Epidemiological Features of Leptospirosis and Identification of Leptospira wolffii as a Persistently Prevailing Species in North–Central Bangladesh
Abstract
:1. Introduction
2. Materials and Methods
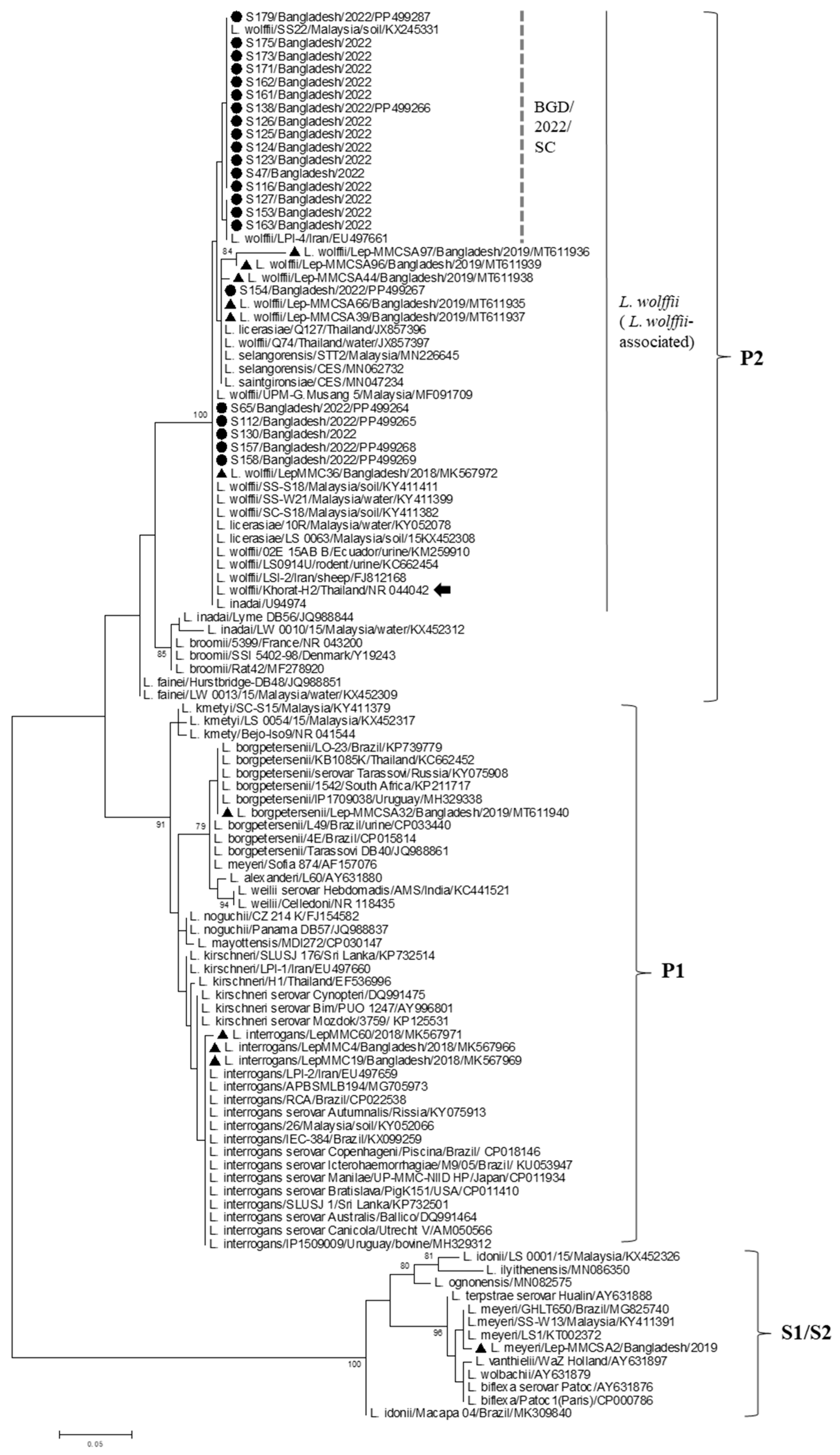
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Costa, F.; Hagan, J.E.; Calcagno, J.; Kane, M.; Torgerson, P.; Martinez-Silveira, M.S.; Stein, C.; Abela-Ridder, B.; Ko, A.I. Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl. Trop. Dis. 2015, 9, e0003898. [Google Scholar] [CrossRef] [PubMed]
- Karpagam, K.B.; Ganesh, B. Leptospirosis: A neglected tropical zoonotic infection of public health importance-an updated review. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Guerra, M.A. Leptospirosis: Public health perspectives. Biologicals 2013, 41, 295–297. [Google Scholar] [CrossRef] [PubMed]
- Izurieta, R.; Galwankar, S.; Clem, A. Leptospirosis: The “mysterious” mimic. J. Emerg. Trauma. Shock. 2008, 1, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Vinetz, J.M. Leptospirosis. Curr. Opin. Infect. Dis. 2001, 4, 527–538. [Google Scholar] [CrossRef]
- Fernandes, L.G.V.; Stone, N.E.; Roe, C.C.; Goris, M.G.A.; van der Linden, H.; Sahl, J.W.; Wagner, D.M.; Nally, J.E. Leptospira sanjuanensis sp. nov., a pathogenic species of the genus Leptospira isolated from soil in Puerto Rico. Int. J. Syst. Evol. Microbiol. 2022, 72, 005560. [Google Scholar] [CrossRef]
- Ca Ferreira, L.; de Fa Ferreira Filho, L.; Cosate, M.R.V.; Sakamoto, T. Genetic structure and diversity of the rfb locus of pathogenic species of the genus Leptospira. Life Sci. Alliance 2024, 7, e202302478. [Google Scholar] [CrossRef]
- Vincent, A.T.; Schiettekatte, O.; Goarant, C.; Neela, V.K.; Bernet, E.; Thibeaux, R.; Ismail, N.; Khalid, M.K.N.M.; Amran, F.; Masuzawa, T.; et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019, 13, e0007270. [Google Scholar] [CrossRef]
- Ko, A.I.; Goarant, C.; Picardeau, M. Leptospira: The dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 2009, 7, 736–747. [Google Scholar] [CrossRef]
- Caimi, K.; Ruybal, P. Leptospira spp., a genus in the stage of diversity and genomic data expansion. Infect. Genet. Evol. 2020, 81, 104241. [Google Scholar] [CrossRef]
- Guglielmini, J.; Bourhy, P.; Schiettekatte, O.; Zinini, F.; Brisse, S.; Picardeau, M. Genus-wide Leptospira core genome multilocus sequence typing for strain taxonomy and global surveillance. PLoS Negl. Trop. Dis. 2019, 13, e0007374. [Google Scholar] [CrossRef] [PubMed]
- Morshed, M.G.; Konishi, H.; Terada, Y.; Arimitsu, Y.; Nakazawa, T. Seroprevalence of leptospirosis in a rural flood prone district of Bangladesh. Epidemiol. Infect. 1994, 112, 527–531. [Google Scholar] [CrossRef] [PubMed]
- LaRocque, R.C.; Breiman, R.F.; Ari, M.D.; Morey, R.E.; Janan, F.A.; Hayes, J.M.; Hossain, M.A.; Brooks, W.A.; Levett, P.N. Leptospirosis during dengue outbreak, Bangladesh. Emerg. Infect. Dis. 2005, 11, 766–769. [Google Scholar] [CrossRef] [PubMed]
- Kendall, E.A.; Galloway, R.; Breiman, R.F.; Bui, D.M.; Brooks, W.A.; Goswami, D.; LaRocque, R.C.; Ari, M.D.; Luby, S. Leptospirosis as a cause of fever in urban Bangladesh. Am. J. Trop. Med. Hyg. 2010, 82, 1127–1130. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.; Aung, M.; Paul, S.; Ahmed, S.; Haque, N.; Roy, S.; Al Amin, M.; Paul, A.; Miah, M.; Alam, M.; et al. First molecular identification of two Leptospira species (Leptospira interrogans and Leptospira wolffii) in Bangladesh. New Microbes New Infect. 2019, 31, 100570. [Google Scholar] [CrossRef]
- Rahman, S.; Paul, S.; Aung, M.; Ahmed, S.; Haque, N.; Raisul, M.; Choity, J.; Nila, S.; Ara, H.; Roy, S.; et al. Predominance of Leptospira wolffii in north-central Bangladesh, 2019. New Microbes New Infect. 2020, 38, 100765. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Rahman, M.Z.; Banu, S.; Rahman, M.; Chisti, M.J.; Chowdhury, F.; Akhtar, Z.; Palit, A.; Martin, D.W.; Ul Anwar, M.; et al. Acute febrile illness among outpatients seeking health care in Bangladeshi hospitals prior to the COVID-19 pandemic. PLoS ONE 2022, 17, e0273902. [Google Scholar] [CrossRef] [PubMed]
- Djadid, N.D.; Ganji, Z.F.; Gouya, M.M.; Rezvani, M.; Zakeri, S. A simple and rapid nested polymerase chain reaction-restriction fragment length polymorphism technique for differentiation of pathogenic and nonpathogenic Leptospira spp. Diagn. Microbiol. Infect. Dis. 2009, 63, 251–256. [Google Scholar] [CrossRef]
- Palaniappan, R.U.; Chang, Y.-F.; Chang, C.-F.; Pan, M.; Yang, C.; Harpending, P.; McDonough, S.P.; Dubovi, E.; Divers, T.; Qu, J.; et al. Evaluation of lig-based conventional and real time PCR for the detection of pathogenic leptospires. Mol. Cell Probes 2005, 19, 111–117. [Google Scholar] [CrossRef]
- Vijayachari, P.; Sugunan, A.P.; Shriram, A.N. Leptospirosis: An emerging global public health problem. J. Biosci. 2008, 33, 557–569. [Google Scholar] [CrossRef]
- Gupta, N.; Wilson, W.; Ravindra, P.; Raghu, R.; Saravu, K. Coinfection of leptospirosis and coronavirus disease 2019: A retrospective case series from a coastal region in South India. J. Med. Virol. 2022, 94, 4508–4511. [Google Scholar] [CrossRef] [PubMed]
- Del Valle-Mendoza, J.; Palomares-Reyes, C.; Carrillo-Ng, H.; Tarazona-Castro, Y.; Kym, S.; Aguilar-Luis, M.A.; del Valle, L.J.; Aquino-Ortega, R.; Martins-Luna, J.; Peña-Tuesta, I.; et al. Leptospirosis in febrile patients with suspected diagnosis of dengue fever. BMC Res. Notes 2021, 14, 209. [Google Scholar] [CrossRef] [PubMed]
- Haider, N.; Asaduzzaman; Hasan, M.N.; Rahman, M.; Sharif, A.R.; Ashrafi, S.A.A.; Lee, S.S.; Zumla, A. Bangladesh’s 2023 Dengue outbreak—Age/gender-related disparity in morbidity and mortality and geographic variability of epidemic burdens. Int. J. Infect. Dis. 2023, 136, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, R.; Das, A.; Premlatha, M.; Choudhary, A.; Chourasia, B.; Chandel, D.; Dey, A. Serological & molecular approaches for diagnosis of leptospirosis in a tertiary care hospital in north India: A 10-year study. Indian. J. Med. Res. 2013, 137, 785–790. [Google Scholar] [PubMed]
- Balasundaram, P.K.; Kanakamma, L.G.; Jayageetha, K.; Selvarajan, B. Epidemiological, clinical and laboratory features of leptospirosis compared to other acute febrile illnesses. J. R. Coll. Physicians Edinb. 2020, 50, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Rafizah, A.N.; Aziah, B.; Azwany, Y.; Imran, M.K.; Rusli, A.M.; Nazri, S.M.; Nikman, A.M.; Nabilah, I.; Asma’, H.S.; Zahiruddin, W.; et al. A hospital-based study on seroprevalence of leptospirosis among febrile cases in northeastern Malaysia. Int. J. Infect. Dis. 2013, 17, e394–e397. [Google Scholar] [CrossRef] [PubMed]
- Wangrangsimakul, T.; Althaus, T.; Mukaka, M.; Kantipong, P.; Wuthiekanun, V.; Chierakul, W.; Blacksell, S.D.; Day, N.P.; Laongnualpanich, A.; Paris, D.H. Causes of acute undifferentiated fever and the utility of biomarkers in Chiangrai, northern Thailand. PLoS Negl. Trop. Dis. 2018, 12, e0006477. [Google Scholar] [CrossRef] [PubMed]
- Allan, K.J.; Biggs, H.M.; Halliday, J.E.B.; Kazwala, R.R.; Maro, V.P.; Cleaveland, S.; Crump, J.A. Epidemiology of Leptospirosis in Africa: A Systematic Review of a Neglected Zoonosis and a Paradigm for ‘One Health’ in Africa. PLoS Negl. Trop. Dis. 2015, 9, e0003899. [Google Scholar] [CrossRef] [PubMed]
- Thipmontree, W.; Suputtamongkol, Y.; Tantibhedhyangkul, W.; Suttinont, C.; Wongswat, E.; Silpasakorn, S. Human leptospirosis trends: Northeast Thailand, 2001-2012. Int. J. Environ. Res. Public Health 2014, 11, 8542–8551. [Google Scholar] [CrossRef]
- Samrot, A.V.; Sean, T.C.; Bhavya, K.S.; Sahithya, C.S.; Chan-Drasekaran, S.; Palanisamy, R.; Robinson, E.R.; Subbiah, S.K.; Mok, P.L. Leptospiral Infection, Pathogenesis and Its Diagnosis-A Review. Pathogens 2021, 10, 145. [Google Scholar] [CrossRef]
- Musso, D.; La Scola, B. Laboratory diagnosis of leptospirosis: A challenge. J. Microbiol. Immunol. Infect. 2013, 46, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, C.D.A.; de Freitas, V.L.T.; Romero, E.C.; Spinosa, C.; Sanches, M.C.A.; Da Silva, M.V.; Shikanai-Yasuda, M.A. Polymerase chain reaction in comparison with serological tests for early diagnosis of human leptospirosis. Trop. Med. Int. Health 2006, 11, 1699–1707. [Google Scholar] [CrossRef] [PubMed]
- Krijger, I.M.; Ahmed, A.A.A.; Goris, M.G.A.; Groot Koerkamp, P.W.G.; Meerburg, B.G. Prevalence of Leptospira Infection in Rodents from Bangladesh. Int. J. Environ. Res. Public Health 2019, 16, 2113. [Google Scholar] [CrossRef] [PubMed]
- Slack, A.T.; Kalambaheti, T.; Symonds, M.L.; Dohnt, M.F.; Galloway, R.L.; Steigerwalt, A.G.; Chaicumpa, W.; Bunyaraksyotin, G.; Craig, S.; Harrower, B.J.; et al. Leptospira wolffii sp. nov., isolated from a human with suspected leptospirosis in Thailand. Int. J. Syst. Evol. Microbiol. 2008, 58 Pt 10, 2305–2308. [Google Scholar] [CrossRef] [PubMed]
- Lata, K.S.; Kumar, S.; Vindal, V.; Patel, S.; Das, J. A core and pan gene map of Leptospira genus and its interactions with human host. Microb. Pathog. 2022, 162, 105347. [Google Scholar] [CrossRef] [PubMed]
- Philip, N.; Affendy, N.B.; Ramli, S.N.A.; Arif, M.; Raja, P.; Nagandran, E.; Renganathan, P.; Taib, N.M.; Masri, S.N.; Yuhana, M.Y.; et al. Leptospira interrogans and Leptospira kirschneri are the dominant Leptospira species causing human leptospirosis in Central Malaysia. PLoS Negl. Trop. Dis. 2020, 14, e0008197. [Google Scholar] [CrossRef] [PubMed]
- Philip, N.; Ahmed, K. Leptospirosis in Malaysia: Current status, insights, and future prospects. J. Physiol. Anthropol. 2023, 42, 30. [Google Scholar] [CrossRef]
- Balamurugan, V.; Gangadhar, N.L.; Mohandoss, N.; Thirumalesh, S.R.A.; Dhar, M.; Shome, R.; Krishnamoorthy, P.; Prabhudas, K.; Rahman, H. Characterization of leptospira isolates from animals and humans: Phylogenetic analysis identifies the prevalence of intermediate species in India. Springerplus 2013, 2, 362. [Google Scholar] [CrossRef]
- Zakeri, S.; Sepahian, N.; Afsharpad, M.; Esfandiari, B.; Ziapour, P.; Djadid, N.D. Molecular epidemiology of leptospirosis in northern Iran by nested polymerase chain reaction/restriction fragment length polymorphism and sequencing methods. Am. J. Trop. Med. Hyg. 2010, 82, 899–903. [Google Scholar] [CrossRef]
- Zakeri, S.; Khorami, N.; Ganji, Z.F.; Sepahian, N.; Malmasi, A.-A.; Gouya, M.M.; Djadid, N.D. Leptospira wolffii, a potential new pathogenic Leptospira species detected in human, sheep and dog. Infect. Genet. Evol. 2010, 10, 273–277. [Google Scholar] [CrossRef]
- Chiani, Y.; Jacob, P.; Varni, V.; Landolt, N.; Schmeling, M.F.; Pujato, N.; Caimi, K.; Vanasco, B. Isolation and clinical sample typing of human leptospirosis cases in Argentina. Infect. Genet. Evol. 2016, 37, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Eo, K.Y.; Lee, W.S.; Kimura, J.; Yamamoto, N. DNA-based detection of Leptospira wolffii, Giardia intestinalis and Toxoplasma gondii in environmental feces of wild animals in Korea. J. Vet. Med. Sci. 2021, 83, 850–854. [Google Scholar] [CrossRef] [PubMed]
- Chiriboga, J.; Barragan, V.; Arroyo, G.; Sosa, A.; Birdsell, D.N.; España, K.; Mora, A.; Espín, E.; Mejía, M.E.; Morales, M.; et al. High Prevalence of Intermediate Leptospira spp. DNA in Febrile Humans from Urban and Rural Ecuador. Emerg. Infect. Dis. 2015, 21, 2141–2147. [Google Scholar] [CrossRef] [PubMed]
- Azali, M.A.; Yean Yean, C.; Harun, A.; Aminuddin Baki, N.N.; Ismail, N. Molecular characterization of Leptospira spp. in environmental samples from North-Eastern Malaysia revealed a pathogenic strain, Leptospira alstonii. J. Trop. Med. 2016, 2016, 2060241. [Google Scholar] [CrossRef]
- Pui, C.F.; Bilung, L.M.; Apun, K.; Su’ut, L. Diversity of Leptospira spp. in rats and environment from urban areas of Sarawak, Malaysia. J. Trop. Med. 2017, 2017, 3760674. [Google Scholar] [CrossRef] [PubMed]
- Suwanpakdee, S.; Sangkachai, N.; Chamsai, T.; Taruyanon, K.; Thongdee, M. Potentially Pathogenic Leptospira Species Isolated from a Waterfall in Thailand. Jpn. J. Infect. Dis. 2018, 71, 65–67. [Google Scholar] [CrossRef]
- Thibeaux, R.; Girault, D.; Bierque, E.; Soupé-Gilbert, M.-E.; Rettinger, A.; Douyère, A.; Meyer, M.; Iraola, G.; Picardeau, M.; Goarant, C. Biodiversity of Environmental Leptospira: Improving Identification and Revisiting the Diagnosis. Front. Microbiol. 2018, 9, 816. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Zanzi, C.; Groene, E.; Morawski, B.M.; Bonner, K.; Costa, F.; Bertherat, E.; Schneider, M.C. A systematic literature review of leptospirosis outbreaks worldwide, 1970–2012. Rev. Panam. Salud Publica 2020, 44, e78. [Google Scholar] [CrossRef]
- Gupta, N.; Wilson, W.; Ravindra, P. Leptospirosis in India: A systematic review and meta-analysis of clinical profile, treatment and outcomes. Infez. Med. 2023, 31, 290–305. [Google Scholar]
- Ferdouse, F.; Hossain, M.A.; Paul, S.K.; Ahmed, S.; Mahmud, C.; Ahmed, R.; Haque, A.F.; Khan, M.N.-A.; Ghosh, S.; Urushibara, N.; et al. Rickettsia felis Infection among Humans, Bangladesh, 2012-2013. Emerg Infect Dis 2015, 21, 1483–1485. [Google Scholar] [CrossRef]
- Al Amin, M.; Paul, S.; Aung, M.; Paul, A.; Aziz, M.; Khan, N.; Haque, A.; Ahamed, F.; Melan, A.; Sarker, S.; et al. Molecular characterization of Orientia tsutsugamushi causing scrub typhus among febrile patients in north-central Bangladesh. New Microbes New Infect. 2019, 32, 100595. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S. Recent trends in the climate of Bangladesh. Clim. Res. 2010, 42, 185–193. [Google Scholar] [CrossRef]
- Cunha, M.; Costa, F.; Ribeiro, G.S.; Carvalho, M.S.; Reis, R.B.; Nery, N., Jr.; Pischel, L.; Gouveia, E.L.; Santos, A.C.; Queiroz, A.; et al. Rainfall and other meteorological factors as drivers of urban transmission of leptospirosis. PLoS Negl. Trop. Dis. 2022, 16, e0007507. [Google Scholar] [CrossRef] [PubMed]
- Chadsuthi, S.; Chalvet-Monfray, K.; Geawduanglek, S.; Wongnak, P.; Cappelle, J. Spatial-temporal patterns and risk factors for human leptospirosis in Thailand, 2012-2018. Sci. Rep. 2022, 12, 5066. [Google Scholar] [CrossRef] [PubMed]
- Mojid, M.A. Climate change-induced challenges to sustainable development in Bangladesh. IOP Conf. Ser. Earth Environ. Sci. 2020, 423, 012001. [Google Scholar] [CrossRef]
- Baharom, M.; Ahmad, N.; Hod, R.; Ja’afar, M.H.; Arsad, F.S.; Tangang, F.; Ismail, R.; Mohamed, N.; Radi, M.F.M.; Osman, Y. Environmental and Occupational Factors Associated with Leptospirosis: A Systematic Review. Heliyon 2023, 10, e23473. [Google Scholar] [CrossRef]

| Duration of Fever | Number of Samples | Number of Positive Sample (%) | ||
|---|---|---|---|---|
| IgM LAT | IgM ELISA | Nested PCR | ||
| 5–10 days | 81 | 29 (35.8%) | 31 (38.3%) | 44 (54.3%) * |
| 11–15 days | 67 | 36 (53.7%) | 34 (50.8%) | 32 (47.8%) |
| 16–20 days | 30 | 04 (13.3%) | 04 (13.3%) | 02 (6.7%) |
| >20 days | 8 | 02 (25%) | 0 | 0 |
| Total | 186 | 71 (38.2%) | 69 (37.1%) | 78 (41.9%) |
| Sociodemographic Variables | Number of Cases (%) | ||
|---|---|---|---|
| Total (n = 186) | Leptospirosis (n = 88) | Non-Leptospirosis (n = 98) | |
| Gender | |||
| male/female | 115 (61.8%)/ 71 (38.2%) | 60 (68.2%)/ 28 (31.2%) | 55 (56.1%)/ 43 (43.9%) |
| Age range | |||
| 0–15 | 3 (1.6%) | 1 (1.1%) | 2 (2.0%) |
| 16–30 | 65 (34.9%) | 34 (38.6%) | 31 (31.6%) |
| 31–45 | 54 (29.0%) | 28 (31.8%) | 26 (26.5%) |
| 46–60 | 45 (24.2%) | 15 (17.0%) * | 30 (30.6%) |
| >61 | 19 (10.2%) | 10 (11.4%) | 9 (9.2%) |
| Locality | |||
| Rural | 130 (69.9%) | 71 (80.7%) * | 59 (60.2%) |
| Urban | 56 (30.1%) | 17 (19.3%) | 39 (39.8%) |
| Educational level | |||
| No education | 36 (19.4%) | 15 (17.0%) | 21 (21.4%) |
| Primary education | 70 (37.5%) | 36 (40.9%) | 34 (34.7%) |
| Secondary education | 52 (28.0%) | 25 (28.4%) | 27 (27.6%) |
| Higher education | 28 (15.1%) | 12 (13.6%) | 16 (16.3%) |
| Occupation | |||
| Farmers | 84 (45.2%) | 36 (40.9%) | 48 (49.0%) |
| Home-maker | 28 (15.1%) | 15 (17.0%) | 13 (13.3%) |
| Day laborer | 26 (14.0%) | 18 (20.5%) * | 8 (8.2%) |
| Student | 19 (10.2%) | 8 (9.1%) | 11 (11.2%) |
| Others | 29 (15.6%) | 11 (12.5%) | 18 (18.4%) |
| Seasonal variation | |||
| November–December | 25 (13.4%) | 13 (14.8%) | 12 (12.2%) |
| January–February | 22 (11.8%) | 11 (12.5%) | 11 (11.2%) |
| March–April | 50 (26.9%) | 25 (28.4%) | 25 (25.5%) |
| May–June | 89 (47.8%) | 39 (44.3%) | 50 (51.0%) |
| Clinical Characteristics | Number of Cases (%) | |
|---|---|---|
| Leptospirosis (n = 88) | Non-Leptospirosis (n = 98) | |
| Symptoms | ||
| Fever | 88 (100%) | 98 (100%) |
| Myalgia | 74 (84.1%) | 70 (71.4%) |
| Jaundice | 62 (70.5%) * | 55 (56.1%) |
| Headache | 55 (62.5%) * | 40 (40.8%) |
| Anorexia | 50 (56.8%) | 42 (42.9%) |
| Abdominal pain | 34 (38.6%) | 30 (30.6%) |
| Cough | 30 (34.1%) | 24 (24.5%) |
| Oliguria | 28 (31.8%) * | 17 (17.3%) |
| Hepato-splenomegaly | 18 (20.5%) | 14 (14.3%) |
| Conjunctival suffusion | 14 (15.9%) | 8 (8.2%) |
| Skin rash | 11 (12.5%) | 8 (8.2%) |
| Sign of meningitis | 4 (4.5%) | 2 (2.0%) |
| Laboratory findings | ||
| Raised serum bilirubin (>1 mg/dL) | 62 (70.5%) * | 50 (51.0%) |
| Raised serum creatinine (>1.2 mg/dL) | 30 (34.1%) * | 12 (12.2%) |
| Thrombocytopenia (< 150,000/mL) | 22 (25.0%) | 15 (15.3%) |
| Raised ALT (>40 IU/L) | 45 (51.1%) | 38 (38.8%) |
| Raised AST (>38 IU/L) | 38 (43.2%) | 32 (32.7%) |
| Leukocytosis (>11,000/mm3) | 35 (39.8%) | 30 (30.6%) |
| Proteinuria (>150 mg/day) | 22 (25.0%) | 15 (15.3%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sultana, M.; Paul, S.K.; Nasreen, S.A.; Haque, N.; Hasan, M.K.; Islam, A.; Nila, S.S.; Jahan, A.; Sathi, F.A.; Hossain, T.; et al. Epidemiological Features of Leptospirosis and Identification of Leptospira wolffii as a Persistently Prevailing Species in North–Central Bangladesh. Infect. Dis. Rep. 2024, 16, 638-649. https://doi.org/10.3390/idr16040049
Sultana M, Paul SK, Nasreen SA, Haque N, Hasan MK, Islam A, Nila SS, Jahan A, Sathi FA, Hossain T, et al. Epidemiological Features of Leptospirosis and Identification of Leptospira wolffii as a Persistently Prevailing Species in North–Central Bangladesh. Infectious Disease Reports. 2024; 16(4):638-649. https://doi.org/10.3390/idr16040049
Chicago/Turabian StyleSultana, Monira, Shyamal Kumar Paul, Syeda Anjuman Nasreen, Nazia Haque, Md. Kamrul Hasan, Arup Islam, Sultana Shabnam Nila, Afsana Jahan, Fardousi Akter Sathi, Tasmia Hossain, and et al. 2024. "Epidemiological Features of Leptospirosis and Identification of Leptospira wolffii as a Persistently Prevailing Species in North–Central Bangladesh" Infectious Disease Reports 16, no. 4: 638-649. https://doi.org/10.3390/idr16040049
APA StyleSultana, M., Paul, S. K., Nasreen, S. A., Haque, N., Hasan, M. K., Islam, A., Nila, S. S., Jahan, A., Sathi, F. A., Hossain, T., Ferdaus, S. J., Aung, M. S., & Kobayashi, N. (2024). Epidemiological Features of Leptospirosis and Identification of Leptospira wolffii as a Persistently Prevailing Species in North–Central Bangladesh. Infectious Disease Reports, 16(4), 638-649. https://doi.org/10.3390/idr16040049






