Phage Endolysins as Promising and Effective Candidates for Use Against Uropathogenic Escherichia coli
Abstract
1. Introduction
2. In Silico Analysis of Anti-UPEC Phage Proteins Annotated as Endolysins
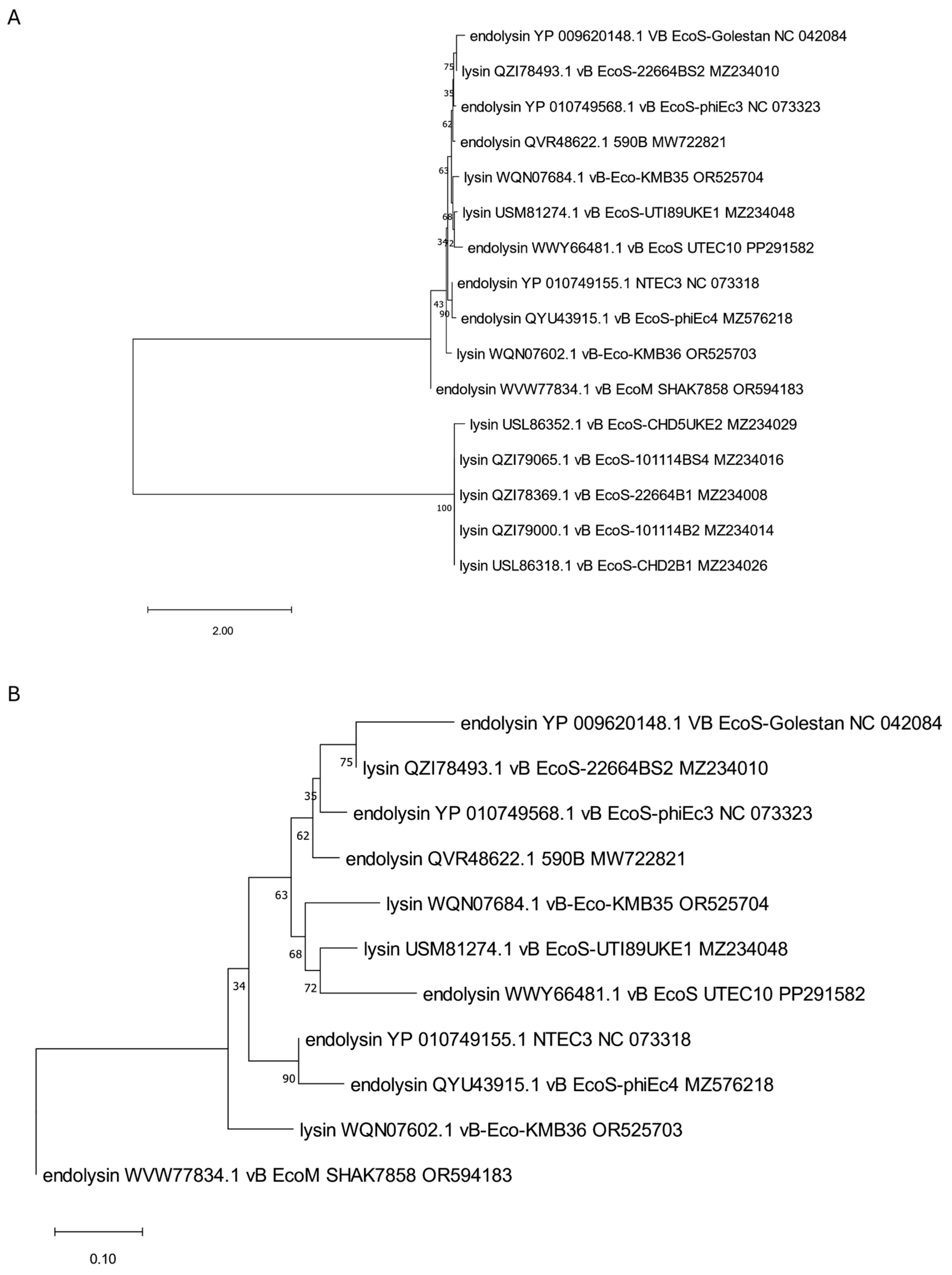
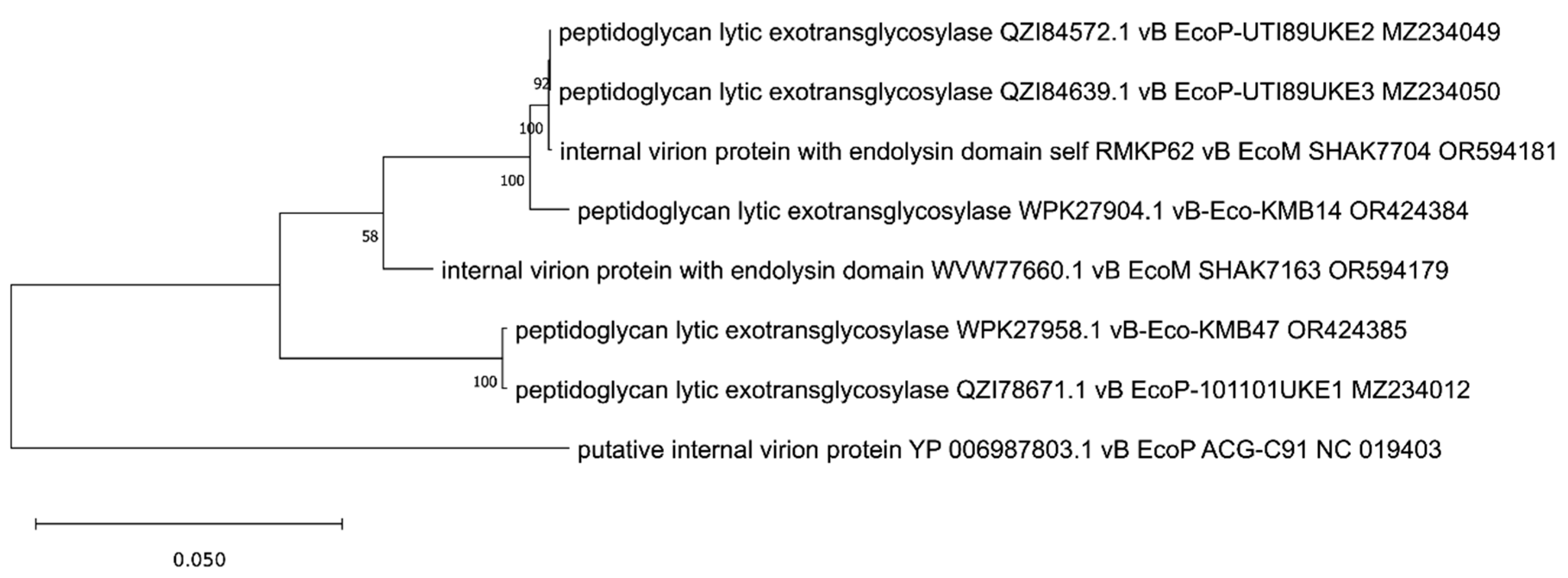
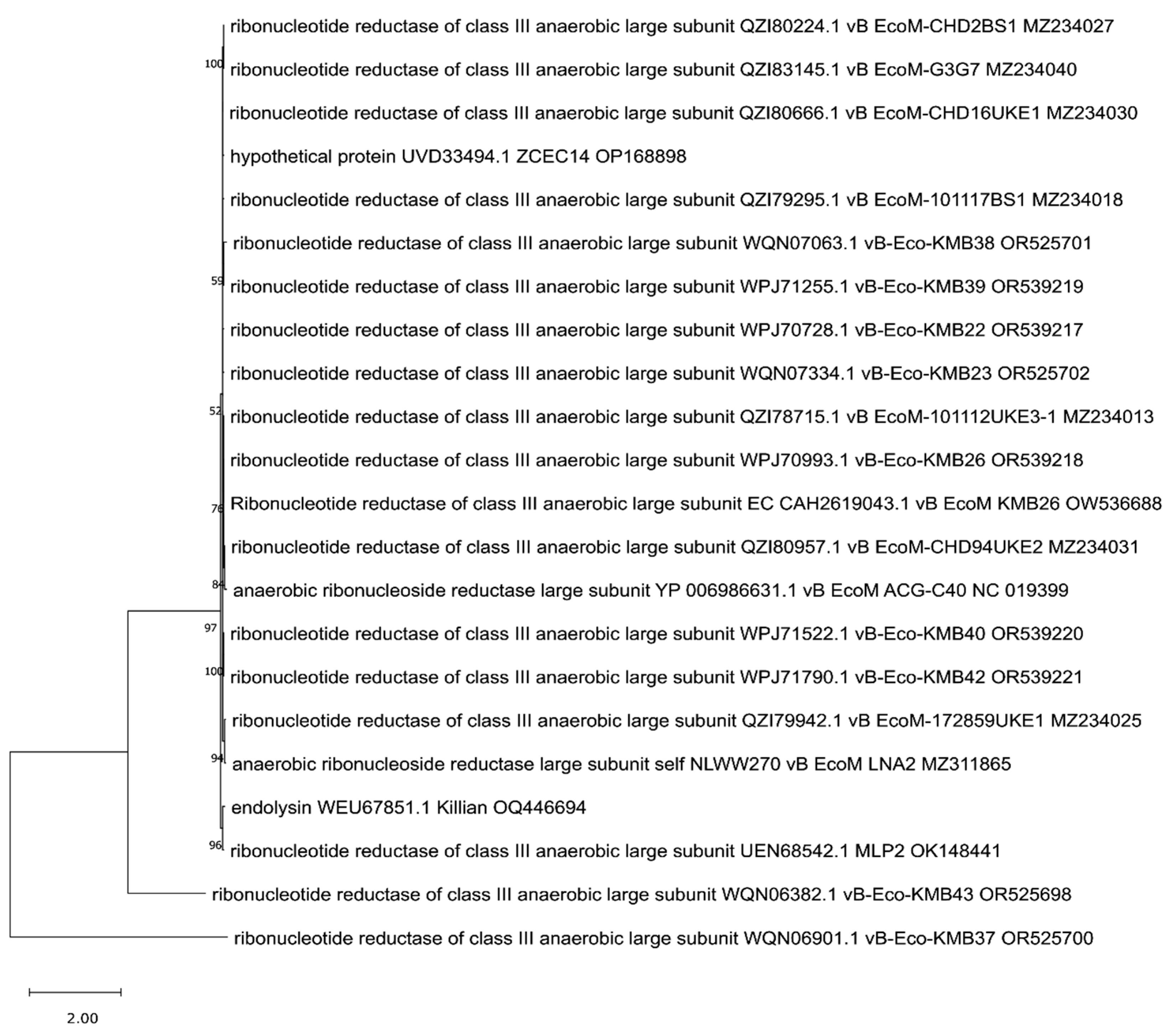
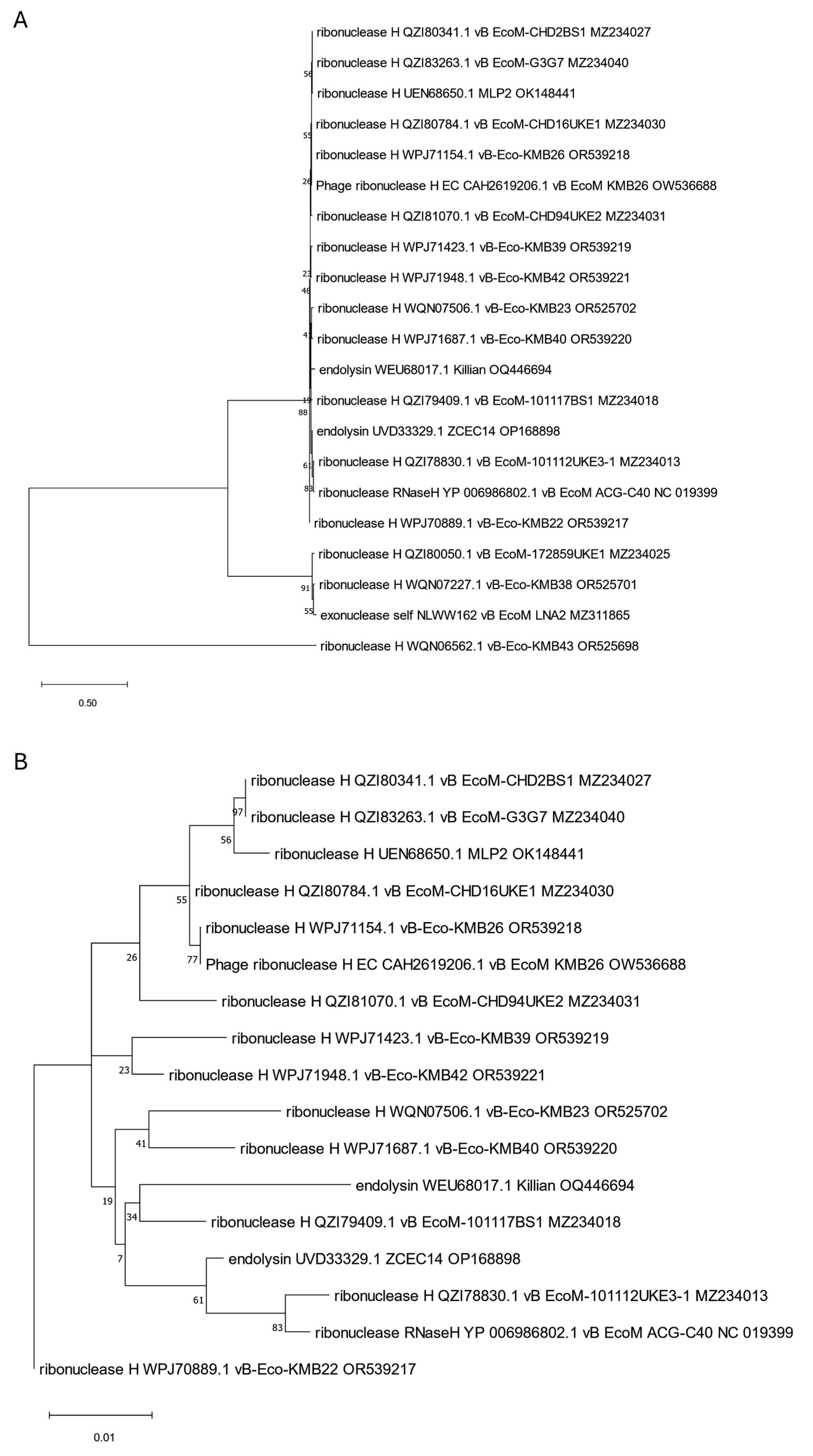
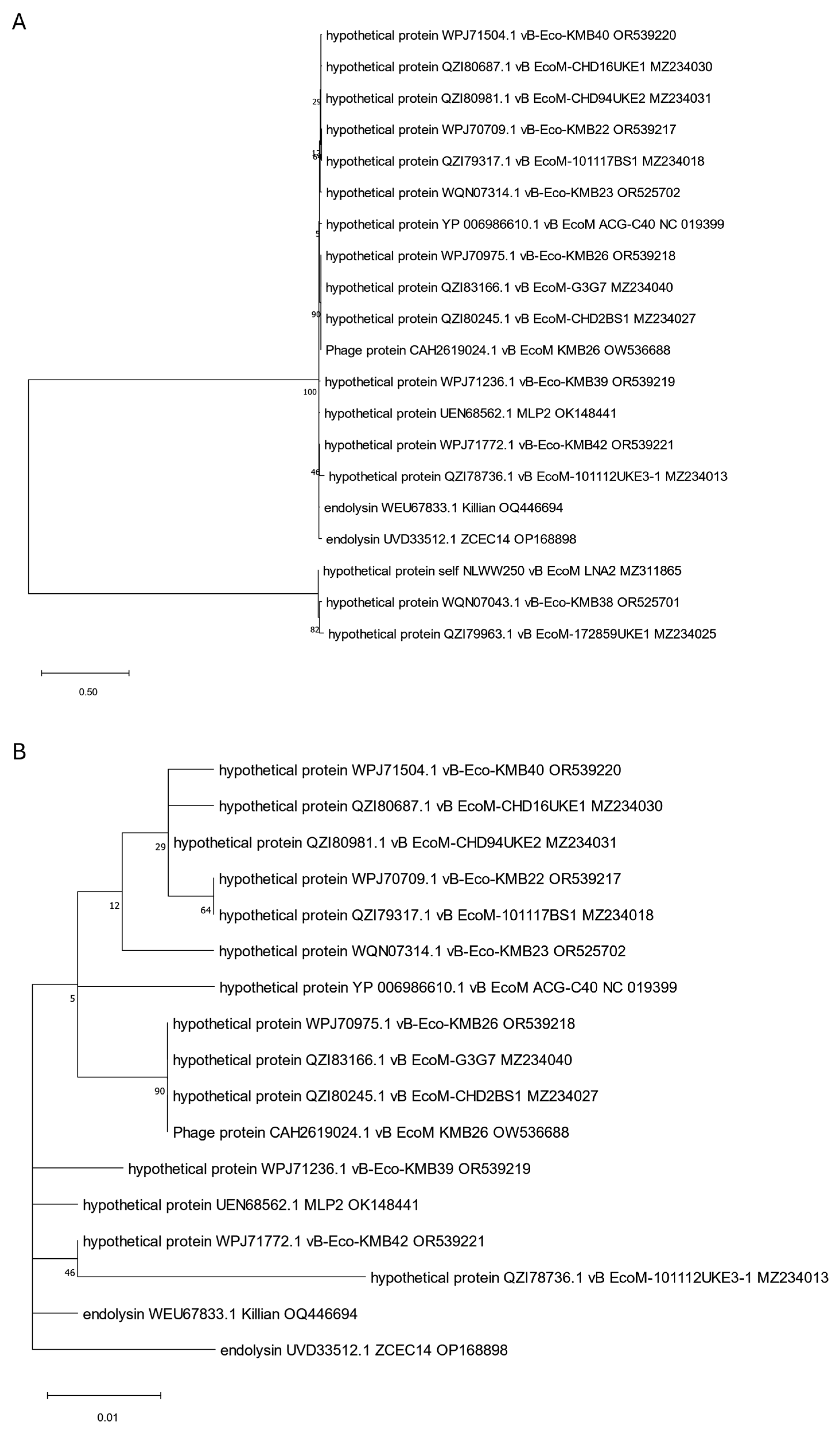
3. Phylogenetic Analysis of Putative Endolysins Annotated in Genomes of Anti-UPEC Phages
4. In Vitro and In Vivo Testing of Endolysin Properties, Including Their Capability to Eradicate Biofilms
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gondil, V.S.; Chhibber, S. Bacteriophage and Endolysin Encapsulation Systems: A Promising Strategy to Improve Therapeutic Outcomes. Front. Pharmacol. 2021, 12, 675440. [Google Scholar] [CrossRef]
- Abdelrahman, F.; Easwaran, M.; Daramola, O.I.; Ragab, S.; Lynch, S.; Oduselu, T.J.; Khan, F.M.; Ayobami, A.; Adnan, F.; Torrents, E.; et al. Phage-Encoded Endolysins. Antibiotics 2021, 10, 124. [Google Scholar] [CrossRef]
- Principi, N.; Silvestri, E.; Esposito, S. Advantages and Limitations of Bacteriophages for the Treatment of Bacterial Infections. Front. Pharmacol. 2019, 10, 513. [Google Scholar] [CrossRef] [PubMed]
- Fischetti, V.A. Development of Phage Lysins as Novel Therapeutics: A Historical Perspective. Viruses 2018, 10, 310. [Google Scholar] [CrossRef] [PubMed]
- Theuretzbacher, U.; Piddock, L.J.V. Non-Traditional Antibacterial Therapeutic Options and Challenges. Cell Host Microbe 2019, 26, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Love, M.J.; Abeysekera, G.S.; Muscroft-Taylor, A.C.; Billington, C.; Dobson, R.C.J. On the Catalytic Mechanism of Bacteriophage Endolysins: Opportunities for Engineering. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140302. [Google Scholar] [CrossRef]
- Li, X.; Zhang, C.; Wei, F.; Yu, F.; Zhao, Z. Bactericidal Activity of a Holin-Endolysin System Derived from Vibrio Alginolyticus Phage HH109. Microb. Pathog. 2021, 159, 105135. [Google Scholar] [CrossRef]
- Nazir, A.; Xu, X.; Liu, Y.; Chen, Y. Phage Endolysins: Advances in the World of Food Safety. Cells 2023, 12, 2169. [Google Scholar] [CrossRef]
- Khan, F.M.; Chen, J.-H.; Zhang, R.; Liu, B. A Comprehensive Review of the Applications of Bacteriophage-Derived Endolysins for Foodborne Bacterial Pathogens and Food Safety: Recent Advances, Challenges, and Future Perspective. Front. Microbiol. 2023, 14, 1259210. [Google Scholar] [CrossRef]
- Oliveira, H.; São-José, C.; Azeredo, J. Phage-Derived Peptidoglycan Degrading Enzymes: Challenges and Future Prospects for In Vivo Therapy. Viruses 2018, 10, 292. [Google Scholar] [CrossRef]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage Endolysins as Novel Antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [PubMed]
- Microbial Products for Health, Environment and Agriculture. Available online: https://www.researchgate.net/publication/354735414_Microbial_Products_for_Health_Environment_and_Agriculture (accessed on 26 February 2025).
- Liu, H.; Hu, Z.; Li, M.; Yang, Y.; Lu, S.; Rao, X. Therapeutic Potential of Bacteriophage Endolysins for Infections Caused by Gram-Positive Bacteria. J. Biomed. Sci. 2023, 30, 29. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.C.B.; Chen, X.; Ho, M.K.Y.; Xia, J.; Leung, S.S.Y. Bacteriophage-Derived Endolysins to Target Gram-Negative Bacteria. Int. J. Pharm. 2020, 589, 119833. [Google Scholar] [CrossRef]
- Oechslin, F.; Zhu, X.; Dion, M.B.; Shi, R.; Moineau, S. Phage Endolysins Are Adapted to Specific Hosts and Are Evolutionarily Dynamic. PLoS Biol. 2022, 20, e3001740. [Google Scholar] [CrossRef]
- Haddad Kashani, H.; Schmelcher, M.; Sabzalipoor, H.; Seyed Hosseini, E.; Moniri, R. Recombinant Endolysins as Potential Therapeutics against Antibiotic-Resistant Staphylococcus Aureus: Current Status of Research and Novel Delivery Strategies. Clin. Microbiol. Rev. 2018, 31, e00071-17. [Google Scholar] [CrossRef]
- Rahman, M.U.; Wang, W.; Sun, Q.; Shah, J.A.; Li, C.; Sun, Y.; Li, Y.; Zhang, B.; Chen, W.; Wang, S. Endolysin, a Promising Solution against Antimicrobial Resistance. Antibiotics 2021, 10, 1277. [Google Scholar] [CrossRef]
- São-José, C. Engineering of Phage-Derived Lytic Enzymes: Improving Their Potential as Antimicrobials. Antibiotics 2018, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Ghose, C.; Euler, C.W. Gram-Negative Bacterial Lysins. Antibiotics 2020, 9, 74. [Google Scholar] [CrossRef]
- Gontijo, M.T.P.; Jorge, G.P.; Brocchi, M. Current Status of Endolysin-Based Treatments against Gram-Negative Bacteria. Antibiotics 2021, 10, 1143. [Google Scholar] [CrossRef]
- Gontijo, M.T.P.; Vidigal, P.M.P.; Lopez, M.E.S.; Brocchi, M. Bacteriophages That Infect Gram-Negative Bacteria as Source of Signal-Arrest-Release Motif Lysins. Res. Microbiol. 2021, 172, 103794. [Google Scholar] [CrossRef]
- Zalewska-Piątek, B.; Piątek, R. Phage Therapy as a Novel Strategy in the Treatment of Urinary Tract Infections Caused by E. coli. Antibiotics 2020, 9, 304. [Google Scholar] [CrossRef]
- Melican, K.; Sandoval, R.M.; Kader, A.; Josefsson, L.; Tanner, G.A.; Molitoris, B.A.; Richter-Dahlfors, A. Uropathogenic Escherichia Coli P and Type 1 Fimbriae Act in Synergy in a Living Host to Facilitate Renal Colonization Leading to Nephron Obstruction. PLoS Pathog. 2011, 7, e1001298. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wen, Y. The Role of Bacterial Biofilm in Persistent Infections and Control Strategies. Int. J. Oral Sci. 2011, 3, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Lila, A.S.A.; Rajab, A.A.H.; Abdallah, M.H.; Rizvi, S.M.D.; Moin, A.; Khafagy, E.-S.; Tabrez, S.; Hegazy, W.A.H. Biofilm Lifestyle in Recurrent Urinary Tract Infections. Life 2023, 13, 148. [Google Scholar] [CrossRef]
- Necel, A.; Bloch, S.; Topka-Bielecka, G.; Janiszewska, A.; Łukasiak, A.; Nejman-Faleńczyk, B.; Węgrzyn, G. Synergistic Effects of Bacteriophage vB_Eco4-M7 and Selected Antibiotics on the Biofilm Formed by Shiga Toxin-Producing Escherichia coli. Antibiotics 2022, 11, 712. [Google Scholar] [CrossRef]
- Ngiam, L.; Schembri, M.A.; Weynberg, K.; Guo, J. Bacteriophage Isolated from Non-Target Bacteria Demonstrates Broad Host Range Infectivity against Multidrug-Resistant Bacteria. Environ. Microbiol. 2021, 23, 5569–5586. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, N.; Maurya, R.K.; Singh, D.; Mohan, B.; Taneja, N. Genome Analysis and Antibiofilm Activity of Phage 590B against Multidrug-Resistant and Extensively Drug-Resistant Uropathogenic Escherichia coli Isolates, India. Pathogens 2022, 11, 1448. [Google Scholar] [CrossRef]
- Rajab, A.A.H.; Fahmy, E.K.; Esmaeel, S.E.; Yousef, N.; Askoura, M. In Vitro and in Vivo Assessment of the Competence of a Novel Lytic Phage vB_EcoS_UTEC10 Targeting Multidrug Resistant Escherichia coli with a Robust Biofilm Eradication Activity. Microb. Pathog. 2024, 197, 107058. [Google Scholar] [CrossRef]
- González-Villalobos, E.; Ribas-Aparicio, R.M.; Montealegre, G.E.R.; Belmont-Monroy, L.; Ortega-García, Y.; Aparicio-Ozores, G.; Balcázar, J.L.; Eslava-Campos, C.A.; Hernández-Chiñas, U.; Molina-López, J. Isolation and Characterization of Novel Bacteriophages as a Potential Therapeutic Option for Escherichia coli Urinary Tract Infections. Appl. Microbiol. Biotechnol. 2021, 105, 5617–5629. [Google Scholar] [CrossRef]
- Chaudhary, N.; Singh, D.; Maurya, R.K.; Mohan, B.; Mavuduru, R.S.; Taneja, N. Whole Genome Sequencing and in Vitro Activity Data of Escherichia Phage NTEC3 against Multidrug-Resistant Uropathogenic and Extensively Drug-Resistant Uropathogenic E. coli Isolates. Data Brief 2022, 43, 108479. [Google Scholar] [CrossRef]
- Yazdi, M.; Bouzari, M.; Ghaemi, E.A.; Shahin, K. Isolation, Characterization and Genomic Analysis of a Novel Bacteriophage VB_EcoS-Golestan Infecting Multidrug-Resistant Escherichia coli Isolated from Urinary Tract Infection. Sci. Rep. 2020, 10, 7690. [Google Scholar] [CrossRef] [PubMed]
- Asgharzadeh Kangachar, S.; Logel, D.Y.; Trofimova, E.; Zhu, H.X.; Zaugg, J.; Schembri, M.A.; Weynberg, K.D.; Jaschke, P.R. Discovery and Characterisation of New Phage Targeting Uropathogenic Escherichia coli. Virology 2024, 597, 110148. [Google Scholar] [CrossRef]
- Chibeu, A.; Lingohr, E.J.; Masson, L.; Manges, A.; Harel, J.; Ackermann, H.-W.; Kropinski, A.M.; Boerlin, P. Bacteriophages with the Ability to Degrade Uropathogenic Escherichia coli Biofilms. Viruses 2012, 4, 471–487. [Google Scholar] [CrossRef]
- Khunti, P.; Chantakorn, K.; Tantibhadrasapa, A.; Htoo, H.H.; Thiennimitr, P.; Nonejuie, P.; Chaikeeratisak, V.A. Novel Coli Myophage and Antibiotics Synergistically Inhibit the Growth of the Uropathogenic E. coli Strain CFT073 in Stoichiometric Niches. Microbiol. Spectr. 2023, 11, e00889-23. [Google Scholar] [CrossRef]
- Loose, M.; Sáez Moreno, D.; Mutti, M.; Hitzenhammer, E.; Visram, Z.; Dippel, D.; Schertler, S.; Tišáková, L.P.; Wittmann, J.; Corsini, L.; et al. Natural Bred Ε2-Phages Have an Improved Host Range and Virulence against Uropathogenic Escherichia coli over Their Ancestor Phages. Antibiotics 2021, 10, 1337. [Google Scholar] [CrossRef]
- Markusková, B.; Elnwrani, S.; Andrezál, M.; Sedláčková, T.; Szemes, T.; Slobodníková, L.; Kajsik, M.; Drahovská, H. Characterization of Bacteriophages Infecting Multidrug-Resistant Uropathogenic Escherichia coli Strains. Arch. Virol. 2024, 169, 142. [Google Scholar] [CrossRef] [PubMed]
- Vera-Mansilla, J.; Sánchez, P.; Silva-Valenzuela, C.A.; Molina-Quiroz, R.C. Isolation and Characterization of Novel Lytic Phages Infecting Multidrug-Resistant Escherichia coli. Microbiol. Spectr. 2022, 10, e01678-21. [Google Scholar] [CrossRef]
- Gu, Y.; Xu, Y.; Xu, J.; Yu, X.; Huang, X.; Liu, G.; Liu, X. Identification of Novel Bacteriophage vB_EcoP-EG1 with Lytic Activity against Planktonic and Biofilm Forms of Uropathogenic Escherichia coli. Appl. Microbiol. Biotechnol. 2019, 103, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Ismael, N.M.; Azzam, M.; Abdelmoteleb, M.; El-Shibiny, A. Phage vB_Ec_ZCEC14 to Treat Antibiotic-Resistant Escherichia Coli Isolated from Urinary Tract Infections. Virol. J. 2024, 21, 44. [Google Scholar] [CrossRef]
- Slobodníková, L.; Markusková, B.; Kajsík, M.; Andrezál, M.; Straka, M.; Liptáková, A.; Drahovská, H. Characterization of Anti-Bacterial Effect of the Two New Phages against Uropathogenic Escherichia coli. Viruses 2021, 13, 1348. [Google Scholar] [CrossRef]
- Bouras, G.; Nepal, R.; Houtak, G.; Psaltis, A.J.; Wormald, P.-J.; Vreugde, S. Pharokka: A Fast Scalable Bacteriophage Annotation Tool. Bioinformatics 2023, 39, btac776. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely Available Python Tools for Computational Molecular Biology and Bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Jung, F.; Frey, K.; Zimmer, D.; Mühlhaus, T. DeepSTABp: A Deep Learning Approach for the Prediction of Thermal Protein Stability. Int. J. Mol. Sci. 2023, 24, 7444. [Google Scholar] [CrossRef] [PubMed]
- Ikai, A. Thermostability and Aliphatic Index of Globular Proteins. J. Biochem. 1980, 88, 1895–1898. [Google Scholar] [PubMed]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-Scale Protein Function Classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro Protein Families and Domains Database: 20 Years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Murray, E.; Draper, L.A.; Ross, R.P.; Hill, C. The Advantages and Challenges of Using Endolysins in a Clinical Setting. Viruses 2021, 13, 680. [Google Scholar] [CrossRef]
- Fenton, M.; Ross, P.; McAuliffe, O.; O’Mahony, J.; Coffey, A. Recombinant Bacteriophage Lysins as Antibacterials. Bioeng. Bugs 2010, 1, 9–16. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional Classification of Proteins via Subfamily Domain Architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The Conserved Domain Database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Turnau, K.; Fiałkowska, E.; Ważny, R.; Rozpądek, P.; Tylko, G.; Bloch, S.; Nejman-Faleńczyk, B.; Grabski, M.; Węgrzyn, A.; Węgrzyn, G. Extraordinary Multi-Organismal Interactions Involving Bacteriophages, Bacteria, Fungi, and Rotifers: Quadruple Microbial Trophic Network in Water Droplets. Int. J. Mol. Sci. 2021, 22, 2178. [Google Scholar] [CrossRef]
- Altenhoff, A.M.; Levy, J.; Zarowiecki, M.; Tomiczek, B.; Warwick Vesztrocy, A.; Dalquen, D.A.; Müller, S.; Telford, M.J.; Glover, N.M.; Dylus, D.; et al. OMA Standalone: Orthology Inference among Public and Custom Genomes and Transcriptomes. Genome Res. 2019, 29, 1152–1163. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; Martínez, B.; Donovan, D.M.; Rodríguez, A.; García, P. Bacteriophage virion-associated peptidoglycan hydrolases: Potential new enzybiotics. Crit. Rev. Microbiol. 2013, 39, 427–434. [Google Scholar] [CrossRef]
- Zhao, A.; Sun, J.; Liu, Y. Understanding Bacterial Biofilms: From Definition to Treatment Strategies. Front. Cell. Infect. Microbiol. 2023, 13, 1137947. [Google Scholar] [CrossRef] [PubMed]
- Gondil, V.S.; Harjai, K.; Chhibber, S. Endolysins as Emerging Alternative Therapeutic Agents to Counter Drug-Resistant Infections. Int. J. Antimicrob. Agents 2020, 55, 105844. [Google Scholar] [CrossRef]
- Soontarach, R.; Srimanote, P.; Voravuthikunchai, S.P.; Chusri, S. Antibacterial and Anti-Biofilm Efficacy of Endolysin LysAB1245 against a Panel of Important Pathogens. Pharmaceuticals 2024, 17, 155. [Google Scholar] [CrossRef]
- Carratalá, J.V.; Ferrer-Miralles, N.; Garcia-Fruitós, E.; Arís, A. LysJEP8: A Promising Novel Endolysin for Combating Multidrug-Resistant Gram-Negative Bacteria. Microb. Biotechnol. 2024, 17, e14483. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Feng, C.; Ren, J.; Zhuang, X.; Zhang, Y.; Zhu, Y.; Dong, K.; He, P.; Guo, X.; Qin, J. A Novel Antimicrobial Endolysin, LysPA26, against Pseudomonas Aeruginosa. Front. Microbiol. 2017, 8, 293. [Google Scholar] [CrossRef]
- Vasina, D.V.; Antonova, N.P.; Grigoriev, I.V.; Yakimakha, V.S.; Lendel, A.M.; Nikiforova, M.A.; Pochtovyi, A.A.; Remizov, T.A.; Usachev, E.V.; Shevlyagina, N.V.; et al. Discovering the Potentials of Four Phage Endolysins to Combat Gram-Negative Infections. Front. Microbiol. 2021, 12, 748718. [Google Scholar] [CrossRef] [PubMed]
- Kajsikova, M.; Kajsik, M.; Bocanova, L.; Papayova, K.; Drahovska, H.; Bukovska, G. Endolysin EN572-5 as an Alternative to Treat Urinary Tract Infection Caused by Streptococcus Agalactiae. Appl. Microbiol. Biotechnol. 2024, 108, 79. [Google Scholar] [CrossRef]
- Fursov, M.V.; Abdrakhmanova, R.O.; Antonova, N.P.; Vasina, D.V.; Kolchanova, A.D.; Bashkina, O.A.; Rubalsky, O.V.; Samotrueva, M.A.; Potapov, V.D.; Makarov, V.V.; et al. Antibiofilm Activity of a Broad-Range Recombinant Endolysin LysECD7: In Vitro and In Vivo Study. Viruses 2020, 12, 545. [Google Scholar] [CrossRef]
- Euler, C.W.; Raz, A.; Hernandez, A.; Serrano, A.; Xu, S.; Andersson, M.; Zou, G.; Zhang, Y.; Fischetti, V.A.; Li, J. PlyKp104, a Novel Phage Lysin for the Treatment of Klebsiella Pneumoniae, Pseudomonas Aeruginosa, and Other Gram-Negative ESKAPE Pathogens. Antimicrob. Agents Chemother. 2023, 67, e0151922. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Yang, R.; Fan, K.; Dong, H.; Gao, C.; Wang, S.; Yu, L.; Cheng, Z.; Lei, L. External Lysis of Escherichia coli by a Bacteriophage Endolysin Modified with Hydrophobic Amino Acids. AMB Express 2019, 9, 106. [Google Scholar] [CrossRef]
- Hasan, M.; Kim, J.; Liao, X.; Ding, T.; Ahn, J. Antibacterial Activity of Bacteriophage-Encoded Endolysins against Planktonic and Biofilm Cells of Pathogenic Escherichia coli. Microb. Pathog. 2024, 193, 106780. [Google Scholar] [CrossRef]
- Park, D.-W.; Park, J.-H. Characterization of Endolysin LysECP26 Derived from rV5-like Phage vB_EcoM-ECP26 for Inactivation of Escherichia coli O157:H7. J. Microbiol. Biotechnol. 2020, 30, 1552–1558. [Google Scholar] [CrossRef]
- Oh, M.; Cevallos-Urena, A.; Kim, B.S. Bacteriophages PECP14, PECP20, and Their Endolysins as Effective Biocontrol Agents for Escherichia coli O157:H7 and Other Foodborne Pathogens. Int. J. Food Microbiol. 2024, 409, 110460. [Google Scholar] [CrossRef]
- Chu, D.; Lan, J.; Liang, L.; Xia, K.; Li, L.; Yang, L.; Liu, H.; Zhang, T. The Antibacterial Activity of a Novel Highly Thermostable Endolysin, LysKP213, against Gram-Negative Pathogens Is Enhanced When Combined with Outer Membrane Permeabilizing Agents. Front. Microbiol. 2024, 15, 1454618. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.J.; Jo, J.; Kim, E.; Yoon, H.; Hong, H.; Kim, M.S.; Myung, H. Motility Increase of Adherent Invasive Escherichia coli (AIEC) Induced by a Sub-Inhibitory Concentration of Recombinant Endolysin LysPA90. Front. Microbiol. 2022, 13, 1093670. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Gu, J.; Lv, M.; Guo, Z.; Yan, G.; Yu, L.; Du, C.; Feng, X.; Han, W.; Sun, C. The antibacterial activity of E. coli bacteriophage lysin lysep3 is enhanced by fusing the Bacillus amyloliquefaciens bacteriophage endolysin binding domain D8 to the C-terminal region. J. Microbiol. 2017, 55, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Sisson, H.M.; Jackson, S.A.; Fagerlund, R.D.; Warring, S.L.; Fineran, P.C. Gram-negative endolysins: Overcoming the outer membrane obstacle. Curr. Opin. Microbiol. 2024, 78, 102433. [Google Scholar] [CrossRef]
- Kim, J.; Wang, J.; Ahn, J. Combined antimicrobial effect of phage-derived endolysin and depolymerase against biofilm-forming Salmonella Typhimurium. Biofouling 2023, 39, 763–774. [Google Scholar] [CrossRef]






| Phage name [References] | Protein Name | Position of the Gene in Virus Genome | Gene Length (nt) | Protein Accession Number | Protein Length (aa) | Molecular Weight (kDa) | Instability Index | Isoelectric Point (pI) | Gravy | Aliphatic Index | Melting Point (Tm) | Name of the Predicted Domain * | Position of the Domain in Protein Sequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| vB_EcoM_LNA1 [27] | endolysin | 77244–77708 | 464 | self_ZKRH133 | 154 | 17.21 | 33.482 | 9.36 | −0.381 | 81.104 | 47.0 °C | lyz_endolysin_autolysin (cd00737) Phage lysozyme (PF00959) Lysozyme-like (SSF53955) Endolysin (MF_04110) | 7–148 13–143 1–152 1–150 |
| 590B [28] | endolysin | 39903–40391 | 488 | QVR48622.1 | 162 | 17.11 | 20.515 | 9.86 | −0.125 | 87.531 | 51.0 °C | lyz_endolysin_autolysin (cd00737) Phage lysozyme (PF00959) Lysozyme-like (SSF53955) | 9–146 15–138 3–149 |
| vB_EcoS_UTEC10 [29] | endolysin | 1847–2332 | 485 | WWY66481.1 | 161 | 17.03 | 20.046 | 9.81 | −0.12 | 88.696 | 50.0 °C | lyz_endolysin_autolysin (cd00737) Phage lysozyme (PF00959) Lysozyme-like (SSF53955) | 9–146 15–138 3–149 |
| vB_EcoS-phiEc3 [30] | endolysin | 41533–42021 | 488 | YP_010749568.1 | 162 | 17.05 | 19.526 | 9.81 | −0.101 | 88.148 | 51.0 °C | lyz_endolysin_autolysin (cd00737) Phage lysozyme (PF00959) Lysozyme-like (SSF53955) | 9–146 15–138 3–149 |
| NTEC3 [31] | endolysin | 11540–12028 | 488 | YP_010749155.1 | 162 | 17.14 | 21.851 | 9.86 | −0.137 | 87.531 | 50.0 °C | lyz_endolysin_autolysin (cd00737) Phage lysozyme (PF00959) Lysozyme-like (SSF53955) | 9–146 15–138 3–149 |
| vB_EcoS-phiEc4 [30] | endolysin | 41414–41899 | 485 | QYU43915.1 | 161 | 17.11 | 22.631 | 9.92 | −0.139 | 88.075 | 50.0 °C | lyz_endolysin_autolysin (cd00737) Phage lysozyme (PF00959) Lysozyme-like (SSF53955) | 9–146 15–138 3–149 |
| VB_EcoS-Golestan [32] | endolysin | 41444–41902 | 458 | YP_009620148.1 | 152 | 16.09 | 17.289 | 9.86 | −0.069 | 88.816 | 49.0 °C | lyz_endolysin_autolysin (cd00737) Phage lysozyme (PF00959) Lysozyme-like (SSF53955) | 3–137 6–129 2–141 |
| vB_EcoM_SHAK7858 [33] | endolysin | 40989–41474 | 485 | WVW77834.1 | 161 | 17.11 | 19.781 | 9.62 | −0.141 | 84.472 | 50.0 °C | lyz_endolysin_autolysin (cd00737) Phage lysozyme (PF00959) Lysozyme-like (SSF53955) | 8–145 14–137 4–148 |
| vB_EcoS_ACG-M12 [34] | endolysin | 41386–41871 | 485 | YP_006987885.1 | 161 | 17.63 | 33.838 | 9.83 | −0.108 | 99.938 | 49.0 °C | endolysin_R21-like (cd16900) SAR-endolysin (MF_04136) Phage lysozyme (PF00959) Lysozyme-like (SSF53955) Endolysin (MF_04110) | 20–156 3–158 25–151 6–159 4–158 |
| Killian [35] | endolysin | 77692–78186 | 494 | WEU67899.1 | 164 | 18.59 | 37.155 | 9.59 | −0.37 | 88.049 | 53.0 °C | T4-like_lys (cd00735) Phage lysozyme (PF00959) Lysozyme-like (SSF53955) Endolysin (MF_04110) Endolysin (G3DSA:1.10.530.40:FF:000002) | 2–160 11–141 1–161 1–159 1–164 |
| vB_EcoM_SHAK7693 [33] | endolysin | 42651–42953 | 302 | WVW77754.1 | 100 | 10.36 | 21.694 | 10.21 | 0.061 | 96.9 | 49.0 °C | Phage lysozyme (PF00959) Lysozyme-like (SSF53955) | 2–76 2–86 |
| vB_EcoM_SHAK7693 [33] | endolysin | 42419–42658 | 239 | WVW77753.1 | 79 | 8.73 | 25.656 | 7.91 | −0.537 | 70.38 | 49.0 °C | Phage lysozyme (PF00959) Lysozyme-like (SSF53955) | 20–67 6–68 |
| vB_EcoP-101136BS1 [36] | endolysin | 19728–20186 | 458 | QZI79903.1 | 152 | 17.06 | 44.325 | 8.83 | −0.391 | 78.158 | 51.0 °C | PGRP (cd06583) N-acetylmuramoyl-L-alanine amidase-like (SSF55846) N-acetylmuramoyl-L-alanine amidase (PF01510) Endolysin (MF_04111) | 14–133 11–147 11–134 2–150 |
| vB_EcoP-101120B2 [36] | endolysin | 20895–21353 | 458 | QZI79844.1 | 152 | 17.1 | 44.325 | 8.47 | −0.46 | 76.25 | 51.0 °C | PGRP (cd06583) N-acetylmuramoyl-L-alanine amidase-like (SSF55846) N-acetylmuramoyl-L-alanine amidase (PF01510) Endolysin (MF_04111) | 14–133 11–147 11–134 2–150 |
| vB_EcoP-101120B1-2 [36] | endolysin | 20840–21298 | 458 | QZI79784.1 | 152 | 17.1 | 44.325 | 8.47 | −0.46 | 76.25 | 51.0 °C | PGRP (cd06583) N-acetylmuramoyl-L-alanine amidase-like (SSF55846) N-acetylmuramoyl-L-alanine amidase (PF01510) Endolysin (MF_04111) | 14–133 11–147 11–134 2–150 |
| vB_EcoP-101118UKE1 [36] | endolysin | 20127–20585 | 458 | QZI79721.1 | 152 | 17.04 | 44.76 | 7.79 | −0.38 | 78.158 | 51.0 °C | PGRP (cd06583) N-acetylmuramoyl-L-alanine amidase-like (SSF55846) N-acetylmuramoyl-L-alanine amidase (PF01510) Endolysin (MF_04111) | 14–133 11–147 11–134 2–150 |
| vB_EcoP-22664UKE3-2 [36] | endolysin | 21058–21516 | 458 | QZI78611.1 | 152 | 17.12 | 45.137 | 8.47 | −0.458 | 76.908 | 51.0 °C | PGRP (cd06583) N-acetylmuramoyl-L-alanine amidase-like (SSF55846) N-acetylmuramoyl-L-alanine amidase (PF01510) Endolysin (MF_04111) | 14–133 11–147 11–134 2–150 |
| vB_EcoP-22664BS1 [36] | endolysin | 19873–20226 | 353 | QZI78465.1 | 117 | 13.16 | 50.611 | 7.83 | −0.531 | 77.436 | 49.0 °C | PGRP (cd06583) N-acetylmuramoyl-L-alanine amidase-like (SSF55846) N-acetylmuramoyl-L-alanine amidase (PF01510) | 5–98 2–112 7–99 |
| vB_EcoP_ACG-C91 [34] | endolysin | 39340–39684 | 344 | YP_006987811.1 | 114 | 12.75 | 25.157 | 9.51 | −0.333 | 72.632 | 52.0 °C | Peptidase M15 (PF08291) Hedgehog/DD-peptidase (SSF55166) | 4–106 5–107 |
| vB_EcoM_SHAK7163 [33] | endolysin | 39749–40093 | 344 | WVW77668.1 | 114 | 12.59 | 29.711 | 9.32 | −0.348 | 71.754 | 50.0 °C | Peptidase M15 (PF08291) Hedgehog/DD-peptidase (SSF55166) | 5–106 5–107 |
| vB-EcoP-KMB14 [37] | endolysin | 38777–39121 | 344 | WPK27911.1 | 114 | 12.65 | 40.639 | 9.35 | −0.392 | 67.544 | 49.0 °C | Peptidase M15 (PF08291) Hedgehog/DD-peptidase (SSF55166) | 4–106 5–108 |
| vB_EcoM_SHAK7704 [33] | endolysin | 3327–3671 | 344 | self_RMKP5 | 114 | 12.65 | 25.787 | 9.32 | −0.401 | 70.088 | 51.0 °C | Peptidase M15 (PF08291) Hedgehog/DD-peptidase (SSF55166) | 5–106 5–107 |
| MLP1 [38] | endolysin | 32790–33185 | 395 | YP_010844467.1 | 131 | 14.96 | 47.821 | 8.79 | −0.303 | 84.122 | 57.0 °C | L-Ala-D-Glu_peptidase_like (cd14845) Hedgehog/DD-peptidase (SSF55166) D-alanyl-D-alanine carboxypeptidase (PF13539) | 9–130 4–129 58–110 |
| vB-EcoM-KMB43 [37] | endolysin | 55240–55629 | 389 | WQN06429.1 | 129 | 14.72 | 12.666 | 9.3 | −0.412 | 79.922 | 53.0 °C | L-Ala-D-Glu_peptidase_like (cd14845) Hedgehog/DD-peptidase (SSF55166) D-alanyl-D-alanine carboxypeptidase (PF13539) | 11–120 2–120 55–120 |
| vB_EcoP_EG1 [39] | internal virion protein with endolysin domain | 30249–34223 | 3974 | YP_009795842.1 | 1324 | 145.33 | 36.856 | 6.58 | −0.397 | 78.52 | 41.0 °C | Transglycosylase SLT domain (PF01464) Peptidoglycan transglycosylase gp16. (MF_04121) Lysozyme-like (SSF53955) LT-like (cd00254) | 15–127 2–1314 5–146 28–144 |
| vB_EcoM_LNA4 [27] | endolysin | 21617–22171 | 554 | self_PFXJ59 | 184 | 20.08 | 23.118 | 9.3 | −0.335 | 88.098 | 53.0 °C | Lysozyme-like (SSF53955) Chitinase class I (PF00182) | 3–182 41–146 |
| vB_EcoM_LNA4 [27] | endolysin | 117557–118129 | 572 | self_PFXJ204 | 190 | 21.65 | 30.802 | 9.67 | −0.374 | 81.105 | 41.0 °C | Cell Wall Hydrolase (PF07486) | 70–181 |
| Killian [35] | endolysin | 54687–56504 | 1817 | WEU67851.1 | 605 | 67.92 | 35.65 | 6.61 | −0.302 | 82.033 | 44.0 °C | anaerobic ribonucleoside-triphosphate reductase (TIGR02487) RNR_III (cd01675) PFL-like glycyl radical enzymes (SSF51998) Anaerobic ribonucleoside-triphosphate reductase (PF13597) | 21–604 21–584 26–585 21–604 |
| vB_Ec_ZCEC14 [40] | endolysin | 13508–14425 | 917 | UVD33329.1 | 305 | 35.56 | 28.885 | 8.61 | −0.448 | 85.967 | 42.0 °C | T4 RNase H, C terminal (PF09293) PIN_53EXO-like (cd00008) PIN domain-like (SSF88723) H3TH_T4-like (cd09899) 5′-3′ exonuclease, N-terminal resolvase-like domain (PF02739) 5′ to 3′ exonuclease, C-terminal subdomain (SSF47807) | 183–305 17–180 13–179 184–259 51–174 183–305 |
| Killian [35] | endolysin | 163120–164037 | 917 | WEU68017.1 | 305 | 35.56 | 28.885 | 8.61 | −0.448 | 85.967 | 42.0 °C | T4 RNase H, C terminal (PF09293) PIN_53EXO-like (cd00008) PIN domain-like (SSF88723) H3TH_T4-like (cd09899) 5′-3′ exonuclease, N-terminal resolvase-like domain (PF02739) 5′ to 3′ exonuclease, C-terminal subdomain (SSF47807) | 183–305 17–180 13–179 184–259 51–174 183–305 |
| vB_Ec_ZCEC14 [40] | endolysin | 51539–52219 | 680 | UVD33388.1 | 226 | 25.57 | 45.489 | 4.43 | −0.378 | 91.947 | 48.0 °C | Bacteriophage T4, inh (IPR016594) | 1–226 |
| Killian [35] | endolysin | 46919–47182 | 263 | WEU67833.1 | 87 | 10.24 | 48.485 | 4.21 | −0.197 | 74.943 | 40.0 °C | - | - |
| vB_EcoM_SHAK7163 [33] | internal virion protein with endolysin domain | 31648–34968 | 3320 | WVW77660.1 | 1106 | 121.66 | 35.115 | 5.61 | −0.423 | 79.322 | 42.0 °C | - | - |
| vB_EcoM_SHAK7704 [33] | internal virion protein with endolysin domain | 39690–42998 | 3308 | self_RMKP62 | 1102 | 121.26 | 35.421 | 5.69 | −0.429 | 78.63 | 42.0 °C | - | - |
| vB_EcoM_SHAK7854 [33] | internal virion protein with endolysin domain | 32030–35917 | 3887 | self_DNYN53 | 1295 | 140.9 | 37.494 | 5.67 | −0.396 | 79.012 | 41.0 °C | - | - |
| vB_EcoM_SHAK9454 [33] | internal virion protein with endolysin domain | 32160–36047 | 3887 | self_ZENB53 | 1295 | 140.83 | 37.467 | 5.87 | −0.39 | 78.942 | 41.0 °C | - | - |
| vB_EcoP_LNA3 [27] | internal virion protein with endolysin domain | 5943–9830 | 3887 | self_GXIJ6 | 1295 | 140.96 | 36.837 | 5.68 | −0.399 | 79.459 | 41.0 °C | - | - |
| vB_EcoP_LNA5 [27] | internal virion protein with endolysin domain | 23530–27417 | 3887 | self_TCEH45 | 1295 | 140.81 | 37.737 | 5.93 | −0.398 | 77.892 | 41.0 °C | - | - |
| vB_Ec_ZCEC14 [40] | endolysin | 127670–127933 | 263 | UVD33512.1 | 87 | 10.29 | 48.816 | 4.3 | −0.277 | 74.943 | 40.0 °C | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wesołowski, W.; Łukasiak, A.; Bloch, S.; Kuligowska, K.; Neumann, J.; Lewandowska, N.; Węglińska, E.; Węgrzyn, G.; Nejman-Faleńczyk, B. Phage Endolysins as Promising and Effective Candidates for Use Against Uropathogenic Escherichia coli. Viruses 2025, 17, 560. https://doi.org/10.3390/v17040560
Wesołowski W, Łukasiak A, Bloch S, Kuligowska K, Neumann J, Lewandowska N, Węglińska E, Węgrzyn G, Nejman-Faleńczyk B. Phage Endolysins as Promising and Effective Candidates for Use Against Uropathogenic Escherichia coli. Viruses. 2025; 17(4):560. https://doi.org/10.3390/v17040560
Chicago/Turabian StyleWesołowski, Wojciech, Aleksandra Łukasiak, Sylwia Bloch, Kaja Kuligowska, Julia Neumann, Natalia Lewandowska, Emilia Węglińska, Grzegorz Węgrzyn, and Bożena Nejman-Faleńczyk. 2025. "Phage Endolysins as Promising and Effective Candidates for Use Against Uropathogenic Escherichia coli" Viruses 17, no. 4: 560. https://doi.org/10.3390/v17040560
APA StyleWesołowski, W., Łukasiak, A., Bloch, S., Kuligowska, K., Neumann, J., Lewandowska, N., Węglińska, E., Węgrzyn, G., & Nejman-Faleńczyk, B. (2025). Phage Endolysins as Promising and Effective Candidates for Use Against Uropathogenic Escherichia coli. Viruses, 17(4), 560. https://doi.org/10.3390/v17040560






