A Novel Toolkit of SARS-CoV-2 Sub-Genomic Replicons for Efficient Antiviral Screening
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria and Yeast
2.2. Generation of SARS-CoV-2 cDNA Fragments for TAR
- (a)
- Synthetic DNA: Initially, nine cDNA fragments with 70 bp end-terminal overlaps were used to assemble a SARS-CoV-2 replicon clone based on the Wuhan-Hu-1 genome sequence (GenBank accession: NC_045512, Table S1). The cDNA fragments were produced by GeneArt™ synthesis (Invitrogen™, Thermo Fisher Scientific) as cDNA inserts in sequence-verified, stable plasmid clones. The 5′ terminal cDNA fragment was modified to contain 70 nucleotides corresponding to nucleotides (nts) 9311–9380 of the pYES1L vector, a T7 RNA polymerase promoter and an extra “G” nucleotide immediately upstream of the SARS-CoV-2 5′-terminal genome sequence, whilst the 3′-terminal cDNA fragment was modified such that the 3′ end of the SARS-CoV-2 genome was followed by a stretch of 33 “A”s followed by the unique restriction enzyme site AscI and nts 1–70 of the pYES1L vector. The first seven cDNA fragments (from the 5’end) spanned nts 1–20,090 of the SARS-CoV-2 genome. The two remaining cDNA fragments spanned nts 20,021–29,903 of the SARS-CoV-2 sequence with the following exceptions: nts 21,653–25,384, encoding the S protein, were replaced with a 1359 nt sequence encoding an enhanced green fluorescence protein (eGFP)-puromycin N-acetyl transferase (pac) fusion protein, and nts 26,523–27,191 encoding the M protein, were replaced with a 936 nt sequence encoding Renilla luciferase (RLuc). The cDNA fragment contained in each clone was PCR amplified using gene specific primer pairs and the Platinum SuperFi II mastermix (Invitrogen™, Thermo Fisher Scientific) following the manufacturer’s instructions. The location of the fragments and primers and sequences of introduced genes are shown in Table S2.
- (b)
- Overlap PCR mutagenesis: Modification of the synthetic replicon cDNA clones to introduce site-specific mutations, gene substitutions and a hepatitis delta virus ribozyme sequence followed by a T7 RNA polymerase terminator sequence (see Table S2, a kind gift from Professor Arvind Patel, MRC-University of Glasgow Centre for Virus Research) immediately downstream of the 3’end poly-A tail was performed by overlap-PCR (OL-PCR) mutagenesis. Template DNA fragments for OL-PCR were first produced as overlapping sub-fragments (20–30 nt overlaps) using an outer primer and an internal mutagenesis primer (primers shown in Table S3). The first-round PCR sub-fragments were purified by extraction from an agarose gel using a GeneJET Gel Extraction Kit (Thermo Scientific™, Thermo Fisher Scientific). A total mass of 10–15 ng of sub-fragments at a 1:1 molar ratio was then used as a template for OL-PCR using the forward and reverse outer primers. Assembly of more than two PCR fragments was performed stepwise.
- (c)
- Viral RNA extraction and reverse-transcriptase (RT)-PCR: For production of a SARS-CoV-2 Delta VOC replicon, fragments 2, 4, 5 and 6, corresponding to the Wuhan-Hu-1 virus genome (Figure 1), were swapped for the corresponding cDNA fragments generated by RT-PCR from Delta VOC extracted RNA. For production of a chimeric SARS-CoV-2 containing the S gene from the Delta VOC in the Wuhan-Hu-1 virus genome, fragments 8 and 9 were generated by RT-PCR from Delta VOC extracted RNA. Viral RNA was extracted from 140 µL of virus stock (SARS-CoV-2 Delta VOC, GISAID ID: EPI_ISL_15250227) using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. RT-PCR was performed using 1 µL of eluted RNA and a SuperScript™ IV One-Step RT-PCR System (Invitrogen™, Thermo Fisher Scientific) as described by the manufacturer.
- (d)
- HiFi DNA assembly: To produce SARS-CoV-2 Delta VOC replicon clones, the viral sequence from nts 20,021–29,903 was replaced with either of two complementary DNA (cDNA) fragments in which the S and M gene coding sequences were replaced with those of the pac and Rluc genes, and either the ORF6 or ORF7a coding sequences were replaced with those of the mScarlet and mNeonGreen genes, respectively. Assembly of the two cDNA fragments was performed using five overlapping cDNA fragments containing the VOC lineage-defining mutations and replicon-specific gene replacements (see Tables S4 and S5) using a NEBuilder® HiFi DNA Assembly Master Mix (NEB, Ipswich, MA, USA) according to the manufacturer’s recommendations. They were assembled in the vector pYES1L, the assembly reactions purified and electroporated into One Shot™ TOP10 Electrocomp™ E. coli (Invitrogen™, Thermo Fisher Scientific). Two colonies were picked, screened for correct assembly and used as PCR templates. The resulting fragments were used for TAR assembly.
2.3. TAR Assembly in Yeast
2.4. Yeast Colony Screens
2.5. Transformation of E. coli with BAC/YAC Shuttle Plasmids
2.6. BAC/YAC Purification
2.7. Preparation of In Vitro RNA Transcripts
2.8. Cell Lines
2.9. Mammalian Cell Electroporation and Replicon Transfection
2.10. Luciferase Assay
2.11. Replicon Antiviral Assay
2.12. Rescue of Recombinant SARS-CoV-2
2.13. Viral Stock Preparation
2.14. Immunofluorescence Assay
2.15. Viral Stock Titration
2.16. Viral Growth Analysis
2.17. Methylthiazolyldiphenyl-Tetrazolium Bromide (MTT) Cell Viability Assay
2.18. Interferon Dose–Response Assays
2.19. Single-Molecule Fluorescence In Situ Hybridisation (smFISH) of Replicon RNA
2.20. Ruxolitinib Dose–Response Assays
3. Results
3.1. Construction of a First-Generation Wuhan-Hu-1 SARS-CoV-2 Replicon
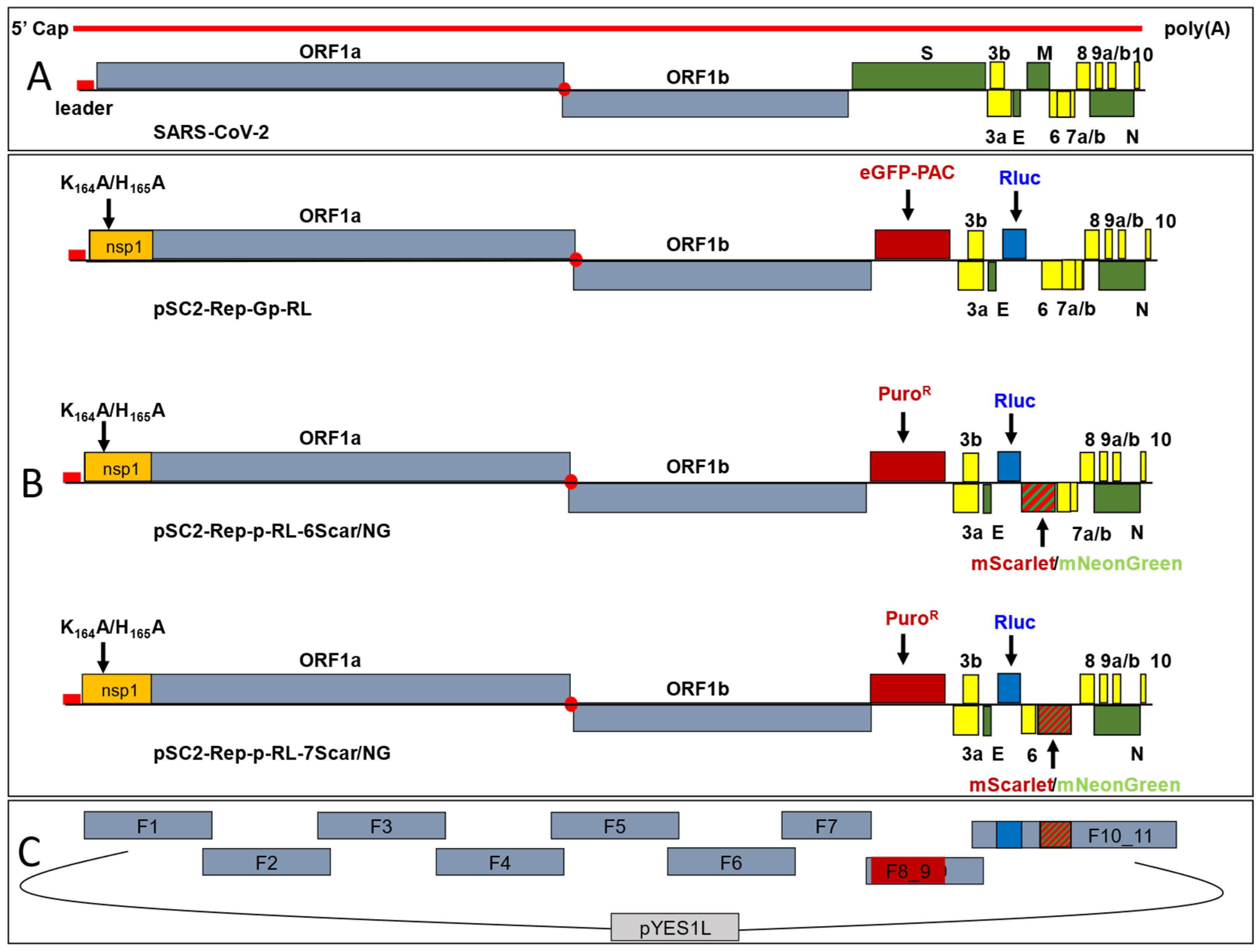
3.2. Construction of Second-Generation Wuhan-Hu-1 SARS-CoV-2 Dual-Reporter Replicons
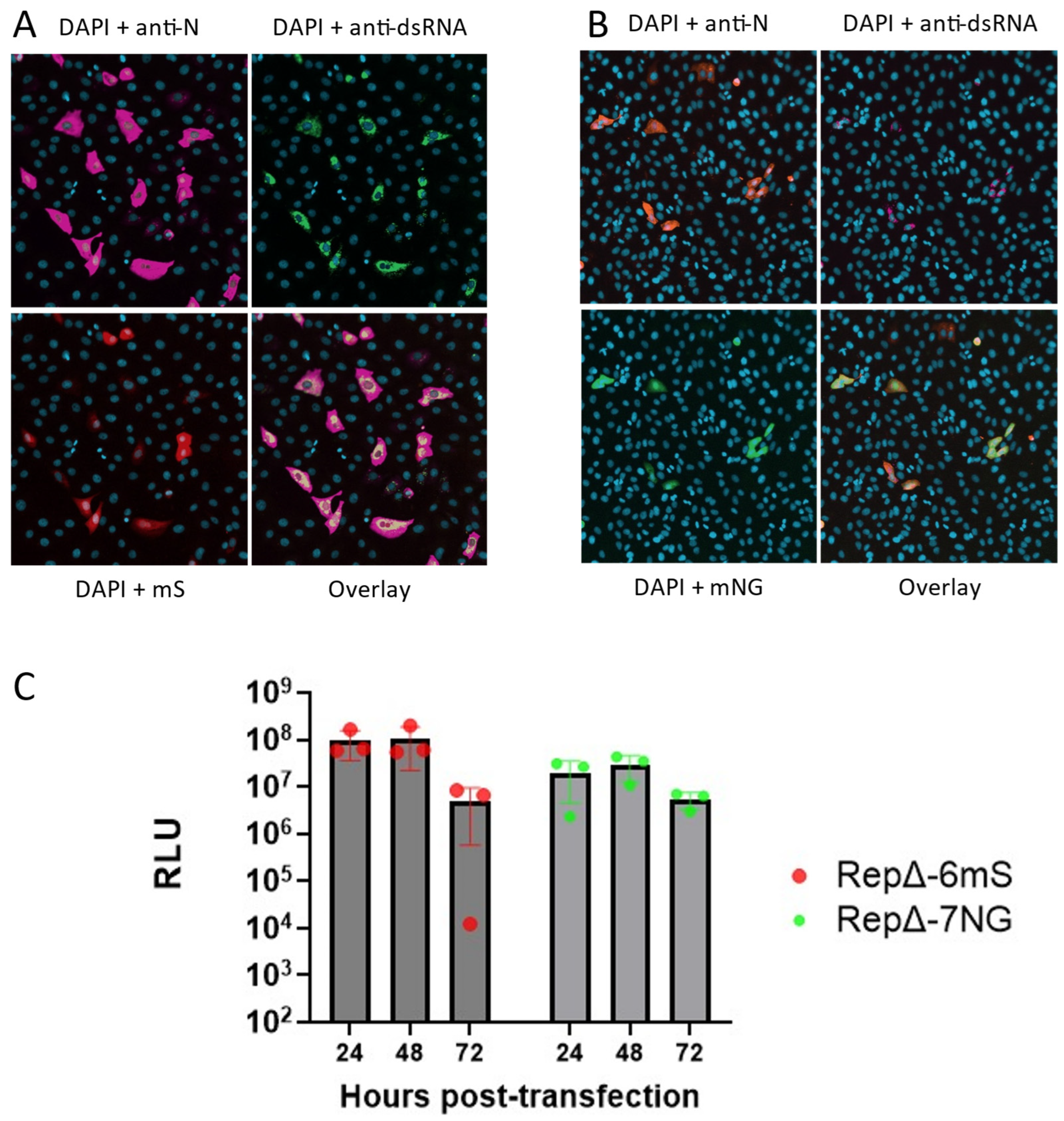
3.3. Construction of a Dual-Reporter Replicon for the SARS-CoV-2 Delta VOC
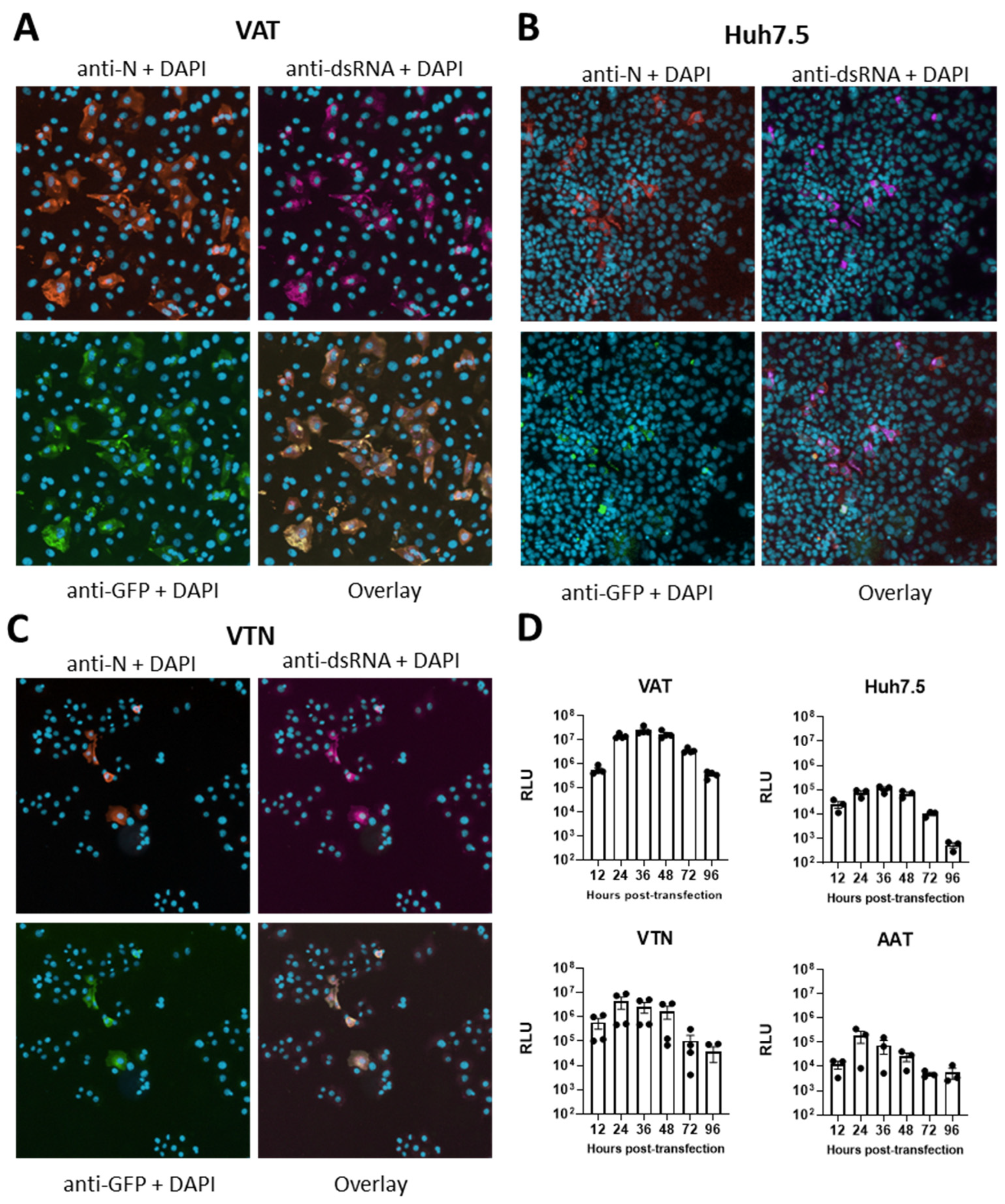
3.4. Comparison of Replicon and Virus RNA Replication
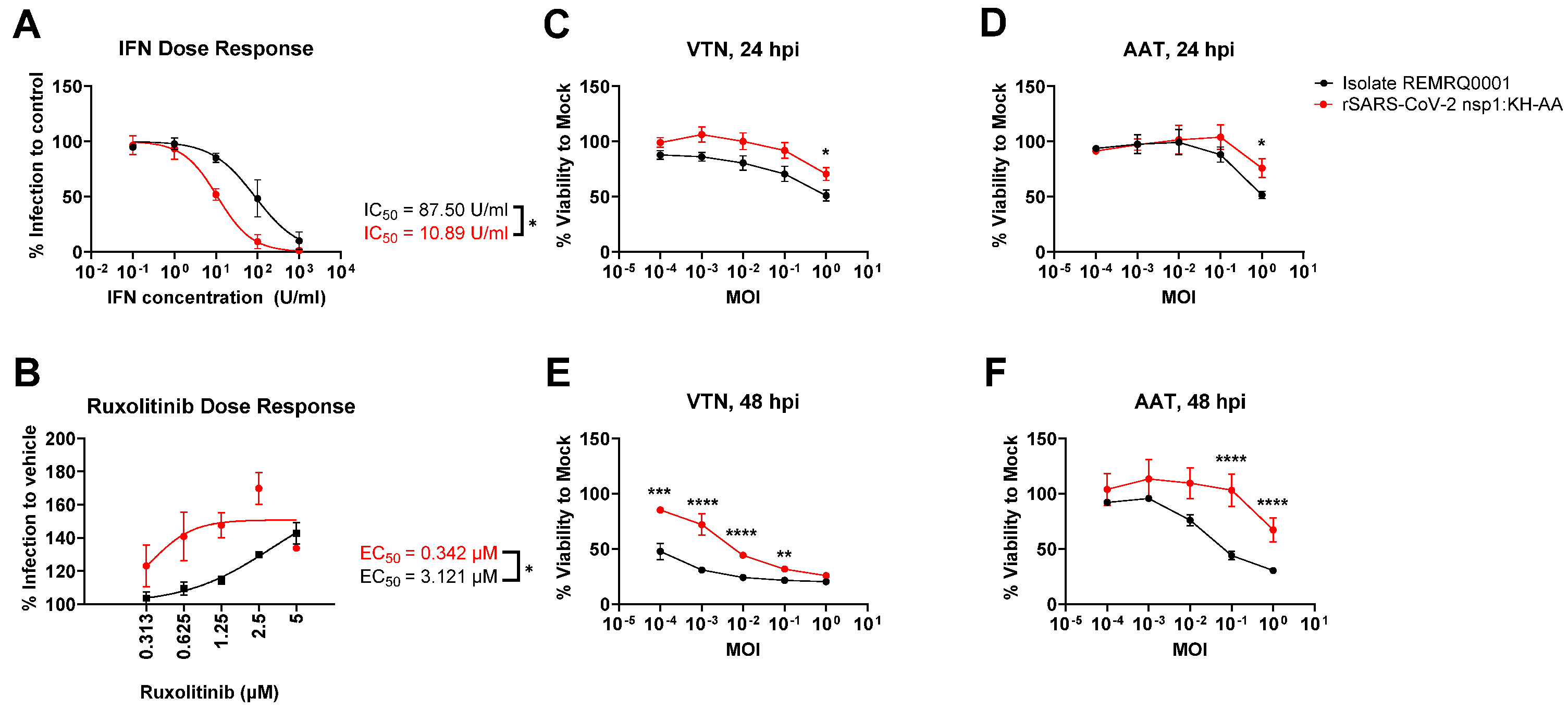
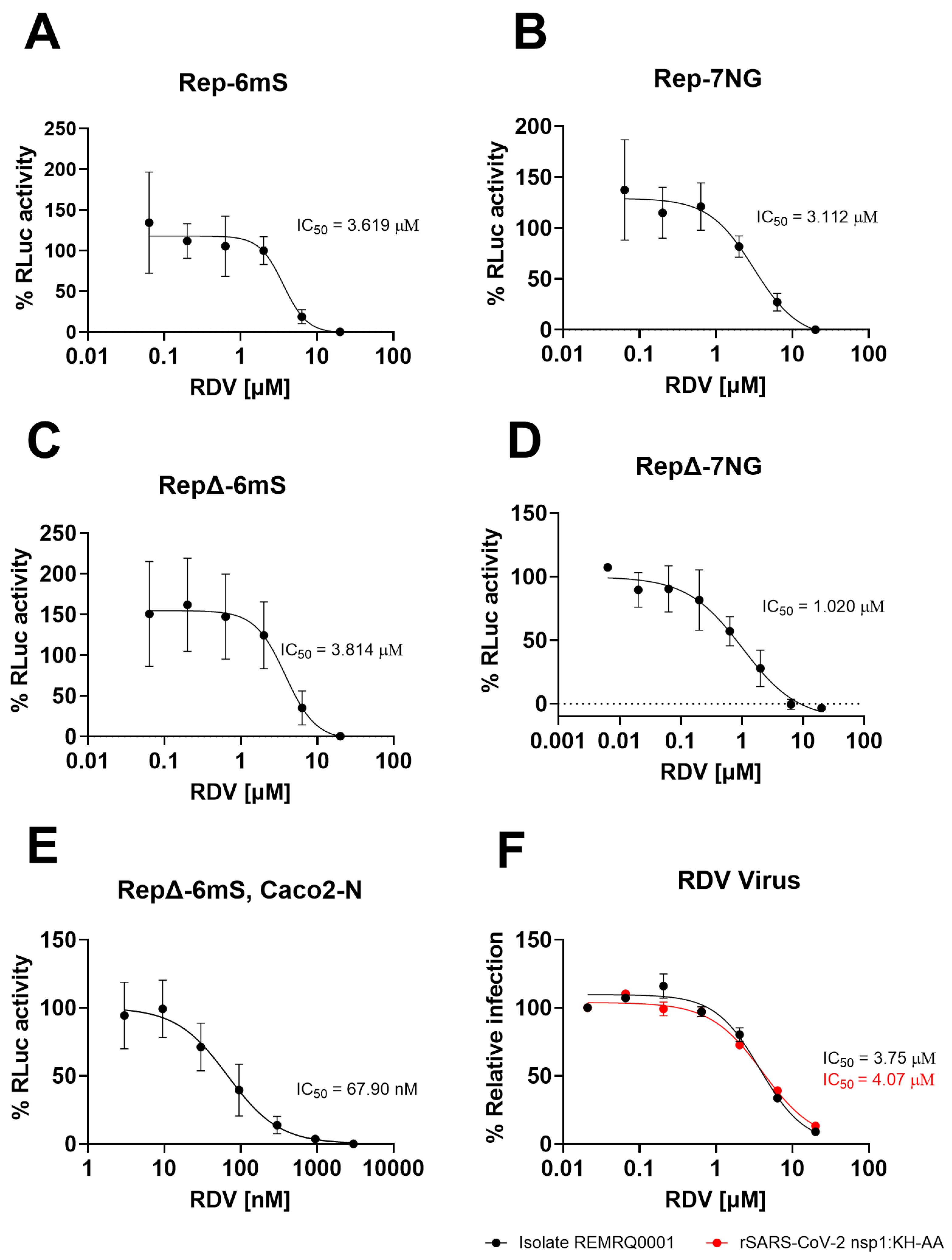
3.5. Replicons Show Comparable Response to Remdesivir Compared to Wild-Type Virus
3.6. Replicons of Wuhan and Delta Lineage Show Differential Drug Responses to Ritonavir and Cobicistat
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CL3 | Containment level 3 |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| CoV | Coronavirus |
| MERS | Middle Eastern respiratory syndrome |
| COVID-19 | Coronavirus disease 19 |
| UTR | Untranslated region |
| ORF | Open reading frame |
| sgRNA | Sub-genomic RNA |
| TRS | Transcriptional regulatory sequences |
| S | Spike |
| M | Membrane |
| E | Envelope |
| N | Nucleocapsid |
| HCoV-229E | Human CoV-229E |
| nsp1 | Nonstructural protein 1 |
| MHV | Murine hepatitis virus |
| ACE2 | Angiotensin-converting enzyme 2 |
| TMPRSS2 | Transmembrane serine protease 2 |
| VTN | VeroE6/TMPRSS2 |
| VAT | VeroE6/ACE2/TMPRSS2 |
| AAT | A549/ACE2/TMPRSS2 |
| BHK-21 | Baby hamster kidney 21 |
| BAN | BHK/ACE2/N |
| BEAS-2BA | BEAS-2BA/ACE2 |
| ALI | Air–liquid interface |
| TAR | Transformation-associated recombination |
| YSM-Trp | Yeast nitrogen base without amino acids supplemented with yeast synthetic drop-out medium minus tryptophan |
| nts | Nucleotides |
| eGFP | Enhanced green fluorescence protein |
| pac | Puromycin N-acetyl transferase |
| RLuc | Renilla luciferase |
| OL-PCR | Overlap-PCR |
| RT-PCR | Reverse-transcriptase PCR |
| VOC | Variant of concern |
| cDNA | Complementary DNA |
| E. coli | Escherichia coli |
| BAC | Bacterial artificial chromosome |
| BAC/YAC | Yeast artificial chromosome |
| LB | Lysogeny broth |
| PBS | Phosphate-buffered saline |
| hpt | Hours post-transfection |
| rSARS-CoV-2 | Recombinant SARS-CoV-2 |
| CPE | Cytopathic effect |
| MOI | Multiplicity of infection |
| MEM | Eagle’s minimal essential medium |
| MTT | Methylthiazolyldiphenyl-tetrazolium bromide |
| DPBS | Dulbecco’s PBS |
| smFISH | Single-molecule fluorescence in situ hybridisation |
| gRNA | Genomic RNA |
| DAPI | 4′,6-diamidino-2-phenylindole |
| RLU | Relative luminescence units |
| dsRNA | Double-stranded RNA |
| hpi | Hours post-infection |
| SEM | Standard error of the mean |
| IFN | Type I interferon |
| IC50 | Half maximal inhibitory concentration |
| JAK-STAT | Janus kinase-signal transducer and activator of transcription |
| DMSO | Dimethyl sulfoxide |
References
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; De Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. Coronaviridae Study Group of the International Committee on Taxonomy of V. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, Y.; Zhang, C.; Huang, F.; Wang, F.; Yuan, J.; Wang, Z.; Li, J.; Li, J.; Feng, C.; et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 2020, 63, 364–374. [Google Scholar] [CrossRef]
- Apolone, G.; Montomoli, E.; Manenti, A.; Boeri, M.; Sabia, F.; Hyseni, I.; Mazzini, L.; Martinuzzi, D.; Cantone, L.; Milanese, G.; et al. Unexpected detection of SARS-CoV-2 antibodies in the prepandemic period in Italy. Tumori J. 2020, 107, 446–451. [Google Scholar] [CrossRef]
- V’kOvski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Miao, Z.; Tidu, A.; Eriani, G.; Martin, F. Secondary structure of the SARS-CoV-2 5-UTR. RNA Biol. 2021, 18, 447–456. [Google Scholar] [CrossRef]
- Snijder, E.; Decroly, E.; Ziebuhr, J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus Res. 2016, 96, 59–126. [Google Scholar] [CrossRef]
- Davidson, A.D.; Williamson, M.K.; Lewis, S.; Shoemark, D.; Carroll, M.W.; Heesom, K.J.; Zambon, M.; Ellis, J.; Lewis, P.A.; Hiscox, J.A.; et al. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020, 12, 68. [Google Scholar] [CrossRef]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Hurtado-Tamayo, J.; Requena-Platek, R.; Enjuanes, L.; Bello-Perez, M.; Sola, I. Contribution to pathogenesis of accessory proteins of deadly human coronaviruses. Front. Cell. Infect. Microbiol. 2023, 13, 1166839. [Google Scholar] [CrossRef]
- Lindenbach, B.D. Reinventing positive-strand RNA virus reverse genetics. Adv. Virus Res. 2022, 112, 1–29. [Google Scholar] [CrossRef]
- Almazán, F.; Sola, I.; Zuñiga, S.; Marquez-Jurado, S.; Morales, L.; Becares, M.; Enjuanes, L. Coronavirus reverse genetic systems: Infectious clones and replicons. Virus Res. 2014, 189, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Thiel, V.; Siddell, S.G. Reverse genetics of coronaviruses using vaccinia virus vectors. Curr. Top. Microbiol. Immunol. 2005, 287, 199–227. [Google Scholar] [CrossRef] [PubMed]
- Thao, T.T.N.; Labroussaa, F.; Ebert, N.; V’kOvski, P.; Stalder, H.; Portmann, J.; Kelly, J.; Steiner, S.; Holwerda, M.; Kratzel, A.; et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature 2020, 582, 561–565. [Google Scholar] [CrossRef]
- Almazán, F.; González, J.M.; Pénzes, Z.; Izeta, A.; Calvo, E.; Plana-Durán, J.; Enjuanes, L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 2000, 97, 5516–5521. [Google Scholar] [CrossRef]
- Casais, R.; Thiel, V.; Siddell, S.G.; Cavanagh, D. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 2001, 75, 12359–12369. [Google Scholar] [CrossRef]
- Thiel, V.; Herold, J.; Schelle, B.; Siddell, S.G. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 2001, 82, 1273–1281. [Google Scholar] [CrossRef]
- Yount, B.; Curtis, K.M.; Baric, R.S. Strategy for systematic assembly of large RNA and DNA genomes: Transmissible gastroenteritis virus model. J. Virol. 2000, 74, 10600–10611. [Google Scholar] [CrossRef]
- Hertzig, T.; Scandella, E.; Schelle, B.; Ziebuhr, J.; Siddell, S.G.; Ludewig, B.; Thiel, V. Rapid identification of coronavirus replicase inhibitors using a selectable replicon RNA. J. Gen. Virol. 2004, 85, 1717–1725. [Google Scholar] [CrossRef]
- Ge, F.; Luo, Y.; Liew, P.X.; Hung, E. Derivation of a novel SARS-coronavirus replicon cell line and its application for anti-SARS drug screening. Virology 2007, 360, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Almazán, F.; Galán, C.; Enjuanes, L. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 2004, 78, 12683–12688. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hu, B.-J.; Zhao, K.; Luo, Y.; Lin, H.-F.; Shi, Z.-L. Development of a MERS-CoV replicon cell line for antiviral screening. Virol. Sin. 2021, 36, 730–735. [Google Scholar] [CrossRef]
- Ng, C.Y.; Gu, F.; Phong, W.Y.; Chen, Y.-L.; Lim, S.P.; Davidson, A.; Vasudevan, S.G. Construction and characterization of a stable subgenomic dengue virus type 2 replicon system for antiviral compound and siRNA testing. Antivir. Res. 2007, 76, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Almazán, F.; DeDiego, M.L.; Galán, C.; Escors, D.; ÁlVarez, E.; Ortego, J.; Sola, I.; Zuñiga, S.; Alonso, S.; Moreno, J.L.; et al. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 2006, 80, 10900–10906. [Google Scholar] [CrossRef]
- Wang, J.-M.; Wang, L.-F.; Shi, Z.-L. Construction of a non-infectious SARS coronavirus replicon for application in drug screening and analysis of viral protein function. Biochem. Biophys. Res. Commun. 2008, 374, 138–142. [Google Scholar] [CrossRef]
- He, X.; Quan, S.; Xu, M.; Rodriguez, S.; Goh, S.L.; Wei, J.; Fridman, A.; Koeplinger, K.A.; Carroll, S.S.; Grobler, J.A.; et al. Generation of SARS-CoV-2 reporter replicon for high-throughput antiviral screening and testing. Proc. Natl. Acad. Sci. USA 2021, 118, e2025866118. [Google Scholar] [CrossRef]
- Xia, H.; Cao, Z.; Xie, X.; Zhang, X.; Chen, J.Y.-C.; Wang, H.; Menachery, V.D.; Rajsbaum, R.; Shi, P.-Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020, 33, 108234. [Google Scholar] [CrossRef]
- Kotaki, T.; Xie, X.; Shi, P.-Y.; Kameoka, M. A PCR amplicon-based SARS-CoV-2 replicon for antiviral evaluation. Sci. Rep. 2021, 11, 2229. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, W.; Chen, S.; Yuan, Z.; Yi, Z. A bacterial artificial chromosome (BAC)-vectored noninfectious replicon of SARS-CoV-2. Antivir. Res. 2021, 185, 104974. [Google Scholar] [CrossRef] [PubMed]
- Ricardo-Lax, I.; Luna, J.M.; Thao, T.T.N.; Pen, J.L.; Yu, Y.; Hoffmann, H.-H.; Schneider, W.M.; Razooky, B.S.; Fernandez-Martinez, J.; Schmidt, F.; et al. Replication and single-cycle delivery of SARS-CoV-2 replicons. Science 2021, 374, 1099–1106. [Google Scholar] [CrossRef]
- Zhang, J.; Ejikemeuwa, A.; Gerzanich, V.; Nasr, M.; Tang, Q.; Simard, J.M.; Zhao, R.Y. Understanding the role of SARS-CoV-2 ORF3a in viral pathogenesis and COVID-19. Front. Microbiol. 2022, 13, 854567. [Google Scholar] [CrossRef]
- Addetia, A.; Lieberman, N.A.P.; Phung, Q.; Hsiang, T.Y.; Xie, H.; Roychoudhury, P.; Shrestha, L.; Loprieno, M.A.; Huang, M.L.; Gale, M., Jr.; et al. SARS-CoV-2 ORF6 disrupts bidirectional nucleocytoplasmic transport through interactions with Rae1 and Nup98. mBio 2021, 12, e00065-21. [Google Scholar] [CrossRef]
- Flower, T.G.; Buffalo, C.Z.; Hooy, R.M.; Allaire, M.; Ren, X.; Hurley, J.H. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc. Natl. Acad. Sci. USA 2021, 118, e2021785118. [Google Scholar] [CrossRef] [PubMed]
- Timilsina, U.; Umthong, S.; Ivey, E.B.; Waxman, B.; Stavrou, S. SARS-CoV-2 ORF7a potently inhibits the antiviral effect of the host factor SERINC5. Nat. Commun. 2022, 13, 2935. [Google Scholar] [CrossRef]
- Tan, Y.-J.; Fielding, B.C.; Goh, P.-Y.; Shen, S.; Tan, T.H.P.; Lim, S.G.; Hong, W. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol. 2004, 78, 14043–14047. [Google Scholar] [CrossRef]
- Schubert, K.; Karousis, E.D.; Jomaa, A.; Scaiola, A.; Echeverria, B.; Gurzeler, L.-A.; Leibundgut, M.; Thiel, V.; Mühlemann, O.; Ban, N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020, 27, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Brockway, S.M.; Denison, M.R. Mutagenesis of the murine hepatitis virus nsp1-coding region identifies residues important for protein processing, viral RNA synthesis, and viral replication. Virology 2005, 340, 209–223. [Google Scholar] [CrossRef]
- Narayanan, K.; Ramirez, S.I.; Lokugamage, K.G.; Makino, S. Coronavirus nonstructural protein 1: Common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015, 202, 89–100. [Google Scholar] [CrossRef]
- Huang, C.; Lokugamage, K.G.; Rozovics, J.M.; Narayanan, K.; Semler, B.L.; Makino, S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: Viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011, 7, e1002433. [Google Scholar] [CrossRef] [PubMed]
- Lokugamage, K.G.; Narayanan, K.; Nakagawa, K.; Terasaki, K.; Ramirez, S.I.; Tseng, C.-T.K.; Makino, S. Middle East respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J. Virol. 2015, 89, 10970–10981. [Google Scholar] [CrossRef] [PubMed]
- Mendez, A.S.; Ly, M.; González-Sánchez, A.M.; Hartenian, E.; Ingolia, N.T.; Cate, J.H.; Glaunsinger, B.A. The N-terminal domain of SARS-CoV-2 nsp1 plays key roles in suppression of cellular gene expression and preservation of viral gene expression. Cell Rep. 2021, 37, 109841. [Google Scholar] [CrossRef]
- Thoms, M.; Buschauer, R.; Ameismeier, M.; Koepke, L.; Denk, T.; Hirschenberger, M.; Kratzat, H.; Hayn, M.; Mackens-Kiani, T.; Cheng, J.; et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 2020, 369, 1249–1255. [Google Scholar] [CrossRef]
- Shen, Z.; Zhang, G.; Yang, Y.; Li, M.; Yang, S.; Peng, G. Lysine 164 is critical for SARS-CoV-2 Nsp1 inhibition of host gene expression. J. Gen. Virol. 2021, 102, jgv001513. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chou, C.-K.; Wu, W.W.; Luan, B.; Wang, T.T. Stable cell clones harboring self-replicating SARS-CoV-2 RNAs for drug screen. J. Virol. 2022, 96, e0221621. [Google Scholar] [CrossRef]
- Blount, B.A.; Driessen, M.R.M.; Ellis, T. GC Preps: Fast and Easy Extraction of Stable Yeast Genomic DNA. Sci. Rep. 2016, 6, 26863. [Google Scholar] [CrossRef]
- Blight, K.J.; McKeating, J.A.; Rice, C.M. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 2002, 76, 13001–13014. [Google Scholar] [CrossRef]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7001–7003. [Google Scholar] [CrossRef]
- Rihn, S.J.; Merits, A.; Bakshi, S.; Turnbull, M.L.; Wickenhagen, A.; Alexander, A.J.T.; Baillie, C.; Brennan, B.; Brown, F.; Brunker, K.; et al. A plasmid DNA-launched SARS-CoV-2 reverse genetics system and coronavirus toolkit for COVID-19 research. PLoS Biol. 2021, 19, e3001091. [Google Scholar] [CrossRef]
- Webb, I.; Erdmann, M.; Milligan, R.; Savage, M.; Matthews, D.A.; Davidson, A.D. Examining the feasibility of replacing ORF3a with fluorescent genes to construct SARS-CoV-2 reporter viruses. J. Gen. Virol. 2025, 106, 2. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.L.; Simonetti, B.; Antón-Plágaro, C.; Williamson, M.K.; Shoemark, D.K.; Simón-Gracia, L.; Klein, K.; Bauer, M.; Hollandi, R.; Greber, U.F.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef]
- Lee, J.Y.; Wing, P.A.; Gala, D.S.; Noerenberg, M.; Järvelin, A.I.; Titlow, J.; Zhuang, X.; Palmalux, N.; Iselin, L.; Thompson, M.K.; et al. Absolute quantitation of individual SARS-CoV-2 RNA molecules provides a new paradigm for infection dynamics and variant differences. eLife 2022, 11, e74153. [Google Scholar] [CrossRef] [PubMed]
- Goldswain, H.; Dong, X.; Penrice-Randal, R.; Alruwaili, M.; Shawli, G.T.; Prince, T.; Williamson, M.K.; Raghwani, J.; Randle, N.; Jones, B.; et al. The P323L substitution in the SARS-CoV-2 polymerase (NSP12) confers a selective advantage during infection. Genome Biol. 2023, 24, 47. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, C.; Lei, X.; Ren, L.; Zhao, Z.; Wang, J.; Huang, H. Construction of non-infectious SARS-CoV-2 replicons and their application in drug evaluation. Virol. Sin. 2021, 36, 890–900. [Google Scholar] [CrossRef] [PubMed]
- Malone, B.; Urakova, N.; Snijder, E.J.; Campbell, E.A. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2022, 23, 21–39. [Google Scholar] [CrossRef]
- Massé, N.; Davidson, A.; Ferron, F.; Alvarez, K.; Jacobs, M.; Romette, J.-L.; Canard, B.; Guillemot, J.-C. Dengue virus replicons: Production of an interserotypic chimera and cell lines from different species, and establishment of a cell-based fluorescent assay to screen inhibitors, validated by the evaluation of ribavirin’s activity. Antivir. Res. 2010, 86, 296–305. [Google Scholar] [CrossRef]
- Mosca, J.D.; Pitha, P.M. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol. Cell. Biol. 1986, 6, 2279–2283. [Google Scholar] [CrossRef]
- Jensen, S.; Thomsen, A.R. Sensing of RNA viruses: A review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol. 2012, 86, 2900–2910. [Google Scholar] [CrossRef]
- Shaner, N.C.; Lambert, G.G.; Chammas, A.; Ni, Y.; Cranfill, P.J.; Baird, M.A.; Sell, B.R.; Allen, J.R.; Day, R.N.; Israelsson, M.; et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 2013, 10, 407–409. [Google Scholar] [CrossRef]
- Bindels, D.S.; Haarbosch, L.; Van Weeren, L.; Postma, M.; Wiese, K.E.; Mastop, M.; Aumonier, S.; Gotthard, G.; Royant, A.; Hink, M.A.; et al. MScarlet: A bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 2017, 14, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Wing, P.A.C.; Schmidt, N.M.; Peters, R.; Erdmann, M.; Brown, R.; Wang, H.; Swadling, L.; COVIDsortium Investigators; Newman, J.; Thakur, N.; et al. An ACAT inhibitor suppresses SARS-CoV-2 replication and boosts antiviral T cell activity. PLoS Pathog. 2023, 19, e1011323. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Binder, J.; Yurgelonis, I.; Rai, D.K.; Lazarro, S.; Costales, C.; Kobylarz, K.; McMonagle, P.; Steppan, C.M.; Aschenbrenner, L.; et al. Generation of a VeroE6 Pgp gene knock out cell line and its use in SARS-CoV-2 antiviral study. Antivir. Res. 2022, 208, 105429. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Falzarano, D.; Gerdts, V.; Liu, Q. Construction of a noninfectious SARS-CoV-2 replicon for antiviral-drug testing and gene function studies. J. Virol. 2021, 95, e0068721. [Google Scholar] [CrossRef]
- Tseng, A.; Hughes, C.A.; Wu, J.; Seet, J.; Phillips, E.J. Cobicistat versus ritonavir: Similar pharmacokinetic enhancers but some important differences. Ann. Pharmacother. 2017, 51, 1008–1022. [Google Scholar] [CrossRef] [PubMed]
- Shytaj, I.L.; Fares, M.; Gallucci, L.; Lucic, B.; Tolba, M.M.; Zimmermann, L.; Adler, J.M.; Xing, N.; Bushe, J.; Gruber, A.D.; et al. The FDA-approved drug cobicistat synergizes with remdesivir to inhibit SARS-CoV-2 replication in vitro and decreases viral titers and disease progression in Syrian hamsters. mBio 2022, 13, e03705-21. [Google Scholar] [CrossRef]
- Gallucci, L.; Bazire, J.; Davidson, A.D.; Shytaj, I.L. Broad-spectrum antiviral activity of two structurally analogous CYP3A inhibitors against pathogenic human coronaviruses in vitro. Antivir. Res. 2023, 221, 105766. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erdmann, M.; Wing, P.A.C.; Webb, I.; Kavanagh Williamson, M.; Jearanaiwitayakul, T.; Sullivan, E.; Bazire, J.; Shytaj, I.L.; McKeating, J.A.; Matthews, D.A.; et al. A Novel Toolkit of SARS-CoV-2 Sub-Genomic Replicons for Efficient Antiviral Screening. Viruses 2025, 17, 597. https://doi.org/10.3390/v17050597
Erdmann M, Wing PAC, Webb I, Kavanagh Williamson M, Jearanaiwitayakul T, Sullivan E, Bazire J, Shytaj IL, McKeating JA, Matthews DA, et al. A Novel Toolkit of SARS-CoV-2 Sub-Genomic Replicons for Efficient Antiviral Screening. Viruses. 2025; 17(5):597. https://doi.org/10.3390/v17050597
Chicago/Turabian StyleErdmann, Maximilian, Peter A. C. Wing, Isobel Webb, Maia Kavanagh Williamson, Tuksin Jearanaiwitayakul, Edward Sullivan, James Bazire, Iart Luca Shytaj, Jane A. McKeating, David A. Matthews, and et al. 2025. "A Novel Toolkit of SARS-CoV-2 Sub-Genomic Replicons for Efficient Antiviral Screening" Viruses 17, no. 5: 597. https://doi.org/10.3390/v17050597
APA StyleErdmann, M., Wing, P. A. C., Webb, I., Kavanagh Williamson, M., Jearanaiwitayakul, T., Sullivan, E., Bazire, J., Shytaj, I. L., McKeating, J. A., Matthews, D. A., & Davidson, A. D. (2025). A Novel Toolkit of SARS-CoV-2 Sub-Genomic Replicons for Efficient Antiviral Screening. Viruses, 17(5), 597. https://doi.org/10.3390/v17050597






