Circulating Cell-Free Nuclear DNA Predicted an Improvement of Systolic Left Ventricular Function in Individuals with Chronic Heart Failure with Reduced Ejection Fraction
Abstract
:1. Introduction
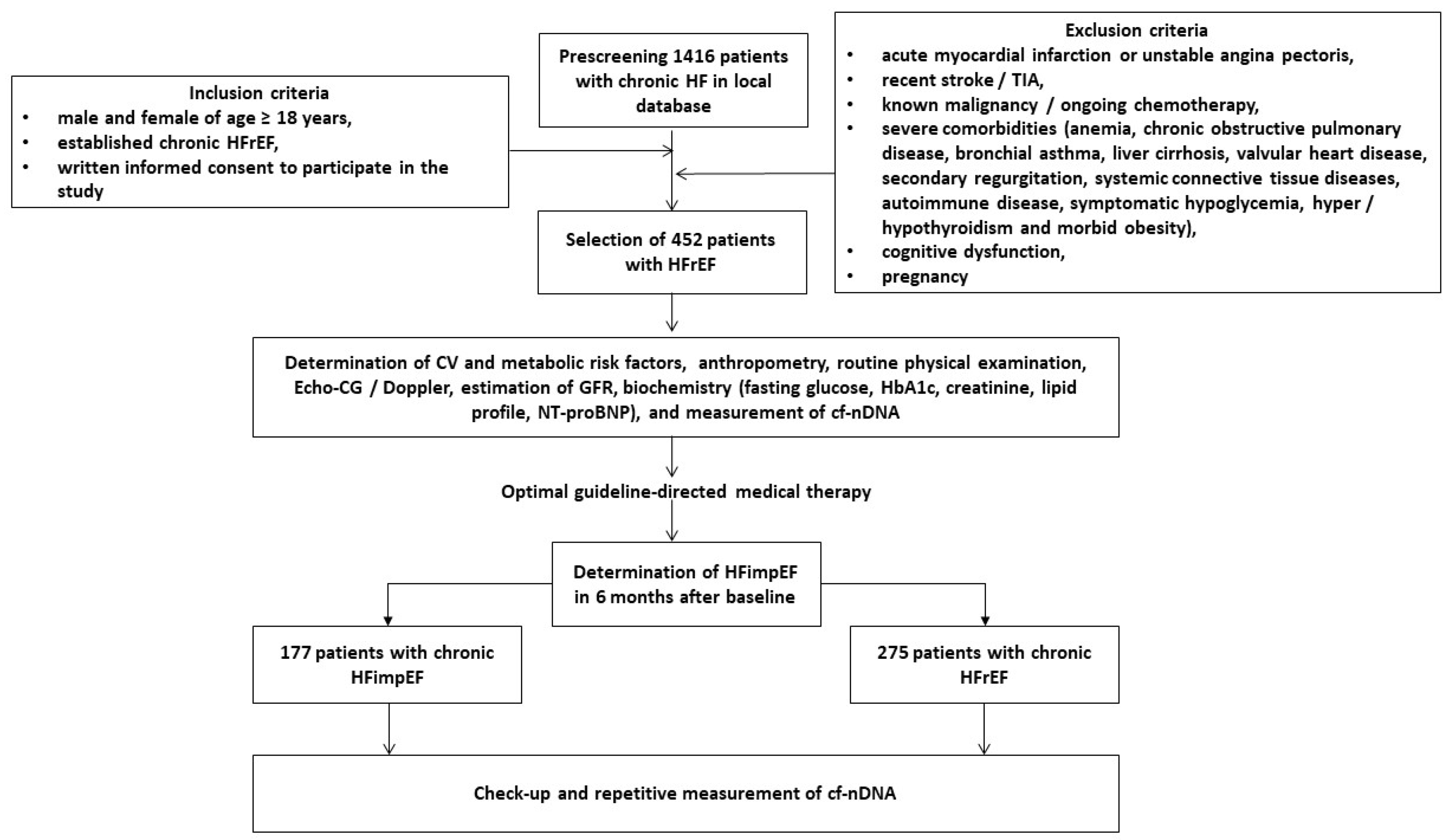
2. Materials and Methods
2.1. Patients’ Characteristics
2.2. Determination of HFimpEF
2.3. Medical Information Collection
2.4. Examination of Hemodynamics
2.5. Glomerular Filtration Rate Calculation
2.6. Blood Sampling
2.7. Biomarker Evaluation
2.8. Cell-Free DNA Extraction
2.9. Measurement of Cell-Free DNA in Plasma Samples
2.10. Statistical Analysis
3. Results
3.1. General Clinical Characteristics of the Patients
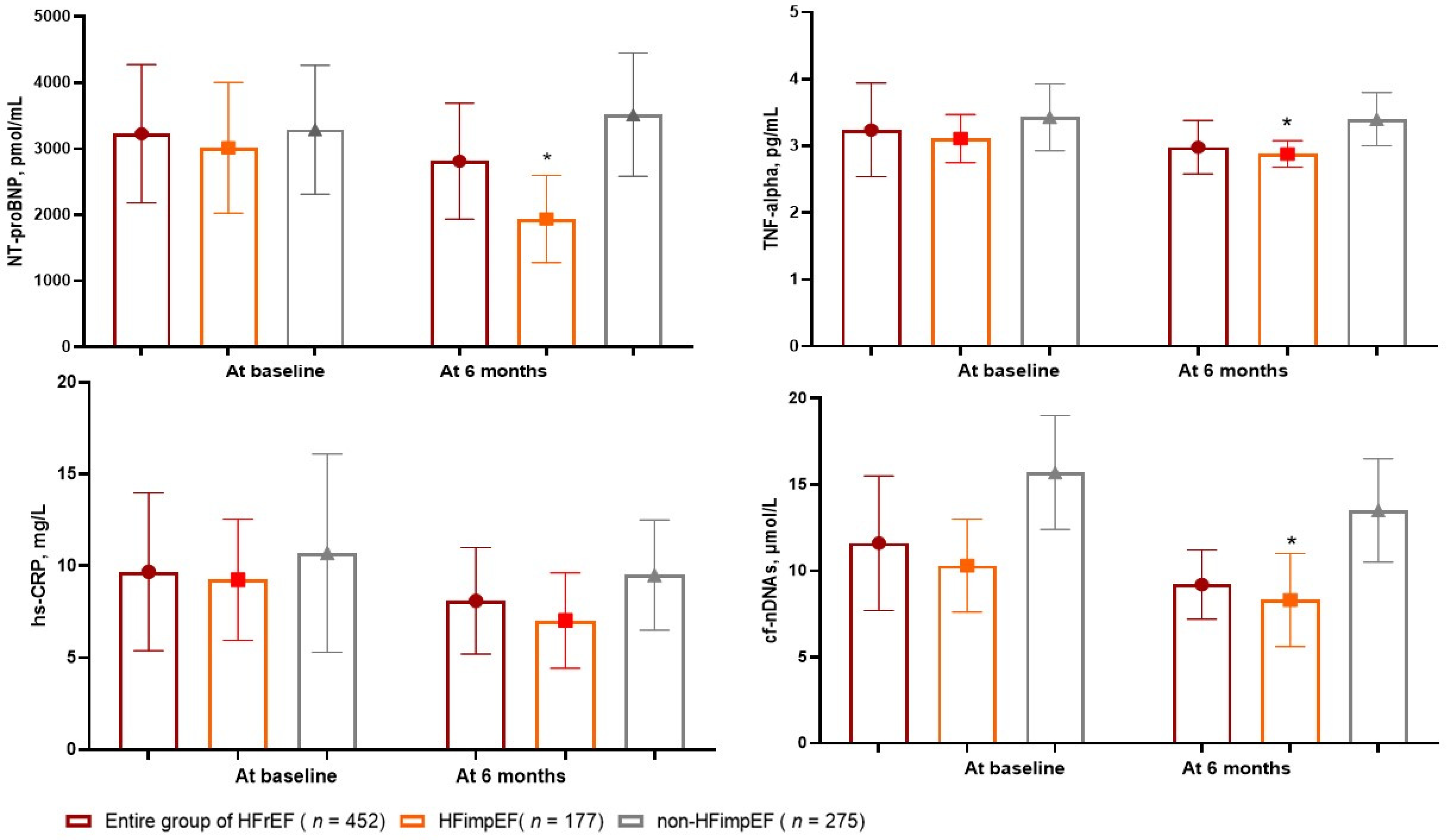
3.2. The Dynamics of Circulating Biomarker Levels
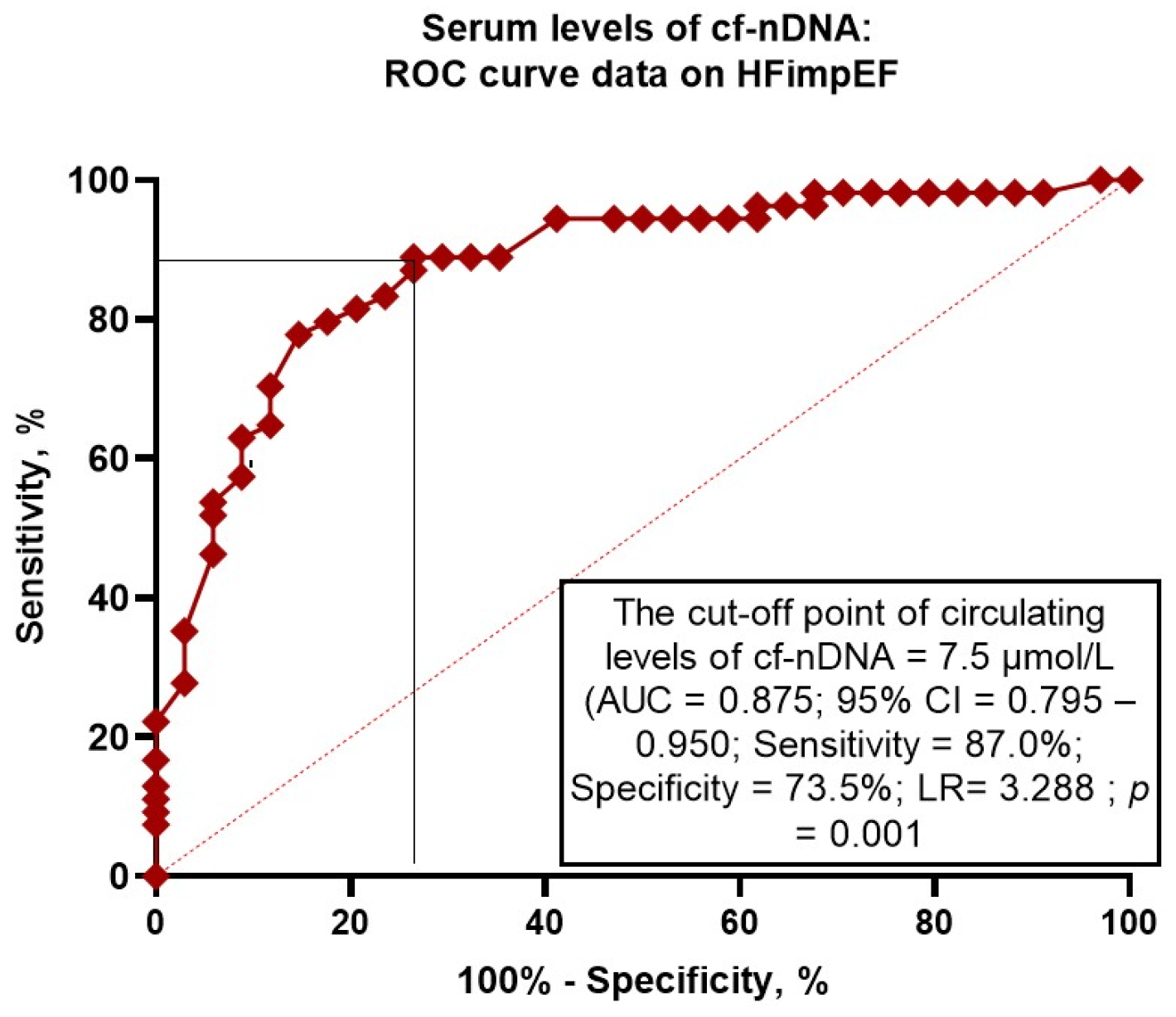
3.3. The Reliability of Circulating Levels of cf-nDNA: The Results of the ROC Curve Analysis
3.4. The Predictors of HFimpEF: The Univariate and Multivariate Logistic Regressions
3.5. Comparison of the Models for HFimpEF
4. Discussion
5. Study Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bozkurt, B.; Coats, A.J.S.; Tsutsui, H.; Abdelhamid, C.M.; Adamopoulos, S.; Albert, N.; Anker, S.D.; Atherton, J.; Böhm, M.; Butler, J.; et al. Universal definition and classification of heart failure: A report of the Heart Failure Society of America, Heart Failure Association of the European Society of Cardiology, Japanese Heart Failure Society and Writing Committee of the Universal Definition of Heart Failure: Endorsed by the Canadian Heart Failure Society, Heart Failure Association of India, Cardiac Society of Australia and New Zealand, and Chinese Heart Failure Association. Eur. J. Heart Fail. 2021, 23, 352–380. [Google Scholar] [CrossRef] [PubMed]
- Keshvani, N.; Shah, S.; Ayodele, I.; Chiswell, K.; Alhanti, B.; Allen, L.A.; Greene, S.J.; Yancy, C.W.; Alonso, W.W.; Van Spall, H.G.; et al. Sex differences in long-term outcomes following acute heart failure hospitalization: Findings from the Get with The Guidelines-Heart Failure registry. Eur. J. Heart Fail. 2023, 25, 1544–1554. [Google Scholar] [CrossRef] [PubMed]
- Chimed, S.; Stassen, J.; Galloo, X.; Meucci, M.C.; van der Bijl, P.; Knuuti, J.; Delgado, V.; Marsan, N.A.; Bax, J.J. Impact of Worsening Heart Failure on Long-Term Prognosis in Patients with Heart Failure with Reduced Ejection Fraction. Am. J. Cardiol. 2022, 184, 63–71. [Google Scholar] [CrossRef]
- Chen, S.; Huang, Z.; Liang, Y.; Zhao, X.; Aobuliksimu, X.; Wang, B.; He, Y.; Kang, Y.; Huang, H.; Li, Q.; et al. Five-year mortality of heart failure with preserved, mildly reduced, and reduced ejection fraction in a 4880 Chinese cohort. ESC Heart Fail. 2022, 9, 2336–2347. [Google Scholar] [CrossRef] [PubMed]
- McDonagh, T.A.; Metra, M.; Adamo, M.; Gardner, R.S.; Baumbach, A.; Böhm, M.; Burri, H.; Butler, J.; Čelutkienė, J.; Chioncel, O.; et al. 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur. Heart J. 2021, 42, 3599–3726, Erratum in: Eur. Heart J. 2021, 42, 4901. [Google Scholar] [CrossRef]
- Shah, K.S.; Xu, H.; Matsouaka, R.A.; Bhatt, D.L.; Heidenreich, P.A.; Hernandez, A.F.; Devore, A.D.; Yancy, C.W.; Fonarow, G.C. Heart Failure with Preserved, Borderline, and Reduced Ejection Fraction: 5-Year Outcomes. J. Am. Coll. Cardiol. 2017, 70, 2476–2486. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wang, P.; Liu, C.; Peng, J.; Liu, Y.; Ma, Q. Pharmacotherapy in patients with heart failure with reduced ejection fraction: A systematic review and meta-analysis. Chin. Med. J. 2024. Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.; Lu, P.; Yang, X. Efficacy of sacubitril-valsartan and SGLT2 inhibitors in heart failure with reduced ejection fraction: A systematic review and meta-analysis. Clin. Cardiol. 2023, 46, 1137–1145. [Google Scholar] [CrossRef]
- Rosano, G.M.C.; Vitale, C.; Spoletini, I. Precision Cardiology: Phenotype-targeted Therapies for HFmrEF and HFpEF. Int. J. Heart Fail. 2024, 6, 47–55. [Google Scholar] [CrossRef]
- Romero, E.; Baltodano, A.F.; Rocha, P.; Sellers-Porter, C.; Patel, D.J.; Soroya, S.; Bidwell, J.; Ebong, I.; Gibson, M.; Liem, D.A.; et al. Clinical, Echocardiographic, and Longitudinal Characteristics Associated with Heart Failure with Improved Ejection Fraction. Am. J. Cardiol. 2024, 211, 143–152. [Google Scholar] [CrossRef]
- Solymossi, B.; Muk, B.; Sepp, R.; Habon, T.; Borbély, A.; Heltai, K.; Majoros, Z.; Járai, Z.; Vágány, D.; Szatmári, Á.; et al. Incidence and predictors of heart failure with improved ejection fraction category in a HFrEF patient population. ESC Heart Fail. 2024, 11, 783–794. [Google Scholar] [CrossRef]
- Su, K.; Li, M.; Wang, L.; Tian, S.; Su, J.; Gu, J.; Chen, S. Clinical characteristics, predictors, and outcomes of heart failure with improved ejection fraction. Int. J. Cardiol. 2022, 357, 72–80. [Google Scholar] [CrossRef]
- Ho, L.T.; Juang, J.J.; Chen, Y.H.; Chen, Y.S.; Hsu, R.B.; Huang, C.C.; Lee, C.M.; Chien, K.L. Predictors of Left Ventricular Ejection Fraction Improvement in Patients with Early-Stage Heart Failure with Reduced Ejection Fraction. Acta Cardiol. Sin. 2023, 39, 854–861. [Google Scholar] [CrossRef]
- Segev, A.; Avrahamy, B.; Fardman, A.; Matetzky, S.; Freimark, D.; Regev, O.; Kuperstein, R.; Grupper, A. Heart failure with improved ejection fraction: Patient characteristics, clinical outcomes and predictors for improvement. Front. Cardiovasc. Med. 2024, 11, 1378955. [Google Scholar] [CrossRef]
- Si, J.; Ding, Z.; Hu, Y.; Zhang, X.; Zhang, Y.; Cao, H.; Liu, Y. Predictors and prognostic implications of left ventricular ejection fraction trajectory improvement in the spectrum of heart failure with reduced and mildly reduced ejection fraction. J. Cardiol. 2024, 83, 250–257. [Google Scholar] [CrossRef]
- Lo, Y.M.D.; Han, D.S.C.; Jiang, P.; Chiu, R.W.K. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science 2021, 372, eaaw3616. [Google Scholar] [CrossRef]
- Cahilog, Z.; Zhao, H.; Wu, L.; Alam, A.; Eguchi, S.; Weng, H.; Ma, D. The Role of Neutrophil NETosis in Organ Injury: Novel Inflammatory Cell Death Mechanisms. Inflammation 2020, 43, 2021–2032. [Google Scholar] [CrossRef]
- Stanley, K.E.; Jatsenko, T.; Tuveri, S.; Sudhakaran, D.; Lannoo, L.; Van Calsteren, K.; de Borre, M.; Van Parijs, I.; Van Coillie, L.; Van Den Bogaert, K.; et al. Cell type signatures in cell-free DNA fragmentation profiles reveal disease biology. Nat. Commun. 2024, 15, 2220. [Google Scholar] [CrossRef]
- Oellerich, M.; Sherwood, K.; Keown, P.; Schütz, E.; Beck, J.; Stegbauer, J.; Rump, L.C.; Walson, P.D. Liquid biopsies: Donor-derived cell-free DNA for the detection of kidney allograft injury. Nat. Rev. Nephrol. 2021, 17, 591–603. [Google Scholar] [CrossRef]
- Tan, E.; Liu, D.; Perry, L.; Zhu, J.; Cid-Serra, X.; Deane, A.; Yeo, C.; Ajani, A. Cell-free DNA as a potential biomarker for acute myocardial infarction: A systematic review and meta-analysis. Int. J. Cardiol. Heart Vasc. 2023, 47, 101246. [Google Scholar] [CrossRef]
- Antonatos, D.; Patsilinakos, S.; Spanodimos, S.; Korkonikitas, P.; Tsigas, D. Cell-free DNA levels as a prognostic marker in acute myocardial infarction. Ann. N. Y. Acad. Sci. 2006, 1075, 278–281. [Google Scholar] [CrossRef]
- Medina, J.E.; Dracopoli, N.C.; Bach, P.B.; Lau, A.; Scharpf, R.B.; Meijer, G.A.; Andersen, C.L.; Velculescu, V.E. Cell-free DNA approaches for cancer early detection and interception. J. Immunother. Cancer 2023, 11, e006013. [Google Scholar] [CrossRef]
- Berezina, T.A.; Berezin, A.E. Cell-free DNA as a plausible biomarker of chronic kidney disease. Epigenomics 2023, 15, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Mansueto, G.; Benincasa, G.; Della Mura, N.; Nicoletti, G.F.; Napoli, C. Epigenetic-sensitive liquid biomarkers and personalised therapy in advanced heart failure: A focus on cell-free DNA and microRNAs. J. Clin. Pathol. 2020, 73, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Yokokawa, T.; Misaka, T.; Kimishima, Y.; Shimizu, T.; Kaneshiro, T.; Takeishi, Y. Clinical Significance of Circulating Cardiomyocyte-Specific Cell-Free DNA in Patients with Heart Failure: A Proof-of-Concept Study. Can. J. Cardiol. 2020, 36, 931–935. [Google Scholar] [CrossRef] [PubMed]
- Berezina, T.A.; Kopytsya, M.P.; Petyunina, O.V.; Berezin, A.A.; Obradovic, Z.; Schmidbauer, L.; Lichtenauer, M.; Berezin, A.E. Lower Circulating Cell-Free Mitochondrial DNA Is Associated with Heart Failure in Type 2 Diabetes Mellitus Patients. Cardiogenetics 2023, 13, 15–30. [Google Scholar] [CrossRef]
- American Diabetes Association. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2021. Diabetes Care 2021, 44 (Suppl. S1), S15–S33. [Google Scholar] [CrossRef]
- Williams, B.; Mancia, G.; Spiering, W.; Agabiti Rosei, E.; Azizi, M.; Burnier, M.; ESC Scientific Document Group. 2018 ESC/ESH Guidelines for the management of arterial hypertension. Eur. Heart J. 2018, 39, 3021–3104. [Google Scholar] [CrossRef]
- Mach, F.; Baigent, C.; Catapano, A.L.; Koskinas, K.C.; Casula, M.; Badimon, L.; ESC Scientific Document Group. 2019 ESC/EAS Guidelines for the management of dyslipidaemias: Lipid modification to reduce cardiovascular risk. Eur. Heart J. 2020, 41, 111–188. [Google Scholar] [CrossRef]
- Knuuti, J.; Wijns, W.; Saraste, A.; Capodanno, D.; Barbato, E.; Funck-Brentano, C.; Prescott, E.; Storey, R.F.; Deaton, C.; Cuisset, T.; et al. 2019 ESC Guidelines for the diagnosis and management of chronic coronary syndromes. Eur. Heart J. 2020, 41, 407–477, Erratum in: Eur. Heart J. 2020, 41, 4242. [Google Scholar] [CrossRef] [PubMed]
- Inker, L.A.; Astor, B.C.; Fox, C.H.; Isakova, T.; Lash, J.P.; Peralta, C.A.; Kurella Tamura, M.; Feldman, H.I. KDOQI US commentary on the 2012 KDIGO clinical practice guideline for the evaluation and management of CKD. Am. J. Kidney Dis. 2014, 63, 713–735. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, C.; Rahko, P.S.; Blauwet, L.A.; Canaday, B.; Finstuen, J.A.; Foster, M.C.; Horton, K.; Ogunyankin, K.O.; Palma, R.A.; Velazquez, E.J. Guidelines for Performing a Comprehensive Transthoracic Echocardiographic Examination in Adults: Recommendations from the American Society of Echocardiography. J. Am. Soc. Echocardiogr. 2018, 32, 1–64. [Google Scholar] [CrossRef]
- Levey, A.S.; Stevens, L.A.; Schmid, C.H.; Zhang, Y.L.; Castro, A.F., 3rd; Feldman, H.I.; CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration). A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009, 150, 604–612. [Google Scholar] [CrossRef]
- Hasenleithner, S.O.; Speicher, M.R. A clinician’s handbook for using ctDNA throughout the patient journey. Mol. Cancer 2022, 21, 81. [Google Scholar] [CrossRef]
- He, Y.; Ling, Y.; Guo, W.; Li, Q.; Yu, S.; Huang, H.; Zhang, R.; Gong, Z.; Liu, J.; Mo, L.; et al. Prevalence and Prognosis of HFimpEF Developed From Patients with Heart Failure with Reduced Ejection Fraction: Systematic Review and Meta-Analysis. Front. Cardiovasc. Med. 2021, 8, 757596. [Google Scholar] [CrossRef]
- Heidenreich, P.A.; Bozkurt, B.; Aguilar, D.; Allen, L.A.; Byun, J.J.; Colvin, M.M.; Deswal, A.; Drazner, M.H.; Dunlay, S.M.; Evers, L.R.; et al. 2022 AHA/ACC/HFSA Guideline for the Management of Heart Failure: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 2022, 145, e895–e1032. [Google Scholar] [CrossRef]
- Zamora, E.; González, B.; Lupón, J.; Borrellas, A.; Domingo, M.; Santiago-Vacas, E.; Cediel, G.; Codina, P.; Rivas, C.; Pulido, A.; et al. Quality of life in patients with heart failure and improved ejection fraction: One-year changes and prognosis. ESC Heart Fail. 2022, 9, 3804–3813. [Google Scholar] [CrossRef]
- Yoshimura, R.; Hayashi, O.; Horio, T.; Fujiwara, R.; Matsuoka, Y.; Yokouchi, G.; Sakamoto, Y.; Matsumoto, N.; Fukuda, K.; Shimizu, M.; et al. The E/e’ ratio on echocardiography as an independent predictor of the improvement of left ventricular contraction in patients with heart failure with reduced ejection fraction. J. Clin. Ultrasound. 2023, 51, 1131–1138. [Google Scholar] [CrossRef]
- Cao, T.H.; Tay, W.T.; Jones, D.J.L.; Cleland, J.G.F.; Tromp, J.; Emmens, J.E.; Teng, T.K.; Chandramouli, C.; Slingsby, O.C.; Anker, S.D.; et al. Heart failure with improved versus persistently reduced left ventricular ejection fraction: A comparison of the BIOSTAT-CHF (European) study with the ASIAN-HF registry. Eur. J. Heart Fail. 2024. Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Lam, C.S.P.; Li, Y.H.; Bayes-Genis, A.; Ariyachaipanich, A.; Huan, D.Q.; Sato, N.; Kahale, P.; Cuong, T.M.; Dong, Y.; Li, X.; et al. The role of N-terminal pro-B-type natriuretic peptide in prognostic evaluation of heart failure. J. Chin. Med. Assoc. 2019, 82, 447–451. [Google Scholar] [CrossRef]
- Liu, D.; Hu, K.; Schregelmann, L.; Hammel, C.; Lengenfelder, B.D.; Ertl, G.; Frantz, S.; Nordbeck, P. Determinants of ejection fraction improvement in heart failure patients with reduced ejection fraction. ESC Heart Fail. 2023, 10, 1358–1371. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Ishizu, T.; Sato, K.; Minami, K.; Terauchi, T.; Nakatsukasa, T.; Kawamatsu, N.; Machino-Ohtsuka, T.; Ieda, M. Longitudinal Changes in Natriuretic Peptides and Reverse Cardiac Remodeling in Patients with Heart Failure Treated with Sacubitril/Valsartan Across the Left Ventricular Ejection Traction Spectrum. Int. Heart J. 2023, 64, 1071–1078. [Google Scholar] [CrossRef]
- Butt, J.H.; Adamson, C.; Docherty, K.F.; de Boer, R.A.; Petrie, M.C.; Inzucchi, S.E.; Kosiborod, M.N.; Maria Langkilde, A.; Lindholm, D.; Martinez, F.A.; et al. Efficacy and Safety of Dapagliflozin in Heart Failure with Reduced Ejection Fraction According to N-Terminal Pro-B-Type Natriuretic Peptide: Insights From the DAPA-HF Trial. Circ. Heart Fail. 2021, 14, e008837. [Google Scholar] [CrossRef]
- Nassif, M.E.; Windsor, S.L.; Tang, F.; Khariton, Y.; Husain, M.; Inzucchi, S.E.; McGuire, D.K.; Pitt, B.; Scirica, B.M.; Austin, B.; et al. Dapagliflozin Effects on Biomarkers, Symptoms, and Functional Status in Patients with Heart Failure with Reduced Ejection Fraction: The DEFINE-HF Trial. Circulation 2019, 140, 1463–1476. [Google Scholar] [CrossRef]
- Martinsson, A.; Oest, P.; Wiborg, M.B.; Reitan, Ö.; Smith, J.G. Longitudinal evaluation of ventricular ejection fraction and NT-proBNP across heart failure subgroups. Scand. Cardiovasc. J. 2018, 52, 205–210. [Google Scholar] [CrossRef]
- Dutta, A.; Das, M.; Ghosh, A.; Rana, S. Molecular and cellular pathophysiology of circulating cardiomyocyte-specific cell free DNA (cfDNA): Biomarkers of heart failure and potential therapeutic targets. Genes. Dis. 2022, 10, 948–959. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ren, J.; Jiang, L.; Liu, X.; Liao, Y.; Zhao, X.; Tang, F.; Yu, H.; Shao, Y.; Wang, J.; Wen, L.; et al. Heart-specific DNA methylation analysis in plasma for the investigation of myocardial damage. J. Transl. Med. 2022, 20, 36. [Google Scholar] [CrossRef]
- Berezin, A. Neutrophil extracellular traps: The core player in vascular complications of diabetes mellitus. Diabetes Metab. Syndr. 2019, 13, 3017–3023. [Google Scholar] [CrossRef]
- Thorsen, S.U.; Moseholm, K.F.; Clausen, F.B. Circulating cell-free DNA and its association with cardiovascular disease: What we know and future perspectives. Curr. Opin. Lipidol. 2024, 35, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Vulesevic, B.; Lavoie, S.S.; Neagoe, P.E.; Dumas, E.; Räkel, A.; White, M.; Sirois, M.G. CRP Induces NETosis in Heart Failure Patients with or without Diabetes. Immunohorizons 2019, 3, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.P.; Cheng, K.; Ning, M.A.; Li, H.H.; Wang, H.C.; Li, F.; Chen, S.Y.; Qu, F.L.; Guo, W.Y. Association between peripheral blood cells mitochondrial DNA content and severity of coronary heart disease. Atherosclerosis 2017, 261, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N.G. The inflammatory response in myocardial injury, repair, and remodelling. Nat. Rev. Cardiol. 2014, 11, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wu, J.; Zhang, X.; Li, X.; Wu, X.; Zhao, Y.; Ren, J. Circulating mitochondrial DNA-triggered autophagy dysfunction via STING underlies sepsis-related acute lung injury. Cell Death Dis. 2021, 12, 673. [Google Scholar] [CrossRef]
- Oommen, S.G.; Man, R.K.; Talluri, K.; Nizam, M.; Kohir, T.; Aviles, M.A.; Nino, M.; Jaisankar, L.G.; Jaura, J.; Wannakuwatte, R.A.; et al. Heart Failure with Improved Ejection Fraction: Prevalence, Predictors, and Guideline-Directed Medical Therapy. Cureus 2024, 16, e61790. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.A.; Kim, S.H.; Lee, K.Y.; Yoon, A.H.; Hwang, B.H.; Choo, E.H.; Kim, J.J.; Choi, I.J.; Kim, C.J.; Lim, S.; et al. Predictors and Long-Term Clinical Impact of Heart Failure with Improved Ejection Fraction After Acute Myocardial Infarction. J. Am. Heart Assoc. 2024, 13, e034920. [Google Scholar] [CrossRef]
| Variables | Entire Patient Cohort (n = 452) | Patients with HFimpEF (n = 177) | Patients with Persistent HFrEF (n = 275) | p-Value |
|---|---|---|---|---|
| Demographic and anthropomorphic parameters | ||||
| Age, year | 59 (50–68) | 59 (52–65) | 60 (49–72) | 0.48 |
| Male/female n (%) | 266 (58.9)/186 (41.2) | 102 (57.6)/75 (42.3) | 164 (59.6)/111 (40.4) | 0.36 |
| BMI, kg/m2 | 25.8 ± 3.5 | 25.1 ± 2.9 | 26.1 ± 2.7 | 0.44 |
| Comorbidities and CV risk factors | ||||
| Dyslipidemia, n (%) | 286 (63.2) | 115 (64.5) | 171 (62.2) | 0.77 |
| Hypertension, n (%) | 71 (15.7) | 28 (15.8) | 43 (15.6) | 0.88 |
| Ischemia-induced cardiomyopathy, n (%) | 141 (31.2) | 44 (24.9) | 97 (35.3) | 0.04 |
| Dilated cardiomyopathy, n (%) | 68 (15.0) | 21 (11.9) | 47 (17.1) | 0.52 |
| AF, n (%) | 137 (30.3) | 47 (26.6) | 90 (32.7) | 0.28 |
| Smoking, n (%) | 168 (37.2) | 65 (36.7) | 103 (37.5) | 0.88 |
| Abdominal obesity, n (%) | 112 (24.8) | 46 (26.0) | 66 (24.0) | 0.87 |
| T2DM, n (%) | 146 (32.3) | 54 (30.5) | 92 (33.5) | 0.26 |
| LVH, n (%) | 316 (69.9) | 120 (67.8) | 196 (71.3) | 0.44 |
| CKD 1–3 grades, n (%) | 132 (29.2) | 45 (25.4) | 87 (31.6) | 0.42 |
| Complete LBBB/RBBB on ECG, n (%) | 98 (21.7) | 35 (19.8) | 63 (22.9) | 0.18 |
| CRT, n (%) | 13 (2.9%) | 5 (2.8%) | 8 (2.9%) | 0.94 |
| NYHA functional classification | ||||
| I/II HF NYHA classes, n (%) | 144 (31.9) | 71 (40.1) | 73 (26.6) | 0.001 |
| III HF NYHA class, n (%) | 230 (50.8) p * = 0.022 | 85 (48.0) p * = 0.48 | 145 (52.7) p * = 0.018 | 0.06 |
| IV HF NYHA class, n (%) | 78 (17.3) p * = 0.01; p ** = 0.024 | 21 (11.9) p * = 0.001; p ** = 0.001 | 57 (20.7) p * = 0.012; p ** = 0.46 | 0.036 |
| Hemodynamic performances | ||||
| SBP, mm Hg | 128 ± 11 | 129 ± 9 | 125 ± 10 | 0.22 |
| DBP, mm Hg | 78 ± 10 | 77 ± 8 | 74 ± 9 | 0.64 |
| LVEDV, mL | 171 (149–192) | 168 (136–188) | 181 (150–202) | 0.04 |
| LVESV, mL | 115 (89–127) | 109 (87–124) | 126 (90–131) | 0.01 |
| LVEF, % | 32 (29–39) | 35 (31–39) | 30 (27–34) | 0.02 |
| LVMMI, g/m2 | 226 ± 15 | 218 ± 15 | 234 ± 13 | 0.46 |
| LAVI, mL/m2 | 46 (39–52) | 44 (35–51) | 47 (39–54) | 0.12 |
| E/e`, unit | 17.3 ± 5.4 | 16.6 ± 4.1 | 19.1 ± 3.3 | 0.56 |
| Biochemistry parameters | ||||
| eGFR, mL/min/1.73 m2 | 72 ± 11 | 80 ± 9 | 65 ± 7 | 0.04 |
| Fasting glucose, mmol/L | 5.11 ± 0.77 | 5.06 ± 0.60 | 5.19 ± 1.1 | 0.66 |
| Creatinine, µmol/L | 99.6 ± 12.8 | 78.9 ± 9.1 | 115.2 ± 8.2 | 0.04 |
| TC, mmol/L | 5.88 ± 0.90 | 5.61 ± 0.52 | 5.92 ± 0.70 | 0.62 |
| HDL-C, mmol/L | 0.97 ± 0.14 | 0.97 ± 0.15 | 0.98 ± 0.18 | 0.68 |
| LDL-C, mmol/L | 3.93 ± 0.18 | 3.80 ± 0.17 | 4.00 ± 0.12 | 0.02 |
| TGs, mmol/L | 1.98 ± 0.17 | 1.90 ± 0.12 | 2.03 ± 0.15 | 0.64 |
| hs-CRP, mg/L | 9.68 (4.31–13.70) | 9.25 (3.45–12.70) | 10.70 (5.80–17.50) | 0.22 |
| TNF-alpha, pg/mL | 3.24 (2.70–3.98) | 3.11 (2.62–3.69) | 3.43 (2.95–4.12) | 0.04 |
| NT-proBNP, pmol/mL | 3228 (1910–5215) | 3015 (1780–5220) | 3290 (1820–5470) | 0.44 |
| cf-nDNA, μmol/L | 11.6 (7.68–15.7) | 9.8 (7.2–12.2) | 14.1 (11.8–16.5) | 0.02 |
| Concomitant medications | ||||
| ACEI, n (%) | 198 (43.8) | 79 (44.6) | 119 (43.3) | 0.88 |
| ARNI, n (%) | 134 (29.6) | 53 (29.9) | 81 (29.5) | 0.90 |
| ARB, n (%) | 86 (19.0) | 35 (19.7) | 51 (18.5) | 0.82 |
| Ivabradine, n (%) | 78 (17.3) | 28 (15.8) | 50 (18.2) | 0.56 |
| Beta-blockers, n (%) | 426 (94.2) | 165 (93.2) | 261 (94.9) | 0.90 |
| Calcium channel blocker, n (%) | 67 (14.8) | 23 (13.0) | 44 (16.0) | 0.44 |
| MRA, n (%) | 405 (89.6) | 161 (91.0) | 244 (88.7) | 0.86 |
| Digoxin, n (%) | 51 (11.3) | 14 (7.9) | 37 (13.5) | 0.010 |
| Loop diuretic, n (%) | 412 (91.2) | 159 (89.8) | 253 (92.0) | 0.46 |
| Antiplatelet, n (%) | 141 (31.2) | 54 (30.5) | 87 (31.6) | 0.84 |
| Anticoagulants, n (%) | 139 (30.8) | 55 (31.1) | 84 (30.5) | 0.82 |
| Metformin, n (%) | 138 (30.5) | 54 (30.5) | 84 (31.0) | 0.86 |
| SGLT2 inhibitors, n (%) | 434 (96.0) | 175 (98.9) | 259 (94.2) | 0.86 |
| Statins, n (%) | 350 (77.4) | 139 (78.5) | 211 (76.7) | 0.88 |
| Dependent Variable: HFimpEF | ||||
|---|---|---|---|---|
| Variables | Univariate Logistic Regression | Multivariate Logistic Regression | ||
| OR (95% CI) | p-Value | OR (95% CI) | p-Value | |
| Ischemia-induced cardiomyopathy (presence vs. absent) | 0.75 (0.62–0.88) | 0.044 | 0.77 (0.60–0.90) | 0.042 |
| IV HF NYHA class | 0.71 (0.57–0.92) | 0.001 | 0.76 (0.63–0.87) | 0.001 |
| T2DM (presence vs. absent) | 0.77 (0.71–0.82) | 0.040 | 0.84 (0.62–0.92) | 0.042 |
| CKD (presence vs. absent) | 0.89 (0.84–0.96) | 0.048 | 0.88 (0.80–0.10) | 0.050 |
| AF (presence vs. absent) | 0.94 (0.80–1.09) | 0.064 | - | |
| LVEDV | 0.93 (0.90–1.01) | 0.052 | - | |
| LAVI | 0.95 (0.92–0.98) | 0.042 | 0.96 (0.90–1.00) | 0.050 |
| E/e` | 0.92 (0.89–0.97) | 0.080 | - | |
| NT-proBNP (≤1940 pmol/mL vs. >1940 pmol/mL) | 1.42 (1.19–1.98) | 0.001 | 1.35 (1.12–1.76) | 0.001 |
| Relative decrease in NT-proBNP levels (>35% vs. ≤35%) from baseline | 1.67 (1.51–1.82) | 0.001 | 1.70 (1.61–1.83) | 0.001 |
| TNF-alpha (≤2.88 pg/mL vs. >2.88 pg/mL) | 1.06 (1.00–1.12) | 0.48 | - | |
| hs-CRP (≤7.02 mg/L vs. >7.02 mg/L) | 1.08 (1.00–1.17) | 0.60 | - | |
| cf-nDNA (≤7.5 μmol/L vs. >7.5 μmol/L) | 1.56 (1.07–2.94) | 0.001 | 1.64 (1.10–2.07) | 0.001 |
| Digoxin (presence vs. absent) | 0.85 (0.72–0.97) | 0.042 | 0.93 (0.86–1.00) | 0.052 |
| Predictive Models | AUC | NRI | IDI | |||
|---|---|---|---|---|---|---|
| M (95% CI) | p-Value | M (95% CI) | p-Value | M (95% CI) | p-Value | |
| Model 1 (ischemia-induced CMP) | 0.766 (0.712–0.836) | - | Reference | - | Reference | - |
| Model 2 (IV NYHA class) | 0.771 (0.720–0.811) | 0.260 | 0.12 (0.10–0.15) | 0.360 | 0.11 (0.09–0.13) | 0.520 |
| Model 3 (NT-proBNP ≤ 1940 pmol/mL) | 0.783 (0.700–0.840) | 0.144 | 0.18 (0.12–0.23) | 0.196 | 0.17 (0.12–0.23) | 0.280 |
| Model 3 (relative decrease in NT-proBNP levels ≤ 35% from baseline) | 0.795 (0.745–0.861) | 0.06 | 0.23 (0.17–0.30) | 0.170 | 0.21 (0.18–0.25) | 0.240 |
| Model 4 (cf-nDNA ≤ 7.5 μmol/L) | 0.875 (0.795–0.950) | 0.001 | 0.54 (0.43–0.67) | 0.001 | 0.51 (0.45–0.58) | 0.001 |
| Model 5 (NT-proBNP levels ≤ 1940 pmol/mL + cf-nDNA) | 0.872 (0.820–0.941) | 0.001 | 0.48 (0.42–0.55) | 0.001 | 0.49 (0.41–0.56) | 0.001 |
| Model 6 (relative decrease in NT-proBNP levels ≤ 35% from baseline + cf-nDNA) | 0.893 (0.844–0.962) | 0.001 | 0.58 (0.45–0.72) | 0.001 | 0.55 (0.49–0.62) | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berezina, T.; Berezin, O.O.; Lichtenauer, M.; Berezin, A.E. Circulating Cell-Free Nuclear DNA Predicted an Improvement of Systolic Left Ventricular Function in Individuals with Chronic Heart Failure with Reduced Ejection Fraction. Cardiogenetics 2024, 14, 183-197. https://doi.org/10.3390/cardiogenetics14040014
Berezina T, Berezin OO, Lichtenauer M, Berezin AE. Circulating Cell-Free Nuclear DNA Predicted an Improvement of Systolic Left Ventricular Function in Individuals with Chronic Heart Failure with Reduced Ejection Fraction. Cardiogenetics. 2024; 14(4):183-197. https://doi.org/10.3390/cardiogenetics14040014
Chicago/Turabian StyleBerezina, Tetiana, Oleksandr O. Berezin, Michael Lichtenauer, and Alexander E. Berezin. 2024. "Circulating Cell-Free Nuclear DNA Predicted an Improvement of Systolic Left Ventricular Function in Individuals with Chronic Heart Failure with Reduced Ejection Fraction" Cardiogenetics 14, no. 4: 183-197. https://doi.org/10.3390/cardiogenetics14040014
APA StyleBerezina, T., Berezin, O. O., Lichtenauer, M., & Berezin, A. E. (2024). Circulating Cell-Free Nuclear DNA Predicted an Improvement of Systolic Left Ventricular Function in Individuals with Chronic Heart Failure with Reduced Ejection Fraction. Cardiogenetics, 14(4), 183-197. https://doi.org/10.3390/cardiogenetics14040014





