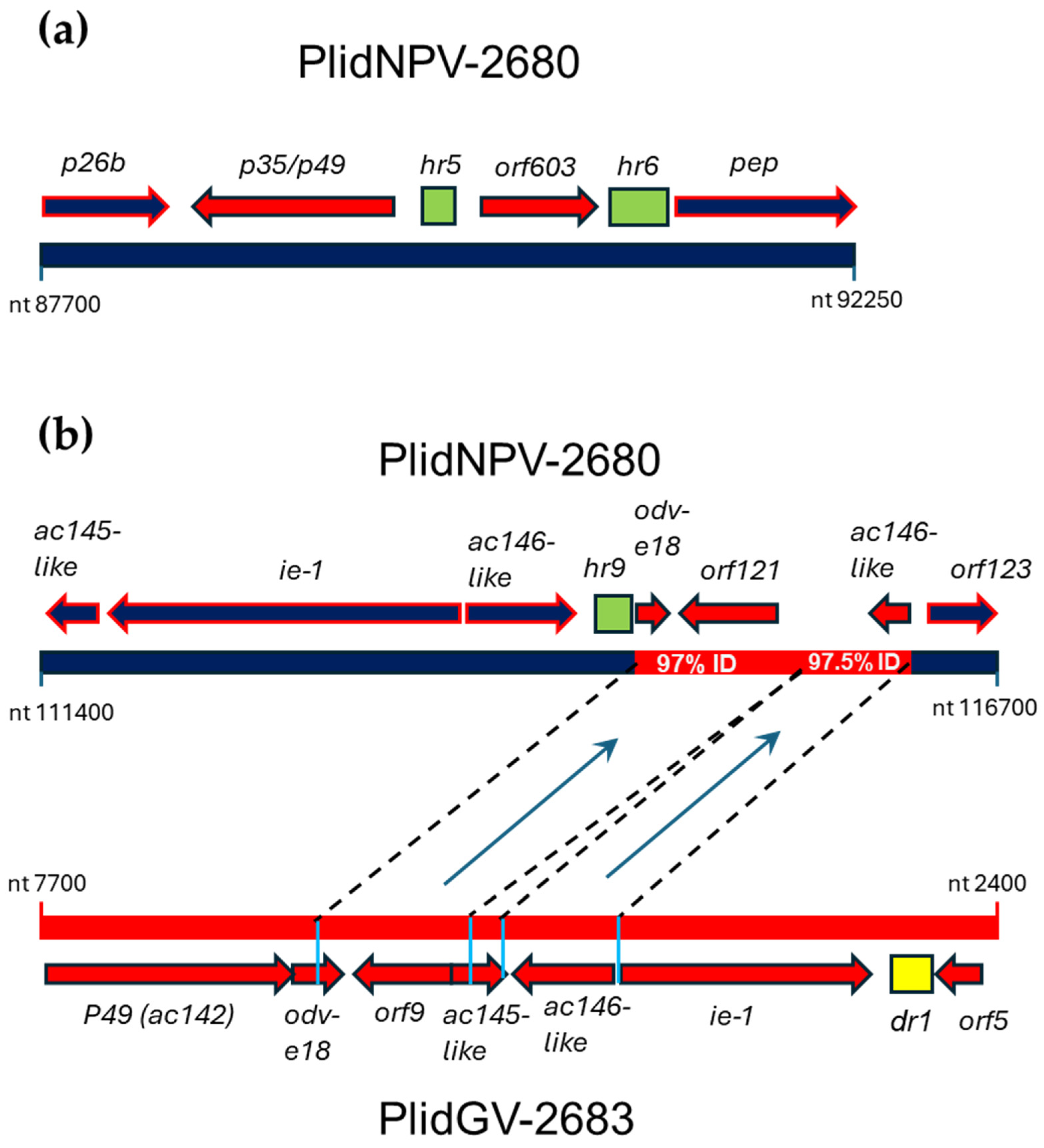
3.3.3. ORFs in PlidNPV-2680 That Appear to Derive from Betabaculoviruses
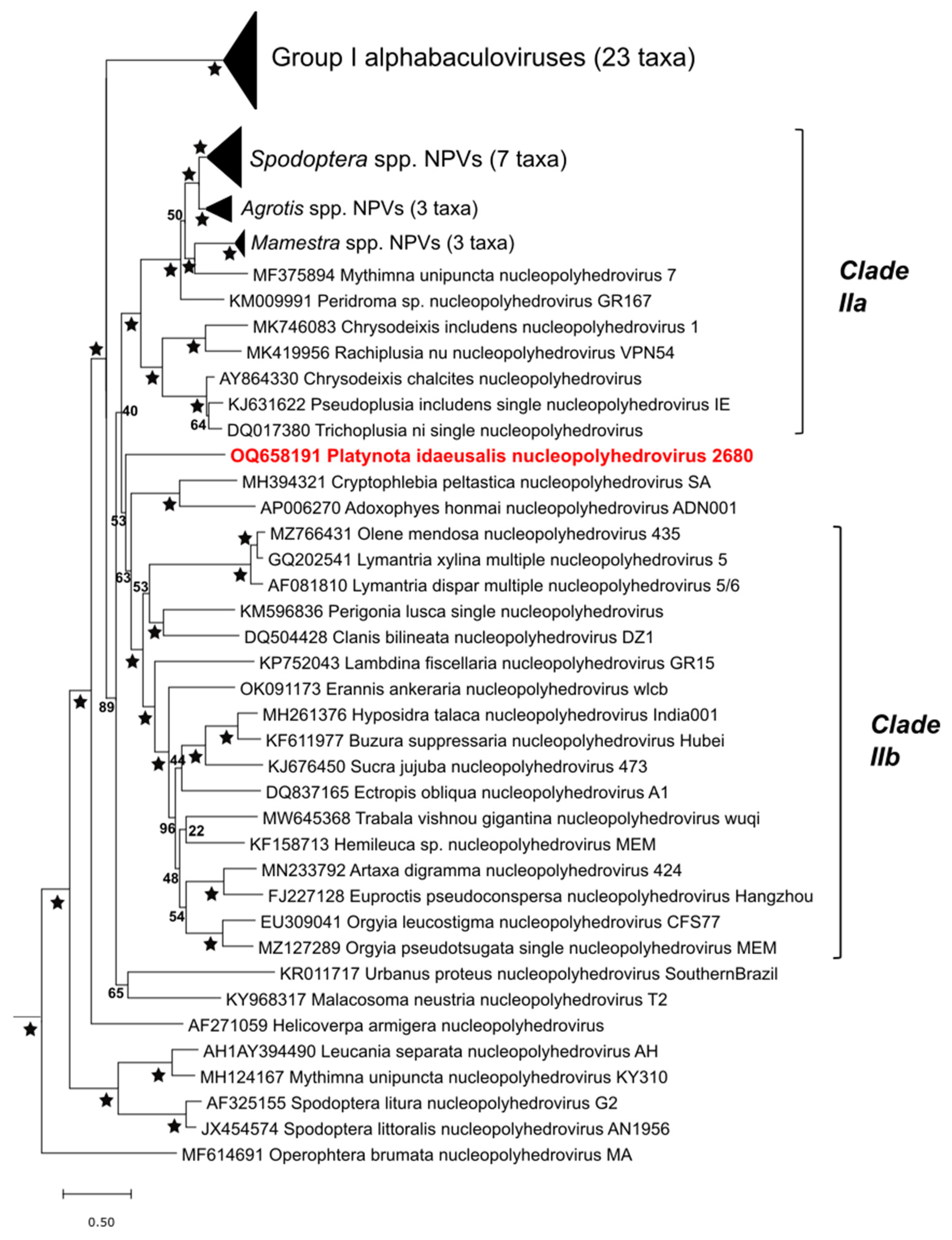
3.4. Relationships with Other Viruses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harrison, R.L.; Hoover, K. Baculoviruses and other occluded insect viruses. In Insect Pathology, 2nd ed.; Vega, F.E., Kaya, H.K., Eds.; Academic Press: Boston, MA, USA, 2012; pp. 73–131. [Google Scholar]
- Van Oers, M.M.; Pijlman, G.P.; Vlak, J.M. Thirty years of baculovirus-insect cell protein expression: From dark horse to mainstream technology. J. Gen. Virol. 2015, 96, 6–23. [Google Scholar] [CrossRef] [PubMed]
- Rohrmann, G.F. Baculovirus Molecular Biology; National Center for Biotechnology Information: Bethesda, MD, USA, 2019.
- Landwehr, A. Benefits of baculovirus use in IPM strategies for open field and protected vegetables. Front. Sustain. Food Syst. 2021, 4, 593796. [Google Scholar] [CrossRef]
- Harrison, R.L.; Herniou, E.A.; Jehle, J.A.; Theilmann, D.A.; Burand, J.P.; Becnel, J.J.; Krell, P.J.; van Oers, M.M.; Mowery, J.D.; Bauchan, G.R.; et al. ICTV Virus Taxonomy Profile: Baculoviridae. J. Gen. Virol. 2018, 99, 1185–1186. [Google Scholar] [CrossRef] [PubMed]
- Van Oers, M.M.; Herniou, E.A.; Jehle, J.A.; Krell, P.J.; Abd-Alla, A.M.M.; Ribeiro, B.M.; Theilmann, D.A.; Hu, Z.; Harrison, R.L. Developments in the classification and nomenclature of arthropod-infecting large DNA viruses that contain pif genes. Arch. Virol. 2023, 168, 182. [Google Scholar] [CrossRef]
- Wang, X.; Liu, X.; Makalliwa, G.A.; Li, J.; Wang, H.; Hu, Z.; Wang, M. Per os infectivity factors: A complicated and evolutionarily conserved entry machinery of baculovirus. Sci. China Life Sci. 2017, 60, 806–815. [Google Scholar] [CrossRef]
- Boogaard, B.; Van Oers, M.M.; van Lent, J.W.M. An advanced view on baculovirus per os infectivity factors. Insects 2019, 9, 84. [Google Scholar] [CrossRef]
- Passarelli, A.L.; Guarino, L.A. Baculovirus late and very late gene regulation. Curr. Drug Targets 2007, 8, 1103–1115. [Google Scholar] [CrossRef]
- Garavaglia, M.J.; Miele, S.A.; Iserte, J.A.; Belaich, M.N.; Ghiringhelli, P.D. The ac53, ac78, ac101, and ac103 genes are newly discovered core genes in the family Baculoviridae. J. Virol. 2012, 86, 12069–12079. [Google Scholar] [CrossRef]
- Javed, M.A.; Biswas, S.; Willis, L.G.; Harris, S.; Pritchard, C.; van Oers, M.M.; Donly, B.C.; Erlandson, M.A.; Hegedus, D.D.; Theilmann, D.A. Autographa californica multiple nucleopolyhedrovirus AC83 is a per os infectivity factor (PIF) protein required for occlusion-derived virus (ODV) and budded virus nucleocapsid assembly as well as assembly of the PIF complex in ODV envelopes. J. Virol. 2017, 91, e02115–e02116. [Google Scholar] [CrossRef]
- Ishimwe, E.; Hodgson, J.J.; Clem, R.J.; Passarelli, A.L. Reaching the melting point: Degradative enzymes and protease inhibitors involved in baculovirus infection and dissemination. Virology 2015, 479–480, 637–649. [Google Scholar] [CrossRef]
- Hodgson, J.J.; Krell, P.J.; Passarelli, A.L. Mature viral cathepsin is required for release of viral occlusion bodies from Autographa californica multiple nucleopolyhedrovirus-infected cells. Virology 2021, 556, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.W.; Mccarthy, W.J. Purification and characterization of the Platynota-idaeusalis baculovirus pathogenic to the tufted apple bud moth. J. Invertebr. Pathol. 1993, 62, 37–46. [Google Scholar] [CrossRef]
- Hogmire, H.W.; Howitt, A.J. Bionomics of the tufted apple budmoth, Platynota-idaeusalis (Lepidopter-Tortricidae) in Michigan. Ann. Entomol. Soc. Am. 1979, 72, 121–126. [Google Scholar] [CrossRef]
- Hogmire, H.W. (Ed.) Mid-Atlantic Orchard Monitoring Guide; Northeast Regional Agricultural Engineering Service: Ithaca, NY, USA, 1995; Volume 75, p. 361. [Google Scholar]
- Adams, J.R.; Wilcox, T.A. Scanning electron microscopical comparisons of insect virus occlusion bodies prepared by several techniques. J. Invert. Path. 1982, 40, 12–20. [Google Scholar] [CrossRef]
- Harrison, R.L.; Mowery, J.D.; Rowley, D.L.; Bauchan, G.R.; Theilmann, D.A.; Rohrmann, G.F.; Erlandson, M.A. The complete genome sequence of a third distinct baculovirus isolated from the true armyworm, Mythimna unipuncta, contains two copies of the lef-7 gene. Virus Genes 2018, 54, 297–310. [Google Scholar] [CrossRef]
- Harrison, R.L.; Rowley, D.L. The Parapoynx stagnalis nucleopolyhedrovirus (PastNPV), a divergent member of the Alphabaculovirus group I clade, encodes a homolog of Ran GTPase. Viruses 2022, 14, 2289. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids. Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kubler, J.; Lozajic, M.; Gabler, F.; Soding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Van Oers, M.M.; Vlak, J.M. Baculovirus genomics. Curr. Drug Targets 2007, 8, 1051–1068. [Google Scholar] [CrossRef]
- Kurtz, S.; Schleiermacher, C. REPuter: Fast computation of maximal repeats in complete genomes. Bioinformatics 1999, 15, 426–427. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nuc. Acids. Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2021, 12, 373–377. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Hu, Z.H.; Arif, B.M.; Jin, F.; Martens, J.W.; Chen, X.W.; Sun, J.S.; Zuidema, D.; Goldbach, R.W.; Vlak, J.M. Distinct gene arrangement in the Buzura suppressaria single-nucleocapsid nucleopolyhedrovirus genome. J. Gen. Virol. 1998, 79 Pt 11, 2841–2851. [Google Scholar] [CrossRef]
- Jehle, J.A.; Lange, M.; Wang, H.; Hu, Z.; Wang, Y.; Hauschild, R. Molecular identification and phylogenetic analysis of baculoviruses from Lepidoptera. Virology 2006, 346, 180–193. [Google Scholar] [CrossRef]
- Tanada, Y.; Hess, R.T. Baculoviridae. Granulosis viruses. In Atlas of Invertebrate Viruses; Adams, J.R., Bonami, J.R., Eds.; CRC Press, Inc.: Boca Raton, FL, USA, 1991. [Google Scholar]
- Oliveira, H.P.; Dos Santos, E.R.; Harrison, R.L.; Ribeiro, B.M.; Ardisson-Araujo, D.M.P. Identification and analysis of putative tRNA genes in baculovirus genomes. Virus Res. 2022, 322, 198949. [Google Scholar] [CrossRef]
- Mikhailov, V.S.; Vanarsdall, A.L.; Rohrmann, G.F. Isolation and characterization of the DNA-binding protein (DBP) of the Autographa californica multiple nucleopolyhedrovirus. Virology 2008, 370, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Means, J.C.; Passarelli, A.L. Viral fibroblast growth factor, matrix metalloproteases, and caspases are associated with enhancing systemic infection by baculoviruses. Proc. Natl. Acad. Sci. USA 2010, 107, 9825–9830. [Google Scholar] [CrossRef]
- Simon, O.; Williams, T.; Caballero, P.; Possee, R.D. Effects of Acp26 on in vitro and in vivo productivity, pathogenesis and virulence of Autographa californica multiple nucleopolyhedrovirus. Virus Res. 2008, 136, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Bideshi, D.K.; Renault, S.; Stasiak, K.; Federici, B.A.; Bigot, Y. Phylogenetic analysis and possible function of bro-like genes, a multigene family widespread among large double-stranded DNA viruses of invertebrates and bacteria. J. Gen. Virol. 2003, 84, 2531–2544. [Google Scholar] [CrossRef] [PubMed]
- Clem, R.J. Viral IAPs, then and now. Semin. Cell. Dev. Biol. 2015, 39, 72–79. [Google Scholar] [CrossRef]
- Slavicek, J.M. Baculovirus enhancins and their role in viral pathogenicity. In Molecular Virology; Adoga, M., Ed.; IntechOpen: Rijeka, Croatia, 2012; pp. 147–168. [Google Scholar]
- Hayakawa, T.; Ko, R.; Okano, K.; Seong, S.I.; Goto, C.; Maeda, S. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology 1999, 262, 277–297. [Google Scholar] [CrossRef]
- Kuzio, J.; Pearson, M.N.; Harwood, S.H.; Funk, C.J.; Evans, J.T.; Slavicek, J.M.; Rohrmann, G.F. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology 1999, 253, 17–34. [Google Scholar] [CrossRef]
- Popham, H.J.; Bischoff, D.S.; Slavicek, J.M. Both Lymantria dispar nucleopolyhedrovirus enhancin genes contribute to viral potency. J. Virol. 2001, 75, 8639–8648. [Google Scholar] [CrossRef]
- Ishihara, G.; Shimada, T.; Katsuma, S. Functional characterization of nucleopolyhedrovirus CG30 protein. Virus Res. 2013, 174, 52–59. [Google Scholar] [CrossRef]
- Imai, N.; Matsumoto, S.; Kang, W. Formation of Bombyx mori nucleopolyhedrovirus IE2 nuclear foci is regulated by the functional domains for oligomerization and ubiquitin ligase activity. J. Gen. Virol. 2005, 86, 637–644. [Google Scholar] [CrossRef]
- Krappa, R.; Roncarati, R.; Knebel-Morsdorf, D. Expression of PE38 and IE2, viral members of the C3HC4 finger family, during baculovirus infection: PE38 and IE2 localize to distinct nuclear regions. J. Virol. 1995, 69, 5287–5293. [Google Scholar] [CrossRef] [PubMed]
- Tenev, T.; Ditzel, M.; Zachariou, A.; Meier, P. The antiapoptotic activity of insect IAPs requires activation by an evolutionarily conserved mechanism. Cell Death Differ. 2007, 14, 1191–1201. [Google Scholar] [CrossRef] [PubMed]
- Vandergaast, R.; Mitchell, J.K.; Byers, N.M.; Friesen, P.D. Insect inhibitor-of-apoptosis (IAP) proteins are negatively regulated by signal-induced N-terminal degrons absent within viral IAP proteins. J. Virol. 2015, 89, 4481–4493. [Google Scholar] [CrossRef] [PubMed]
- Escasa, S.R.; Lauzon, H.A.M.; Mathur, A.C.; Krell, P.J.; Arif, B.M. Sequence analysis of the Choristoneura occidentalis granulovirus genome. J. Gen. Virol. 2006, 87, 1917–1933. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, C.B.; Theilmann, D.A. AcMNPV ac143 (odv-e18) is essential for mediating budded virus production and is the 30th baculovirus core gene. Virology 2008, 375, 277–291. [Google Scholar] [CrossRef]
- Miele, S.A.; Garavaglia, M.J.; Belaich, M.N.; Ghiringhelli, P.D. Baculovirus: Molecular insights on their diversity and conservation. Int. J. Evol. Biol. 2011, 2011, 379424. [Google Scholar] [CrossRef]
- Theze, J.; Lopez-Vaamonde, C.; Cory, J.S.; Herniou, E.A. Biodiversity, Evolution and Ecological Specialization of Baculoviruses: A Treasure Trove for Future Applied Research. Viruses 2018, 10, 366. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, Z.; Zhang, L.; Hou, D.; Wang, M.; Arif, B.; Kou, Z.; Wang, H.; Deng, F.; Hu, Z. Genome sequencing and analysis of Catopsilia pomona nucleopolyhedrovirus: A distinct species in group I Alphabaculovirus. PLoS ONE 2016, 11, e0155134. [Google Scholar] [CrossRef]
- Escasa, S.R.; Harrison, R.L.; Mowery, J.D.; Bauchan, G.R.; Cory, J.S. The complete genome sequence of an alphabaculovirus from Spodoptera exempta, an agricultural pest of major economic significance in Africa. PLoS ONE 2019, 14, e0209937. [Google Scholar] [CrossRef]
- Hajos, J.P.; Pijnenburg, J.; Usmany, M.; Zuidema, D.; Zavodszky, P.; Vlak, J.M. High frequency recombination between homologous baculoviruses in cell culture. Arch. Virol. 2000, 145, 159–164. [Google Scholar] [CrossRef]
- Kamita, S.G.; Maeda, S.; Hammock, B.D. High-frequency homologous recombination between baculoviruses involves DNA replication. J. Virol. 2003, 77, 13053–13061. [Google Scholar] [CrossRef] [PubMed]
- Jehle, J.A.; Fritsch, E.; Huber, J.; Backhaus, H. Intra-specific and inter-specific recombination of tortricid-specific granuloviruses during co-infection in insect larvae. Arch. Virol. 2003, 148, 1317–1333. [Google Scholar] [CrossRef] [PubMed]
- Theze, J.; Takatsuka, J.; Nakai, M.; Arif, B.; Herniou, E.A. Gene acquisition convergence between entomopoxviruses and baculoviruses. Viruses 2015, 7, 1960–1974. [Google Scholar] [CrossRef] [PubMed]
- Winstanley, D.; Crook, N.E. Replication of Cydia pomonella granulosis virus in cell cultures. J. Gen. Virol. 1993, 74 Pt 8, 1599–1609. [Google Scholar] [CrossRef]
- Federici, B.A. Baculovirus pathogenesis. In The Baculoviruses; Miller, L.K., Ed.; Plenum Press: New York, NY, USA, 1997; pp. 33–59. [Google Scholar]
- Ardisson-Araujo, D.M.; Lima, R.N.; Melo, F.L.; Clem, R.J.; Huang, N.; Bao, S.N.; Sosa-Gomez, D.R.; Ribeiro, B.M. Genome sequence of Perigonia lusca single nucleopolyhedrovirus: Insights into the evolution of a nucleotide metabolism enzyme in the family Baculoviridae. Sci. Rep. 2016, 6, 24612. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.L.; Mowery, J.D.; Bauchan, G.R.; Theilmann, D.A.; Erlandson, M.A. The complete genome sequence of a second alphabaculovirus from the true armyworm, Mythimna unipuncta: Implications for baculovirus phylogeny and host specificity. Virus Genes 2019, 55, 104–116. [Google Scholar] [CrossRef]
- Ardisson-Araujo, D.M.; Pereira, B.T.; Melo, F.L.; Ribeiro, B.M.; Bao, S.N.; de Zanotto, P.M.; Moscardi, F.; Kitajima, E.W.; Sosa-Gomez, D.R.; Wolff, J.L. A betabaculovirus encoding a gp64 homolog. BMC Genom. 2016, 17, 94. [Google Scholar] [CrossRef]
- Li, L.; Donly, C.; Li, Q.; Willis, L.G.; Keddie, B.A.; Erlandson, M.A.; Theilmann, D.A. Identification and genomic analysis of a second species of nucleopolyhedrovirus isolated from Mamestra configurata. Virology 2002, 297, 226–244. [Google Scholar] [CrossRef]
- Erlandson, M.; Baldwin, D.; Vlak, J.M.; Theilmann, D. Genomics of alphabaculovirus isolates infecting Mamestra species from North America and Eurasia. J. Invertebr. Pathol. 2024, 203, 108063. [Google Scholar] [CrossRef]
- De Jong, J.G.; Lauzon, H.A.; Dominy, C.; Poloumienko, A.; Carstens, E.B.; Arif, B.M.; Krell, P.J. Analysis of the Choristoneura fumiferana nucleopolyhedrovirus genome. J. Gen. Virol. 2005, 86, 929–943. [Google Scholar] [CrossRef]
- Crouch, E.A.; Passarelli, A.L. Genetic requirements for homologous recombination in Autographa californica nucleopolyhedrovirus. J. Virol. 2002, 76, 9323–9334. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pearson, M.; Bjornson, R.; Pearson, G.; Rohrmann, G. The Autographa californica baculovirus genome: Evidence for multiple replication origins. Science 1992, 257, 1382–1384. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, Y.; Matsumoto, S.; Nagamine, T. Analysis of baculovirus IE1 in living cells: Dynamics and spatial relationships to viral structural proteins. J. Gen. Virol. 2004, 85, 3575–3583. [Google Scholar] [CrossRef] [PubMed]
- Bossert, M.; Carstens, E.B. Sequential deletion of AcMNPV homologous regions leads to reductions in budded virus production and late protein expression. Virus Res. 2018, 256, 125–133. [Google Scholar] [CrossRef]
- Hu, H.; Pan, K.; Shang, Y.; Guo, Y.; Xiao, H.; Deng, F.; Wang, M.; Hu, Z. Multiloci Manipulation of Baculovirus Genome Reveals the Pivotal Role of Homologous Regions in Viral DNA Replication, Progeny Production, and Enhancing Transcription. ACS Synth. Biol. 2022, 11, 144–153. [Google Scholar] [CrossRef]
- Rubnitz, J.; Subramani, S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol. Cell. Biol. 1984, 4, 2253–2258. [Google Scholar] [CrossRef]
- Volodin, A.A.; Bocharova, T.N.; Smirnova, E.A.; Camerini-Otero, R.D. Reversibility, equilibration, and fidelity of strand exchange reaction between short oligonucleotides promoted by RecA protein from escherichia coli and human Rad51 and Dmc1 proteins. J. Biol. Chem. 2009, 284, 1495–1504. [Google Scholar] [CrossRef]
- Nagamine, T. Apoptotic arms races in insect-baculovirus coevolution. Physiol. Entomol. 2022, 47, 1–10. [Google Scholar] [CrossRef]
- Popham, H.J.R.; Pellock, B.J.; Robson, M.; Dierks, P.M.; Miller, L.K. Characterization of a variant of Autographa californica nuclear polyhedrosis virus with a nonfunctional ORF 603. Biol. Control 1998, 12, 223–230. [Google Scholar] [CrossRef]
- Wormleaton, S.; Kuzio, J.; Winstanley, D. The complete sequence of the Adoxophyes orana granulovirus genome. Virology 2003, 311, 350–365. [Google Scholar] [CrossRef]
- Ardisson-Araujo, D.M.; de Melo, F.L.; Andrade Mde, S.; Sihler, W.; Bao, S.N.; Ribeiro, B.M.; de Souza, M.L. Genome sequence of Erinnyis ello granulovirus (ErelGV), a natural cassava hornworm pesticide and the first sequenced sphingid-infecting betabaculovirus. BMC Genom. 2014, 15, 856. [Google Scholar] [CrossRef] [PubMed]
- Cuartas, P.E.; Barrera, G.P.; Belaich, M.N.; Barreto, E.; Ghiringhelli, P.D.; Villamizar, L.F. The complete sequence of the first Spodoptera frugiperda Betabaculovirus genome: A natural multiple recombinant virus. Viruses 2015, 7, 394–421. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.L.; Rowley, D.L.; Funk, C.J. The complete genome sequence of Plodia interpunctella granulovirus: Evidence for horizontal gene transfer and discovery of an unusual Inhibitor-of-Apoptosis gene. PLoS ONE 2016, 11, e0160389. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.L.; Rowley, D.L.; Mowery, J.; Bauchan, G.R.; Theilmann, D.A.; Rohrmann, G.F.; Erlandson, M.A. The Complete Genome Sequence of a Second Distinct Betabaculovirus from the True Armyworm, Mythimna unipuncta. PLoS ONE 2017, 12, e0170510. [Google Scholar] [CrossRef]
- Gueli Alletti, G.; Eigenbrod, M.; Carstens, E.B.; Kleespies, R.G.; Jehle, J.A. The genome sequence of Agrotis segetum granulovirus, isolate AgseGV-DA, reveals a new Betabaculovirus species of a slow killing granulovirus. J. Invertebr. Pathol. 2017, 146, 58–68. [Google Scholar] [CrossRef]
- Kokusho, R.; Koh, Y.; Fujimoto, M.; Shimada, T.; Katsuma, S. Bombyx mori nucleopolyhedrovirus BM5 protein regulates progeny virus production and viral gene expression. Virology 2016, 498, 240–249. [Google Scholar] [CrossRef]
- Nagamine, T.; Inaba, T.; Sako, Y. A nuclear envelop-associated baculovirus protein promotes intranuclear lipid accumulation during infection. Virology 2019, 532, 108–117. [Google Scholar] [CrossRef]
- Chen, X.G.; Yang, X.Q.; Lei, C.F.; Qin, F.J.; Sun, X.L.; Hu, J. Autographa californica multiple nucleopolyhedrovirus is required for efficient nuclear egress of nucleocapsids. Virol. Sin. 2021, 36, 968–980. [Google Scholar] [CrossRef]
- Slack, J.M.; Kuzio, J.; Faulkner, P. Characterization of v-cath, a cathepsin L-like proteinase expressed by the baculovirus Autographa californica multiple nuclear polyhedrosis virus. J. Gen. Virol. 1995, 76, 1091–1098. [Google Scholar] [CrossRef]
- Hawtin, R.E.; Zarkowska, T.; Arnold, K.; Thomas, C.J.; Gooday, G.W.; King, L.A.; Kuzio, J.A.; Possee, R.D. Liquefaction of Autographa californica nucleopolyhedrovirus-infected insects is dependent on the integrity of virus-encoded chitinase and cathepsin genes. Virology 1997, 238, 243–253. [Google Scholar] [CrossRef]
- Oliveira, J.V.; Wolff, J.L.; Garcia-Maruniak, A.; Ribeiro, B.M.; de Castro, M.E.; de Souza, M.L.; Moscardi, F.; Maruniak, J.E.; Zanotto, P.M. Genome of the most widely used viral biopesticide: Anticarsia gemmatalis multiple nucleopolyhedrovirus. J. Gen. Virol. 2006, 87, 3233–3250. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Zhang, Y.; Wu, Y.; Sun, P.; Zhu, S.; Guo, X.; Gao, K.; Xu, A.; Wang, W. Analysis of the genomic sequence of Philosamia cynthia nucleopolyhedrin virus and comparison with Antheraea pernyi nucleopolyhedrin virus. BMC Genom. 2013, 14, 115. [Google Scholar] [CrossRef] [PubMed]
- Aragao-Silva, C.W.; Andrade, M.S.; Ardisson-Araujo, D.M.; Fernandes, J.E.; Morgado, F.S.; Bao, S.N.; Moraes, R.H.; Wolff, J.L.; Melo, F.L.; Ribeiro, B.M. The complete genome of a baculovirus isolated from an insect of medical interest: Lonomia obliqua (Lepidoptera: Saturniidae). Sci. Rep. 2016, 6, 23127. [Google Scholar] [CrossRef] [PubMed]
- Castro, M.E.B.; Melo, F.L.; Tagliari, M.; Inglis, P.W.; Craveiro, S.R.; Ribeiro, Z.M.A.; Ribeiro, B.M.; Bao, S.N. The genome sequence of Condylorrhiza vestigialis NPV, a novel baculovirus for the control of the Alamo moth on Populus spp. in Brazil. J. Invertebr. Pathol. 2017, 148, 152–161. [Google Scholar] [CrossRef]
- Nakai, M.; Goto, C.; Kang, W.; Shikata, M.; Luque, T.; Kunimi, Y. Genome sequence and organization of a nucleopolyhedrovirus isolated from the smaller tea tortrix, Adoxophyes honmai. Virology 2003, 316, 171–183. [Google Scholar] [CrossRef]


| Isolate | Size (bp) | %GC | Annotated ORFs | Repeated Regions | GenBank Accession No. |
|---|---|---|---|---|---|
| PlidNPV-2680 | 121,881 | 39.22% | 128 | 10 hrs | OQ658191 |
| PlidGV-2683 | 106,633 | 31.37% | 125 | 5 drs | PP449363 |
| Isolate | ORF | Nt Position | Size (aa) | BLAST/HHpred Query Results |
|---|---|---|---|---|
| PlidNPV-2680 | 7 | 7197 → 8261 | 354 | - |
| 28 | 30,507 ← 31,052 | 181 | - | |
| 36 | 36,833 → 37,321 | 162 | - | |
| 101 | 99,237 → 99,662 | 141 | - | |
| 108 | 102,642 → 103,100 | 152 | - | |
| 121 | 114,940 ← 115,479 | 179 | Baculovirus U-Box/RING-like domain, residues 122–167; HHpred probability 97.3%. | |
| PlidGV-2683 | 9 | 5428 → 5964 | 178 | Baculovirus U-Box/RING-like domain residues 121–166; HHpred probability 97.51%. |
| 17 | 11,791 ← 12,639 | 282 | RING finger domains in baculovirus CG30, IE2, PE38, residues 3–58, top HHpred probability 99.2% | |
| 18 | 13,005 ← 14,015 | 336 | As for ORF17. | |
| 21 | 17,812 → 17,979 | 55 | - | |
| 30 | 27,087 ← 27,341 | 84 | - | |
| 35 | 28,907 ← 29,260 | 117 | Zinc finger domains in reovirus outer capsid protein and AcMNPV CG30, HHpred probabilities 94.5–94.9%. | |
| 76 | 59,983 → 60,270 | 95 | - | |
| 92 | 75,968 → 76,708 | 246 | baculoviral IAP repeat-containing protein 3 isoform X2 [Nymphalis io], 49.9% identity, e = 3.9 × 10−100. | |
| 110 | 93,221 ← 93,871 | 216 | Putative F-box proteins, residues 1–35, top HHpred probability 92.9% with putative F-box/LRR-repeat protein R542 from Acanthamoeba polyphaga mimivirus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harrison, R.L.; Jansen, M.A.; Fife, A.N.; Rowley, D.L. The Genome Sequences of Baculoviruses from the Tufted Apple Bud Moth, Platynota idaeusalis, Reveal Recombination Between an Alphabaculovirus and a Betabaculovirus from the Same Host. Viruses 2025, 17, 202. https://doi.org/10.3390/v17020202
Harrison RL, Jansen MA, Fife AN, Rowley DL. The Genome Sequences of Baculoviruses from the Tufted Apple Bud Moth, Platynota idaeusalis, Reveal Recombination Between an Alphabaculovirus and a Betabaculovirus from the Same Host. Viruses. 2025; 17(2):202. https://doi.org/10.3390/v17020202
Chicago/Turabian StyleHarrison, Robert L., Michael A. Jansen, Austin N. Fife, and Daniel L. Rowley. 2025. "The Genome Sequences of Baculoviruses from the Tufted Apple Bud Moth, Platynota idaeusalis, Reveal Recombination Between an Alphabaculovirus and a Betabaculovirus from the Same Host" Viruses 17, no. 2: 202. https://doi.org/10.3390/v17020202
APA StyleHarrison, R. L., Jansen, M. A., Fife, A. N., & Rowley, D. L. (2025). The Genome Sequences of Baculoviruses from the Tufted Apple Bud Moth, Platynota idaeusalis, Reveal Recombination Between an Alphabaculovirus and a Betabaculovirus from the Same Host. Viruses, 17(2), 202. https://doi.org/10.3390/v17020202






