Predatory Bacteria in the Treatment of Infectious Diseases and Beyond
Abstract
:1. Introduction
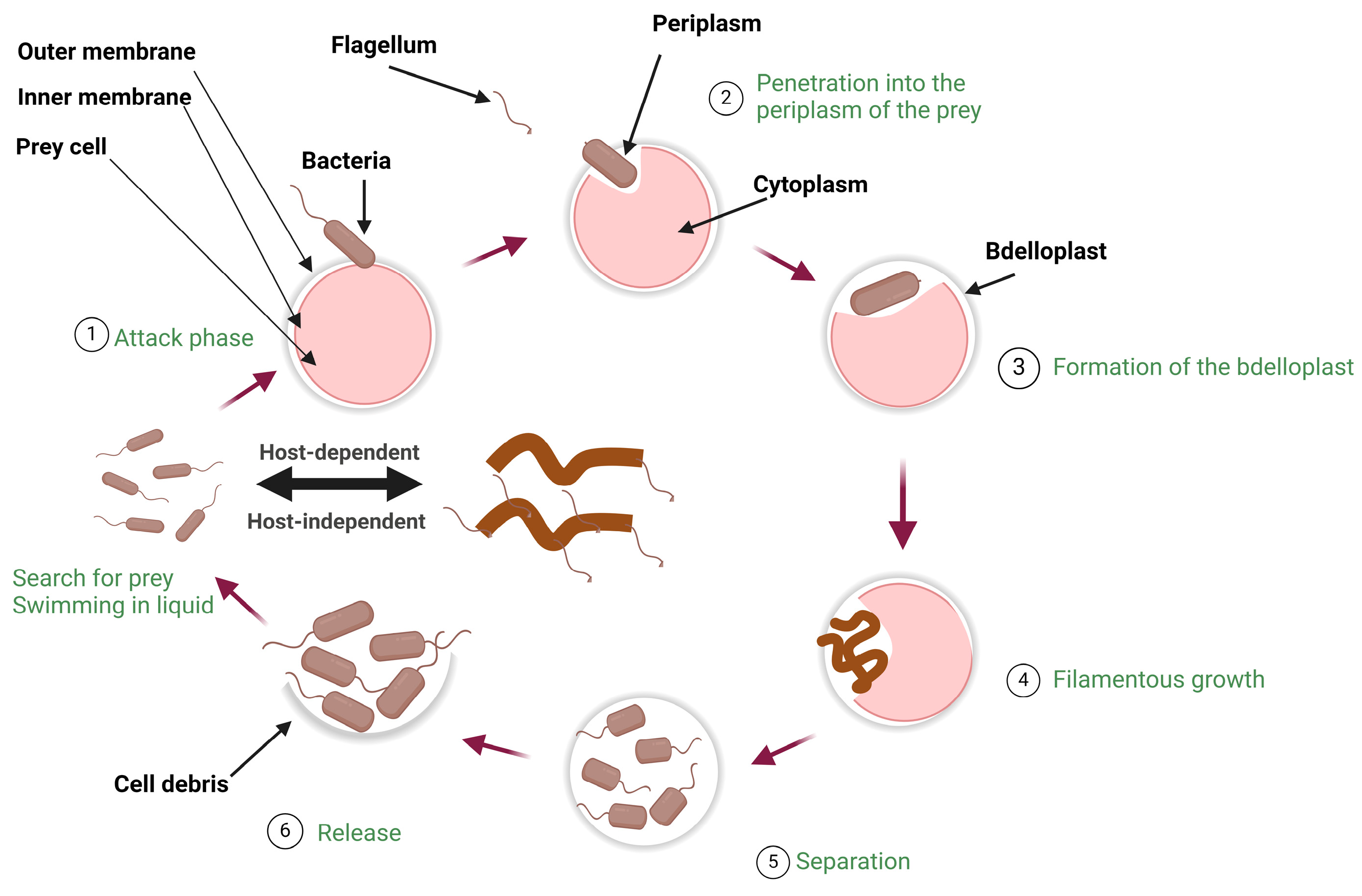
2. Biology of Predatory Bacteria
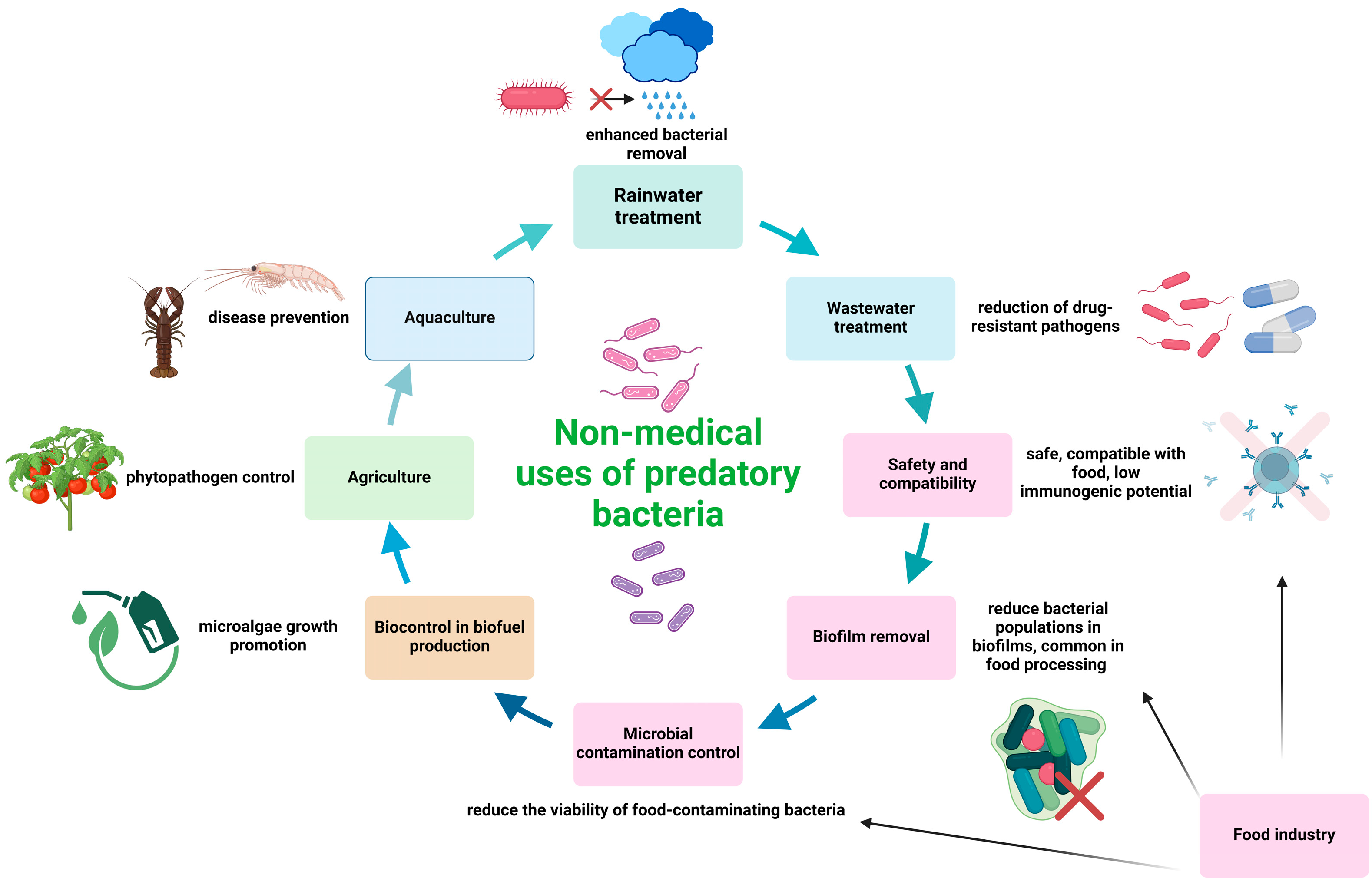
3. Non-Medical Applications of Predatory Bacteria
3.1. Food Industry
3.2. Biocontrol
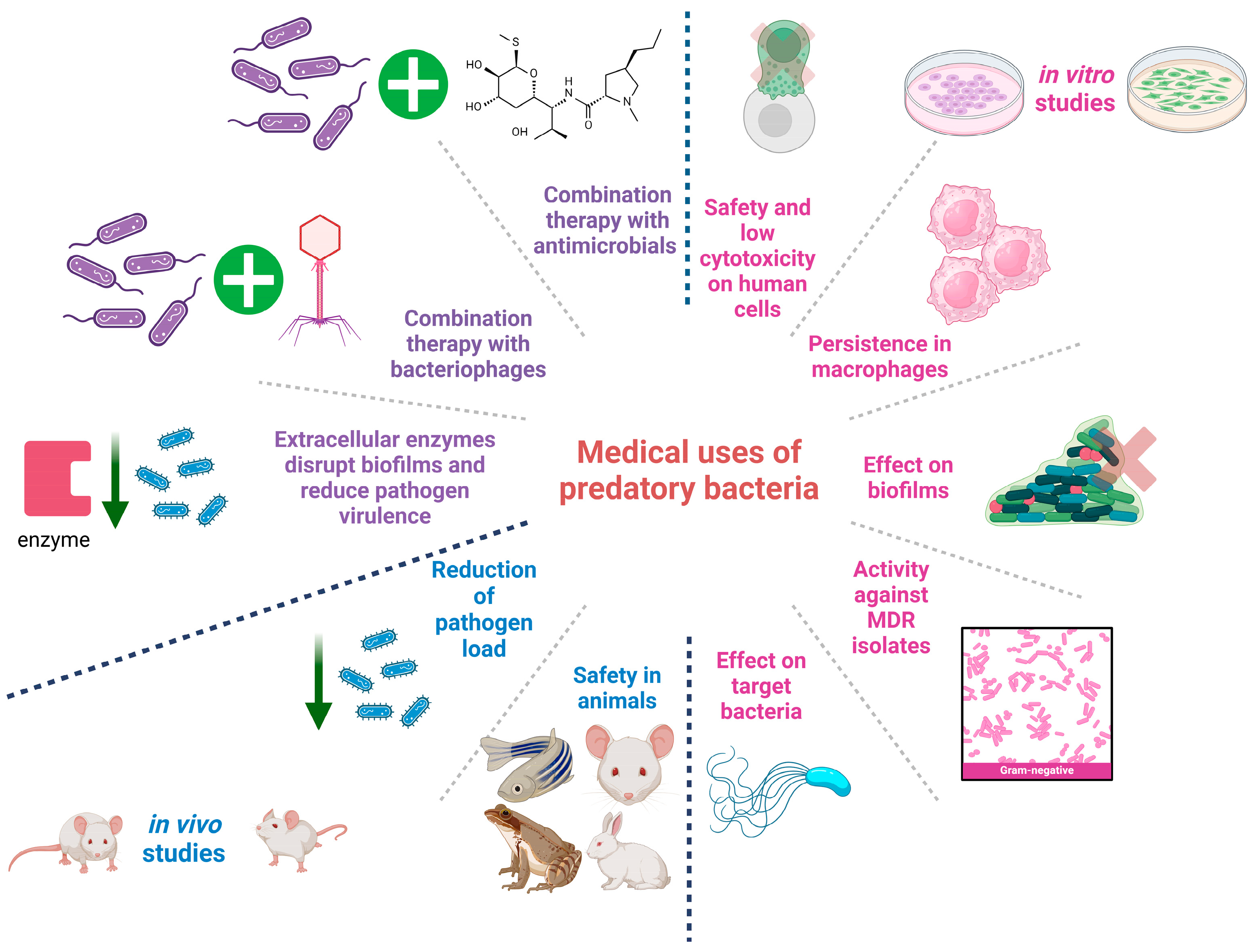
4. Medical Applications of Predatory Bacteria
4.1. In Vitro Studies
4.2. In Vivo Studies
4.3. Predatory Bacteria Metabolites as Antimicrobials
4.4. Combining Treatment with Predatory Bacteria
5. Conclusions and Future Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abraham, E.P.; Chain, E. An Enzyme from Bacteria Able to Destroy Penicillin. 1940. Rev. Infect. Dis. 1988, 10, 677–678. [Google Scholar] [PubMed]
- Lobanovska, M.; Pilla, G. Penicillin’s Discovery and Antibiotic Resistance: Lessons for the Future? Yale J. Biol. Med. 2017, 90, 135–145. [Google Scholar]
- Spellberg, B.; Guidos, R.; Gilbert, D.; Bradley, J.; Boucher, H.W.; Scheld, W.M.; Bartlett, J.G.; Edwards, J.; Infectious Diseases Society of America. The Epidemic of Antibiotic-Resistant Infections: A Call to Action for the Medical Community from the Infectious Diseases Society of America. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2008, 46, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Long, X.; Wang, X.; Li, L.; Mao, D.; Luo, Y.; Ren, H. Global Trend of Antimicrobial Resistance in Common Bacterial Pathogens in Response to Antibiotic Consumption. J. Hazard. Mater. 2023, 442, 130042. [Google Scholar] [CrossRef] [PubMed]
- Karakonstantis, S.; Kritsotakis, E.I.; Gikas, A. Pandrug-Resistant Gram-Negative Bacteria: A Systematic Review of Current Epidemiology, Prognosis and Treatment Options. J. Antimicrob. Chemother. 2020, 75, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Kasiakou, S.K. Colistin: The Revival of Polymyxins for the Management of Multidrug-Resistant Gram-Negative Bacterial Infections. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2005, 40, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Samson, I. A New Class of Antimycobacterial Drugs: The Diarylquinolines. Thorax 2005, 60, 495. [Google Scholar] [CrossRef]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A New Antibiotic Kills Pathogens without Detectable Resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef]
- Hover, B.M.; Kim, S.-H.; Katz, M.; Charlop-Powers, Z.; Owen, J.G.; Ternei, M.A.; Maniko, J.; Estrela, A.B.; Molina, H.; Park, S.; et al. Culture-Independent Discovery of the Malacidins as Calcium-Dependent Antibiotics with Activity against Multidrug-Resistant Gram-Positive Pathogens. Nat. Microbiol. 2018, 3, 415–422. [Google Scholar] [CrossRef]
- Butler, M.S.; Henderson, I.R.; Capon, R.J.; Blaskovich, M.A.T. Antibiotics in the Clinical Pipeline as of December 2022. J. Antibiot. 2023, 76, 431–473. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, P.; Baliou, S.; Samonis, G. Bacteriophages in Infectious Diseases and Beyond—A Narrative Review. Antibiotics 2023, 12, 1012. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, P.; Baliou, S.; Kofteridis, D.P. Antimicrobial Peptides in Infectious Diseases and Beyond-A Narrative Review. Life 2023, 13, 1651. [Google Scholar] [CrossRef]
- Ioannou, P.; Baliou, S.; Samonis, G. Nanotechnology in the Diagnosis and Treatment of Antibiotic-Resistant Infections. Antibiotics 2024, 13, 121. [Google Scholar] [CrossRef] [PubMed]
- Hungate, B.A.; Marks, J.C.; Power, M.E.; Schwartz, E.; van Groenigen, K.J.; Blazewicz, S.J.; Chuckran, P.; Dijkstra, P.; Finley, B.K.; Firestone, M.K.; et al. The Functional Significance of Bacterial Predators. mBio 2021, 12, e00466-21. [Google Scholar] [CrossRef] [PubMed]
- Makowski, Ł.; Trojanowski, D.; Till, R.; Lambert, C.; Lowry, R.; Sockett, R.E.; Zakrzewska-Czerwińska, J. Dynamics of Chromosome Replication and Its Relationship to Predatory Attack Lifestyles in Bdellovibrio bacteriovorus. Appl. Environ. Microbiol. 2019, 85, e00730-19. [Google Scholar] [CrossRef] [PubMed]
- Cohen, Y.; Pasternak, Z.; Müller, S.; Hübschmann, T.; Schattenberg, F.; Sivakala, K.K.; Abed-Rabbo, A.; Chatzinotas, A.; Jurkevitch, E. Community and Single Cell Analyses Reveal Complex Predatory Interactions between Bacteria in High Diversity Systems. Nat. Commun. 2021, 12, 5481. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Mun, W.; Choi, S.Y.; Mitchell, R.J. Use of Resazurin to Rapidly Enumerate Bdellovibrio and like Organisms and Evaluate Their Activities. Microbiol. Spectr. 2022, 10, e0082522. [Google Scholar] [CrossRef] [PubMed]
- Pineiro, S.; Chauhan, A.; Berhane, T.; Athar, R.; Zheng, G.; Wang, C.; Dickerson, T.; Liang, X.; Lymperopoulou, D.S.; Chen, H.; et al. Niche Partition of Bacteriovorax Operational Taxonomic Units along Salinity and Temporal Gradients in the Chesapeake Bay Reveals Distinct Estuarine Strains. Microb. Ecol. 2013, 65, 652–660. [Google Scholar] [CrossRef]
- Mun, W.; Choi, S.Y.; Upatissa, S.; Mitchell, R.J. Predatory Bacteria as Potential Biofilm Control and Eradication Agents in the Food Industry. Food Sci. Biotechnol. 2023, 32, 1729–1743. [Google Scholar] [CrossRef]
- Jurkevitch, E.; Minz, D.; Ramati, B.; Barel, G. Prey Range Characterization, Ribotyping, and Diversity of Soil and Rhizosphere Bdellovibrio spp. Isolated on Phytopathogenic Bacteria. Appl. Environ. Microbiol. 2000, 66, 2365–2371. [Google Scholar] [CrossRef]
- Oyedara, O.O.; De Luna-Santillana, E.D.J.; Olguin-Rodriguez, O.; Guo, X.; Mendoza-Villa, M.A.; Menchaca-Arredondo, J.L.; Elufisan, T.O.; Garza-Hernandez, J.A.; Garcia Leon, I.; Rodriguez-Perez, M.A. Isolation of Bdellovibrio sp. from Soil Samples in Mexico and Their Potential Applications in Control of Pathogens. MicrobiologyOpen 2016, 5, 992–1002. [Google Scholar] [CrossRef] [PubMed]
- Baer, M.L.; Ravel, J.; Piñeiro, S.A.; Guether-Borg, D.; Williams, H.N. Reclassification of Salt-Water Bdellovibrio sp. as Bacteriovorax marinus sp. Nov. and Bacteriovorax litoralis sp. Nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 1011–1016. [Google Scholar] [CrossRef] [PubMed]
- Mun, W.; Upatissa, S.; Lim, S.; Dwidar, M.; Mitchell, R.J. Outer Membrane Porin F in E. coli Is Critical for Effective Predation by Bdellovibrio. Microbiol. Spectr. 2022, 10, e0309422. [Google Scholar] [CrossRef]
- Guo, Y.; Pan, Q.; Yan, S.; Chen, Y.; Li, M.; Chen, D.; Han, H.; Wu, B.; Cai, J. Bdellovibrio and like Organisms Promoted Growth and Survival of Juvenile Abalone Haliotis Discus Hannai Ino and Modulated Bacterial Community Structures in Its Gut. Aquac. Int. 2017, 25, 1625–1643. [Google Scholar] [CrossRef]
- Schwudke, D.; Strauch, E.; Krueger, M.; Appel, B. Taxonomic Studies of Predatory Bdellovibrios Based on 16S rRNA Analysis, Ribotyping and the Hit Locus and Characterization of Isolates from the Gut of Animals. Syst. Appl. Microbiol. 2001, 24, 385–394. [Google Scholar] [CrossRef]
- Seccareccia, I.; Kost, C.; Nett, M. Quantitative Analysis of Lysobacter Predation. Appl. Environ. Microbiol. 2015, 81, 7098–7105. [Google Scholar] [CrossRef]
- Livingstone, P.G.; Morphew, R.M.; Whitworth, D.E. Myxobacteria Are Able to Prey Broadly upon Clinically-Relevant Pathogens, Exhibiting a Prey Range Which Cannot Be Explained by Phylogeny. Front. Microbiol. 2017, 8, 1593. [Google Scholar] [CrossRef]
- Xiao, Y.; Wei, X.; Ebright, R.; Wall, D. Antibiotic Production by Myxobacteria Plays a Role in Predation. J. Bacteriol. 2011, 193, 4626–4633. [Google Scholar] [CrossRef]
- Evans, A.G.L.; Davey, H.M.; Cookson, A.; Currinn, H.; Cooke-Fox, G.; Stanczyk, P.J.; Whitworth, D.E. Predatory Activity of Myxococcus xanthus Outer-Membrane Vesicles and Properties of Their Hydrolase Cargo. Microbiology 2012, 158, 2742–2752. [Google Scholar] [CrossRef]
- McBride, M.J.; Zusman, D.R. Behavioral Analysis of Single Cells of Myxococcus xanthus in Response to Prey Cells of Escherichia coli. FEMS Microbiol. Lett. 1996, 137, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Berleman, J.E.; Kirby, J.R. Deciphering the Hunting Strategy of a Bacterial Wolfpack. FEMS Microbiol. Rev. 2009, 33, 942–957. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Dorado, J.; Marcos-Torres, F.J.; García-Bravo, E.; Moraleda-Muñoz, A.; Pérez, J. Myxobacteria: Moving, Killing, Feeding, and Surviving Together. Front. Microbiol. 2016, 7, 781. [Google Scholar] [CrossRef] [PubMed]
- Shimkets, L.J. Intercellular Signaling during Fruiting-Body Development of Myxococcus xanthus. Annu. Rev. Microbiol. 1999, 53, 525–549. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, R.; Pedros-Alio, C.; Esteve, I.; Mas, J.; Chase, D.; Margulis, L. Predatory Prokaryotes: Predation and Primary Consumption Evolved in Bacteria. Proc. Natl. Acad. Sci. USA 1986, 83, 2138–2142. [Google Scholar] [CrossRef] [PubMed]
- Koval, S.F.; Hynes, S.H.; Flannagan, R.S.; Pasternak, Z.; Davidov, Y.; Jurkevitch, E. Bdellovibrio exovorus sp. Nov., a Novel Predator of Caulobacter crescentus. Int. J. Syst. Evol. Microbiol. 2013, 63, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, Z.; Njagi, M.; Shani, Y.; Chanyi, R.; Rotem, O.; Lurie-Weinberger, M.N.; Koval, S.; Pietrokovski, S.; Gophna, U.; Jurkevitch, E. In and out: An Analysis of Epibiotic vs Periplasmic Bacterial Predators. ISME J. 2014, 8, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Hovde, B.T.; Steichen, S.A.; Starkenburg, S.R.; Brown, J.K. Vampirovibrio chlorellavorus Draft Genome Sequence, Annotation, and Preliminary Characterization of Pathogenicity Determinants. Phycol. Res. 2020, 68, 23–29. [Google Scholar] [CrossRef]
- Moreira, D.; Zivanovic, Y.; López-Archilla, A.I.; Iniesto, M.; López-García, P. Reductive Evolution and Unique Predatory Mode in the CPR Bacterium Vampirococcus lugosii. Nat. Commun. 2021, 12, 2454. [Google Scholar] [CrossRef]
- Seideler, R.J.; Mandel, M.; Baptist, J.N. Molecular Heterogeneity of the Bdellovibrios: Evidence of Two New Species. J. Bacteriol. 1972, 109, 209–217. [Google Scholar] [CrossRef]
- Im, H.; Kwon, H.; Cho, G.; Kwon, J.; Choi, S.Y.; Mitchell, R.J. Viscosity Has Dichotomous Effects on Bdellovibrio bacteriovorus HD100 Predation. Environ. Microbiol. 2019, 21, 4675–4684. [Google Scholar] [CrossRef] [PubMed]
- Sathyamoorthy, R.; Maoz, A.; Pasternak, Z.; Im, H.; Huppert, A.; Kadouri, D.; Jurkevitch, E. Bacterial Predation under Changing Viscosities. Environ. Microbiol. 2019, 21, 2997–3010. [Google Scholar] [CrossRef]
- McCauley, E.P.; Haltli, B.; Kerr, R.G. Description of Pseudobacteriovorax antillogorgiicola Gen. Nov., sp. Nov., a Bacterium Isolated from the Gorgonian Octocoral Antillogorgia elisabethae, Belonging to the Family Pseudobacteriovoracaceae Fam. Nov., within the Order Bdellovibrionales. Int. J. Syst. Evol. Microbiol. 2015, 65, 522–530. [Google Scholar] [CrossRef]
- Fenton, A.K.; Kanna, M.; Woods, R.D.; Aizawa, S.-I.; Sockett, R.E. Shadowing the Actions of a Predator: Backlit Fluorescent Microscopy Reveals Synchronous Nonbinary Septation of Predatory Bdellovibrio inside Prey and Exit through Discrete Bdelloplast Pores. J. Bacteriol. 2010, 192, 6329–6335. [Google Scholar] [CrossRef] [PubMed]
- Rotem, O.; Pasternak, Z.; Shimoni, E.; Belausov, E.; Porat, Z.; Pietrokovski, S.; Jurkevitch, E. Cell-Cycle Progress in Obligate Predatory Bacteria Is Dependent upon Sequential Sensing of Prey Recognition and Prey Quality Cues. Proc. Natl. Acad. Sci. USA 2015, 112, E6028–E6037. [Google Scholar] [CrossRef]
- Oyedara, O.O.; Segura-Cabrera, A.; Guo, X.; Elufisan, T.O.; Cantú González, R.A.; Rodríguez Pérez, M.A. Whole-Genome Sequencing and Comparative Genome Analysis Provided Insight into the Predatory Features and Genetic Diversity of Two Bdellovibrio Species Isolated from Soil. Int. J. Genom. 2018, 2018, 9402073. [Google Scholar] [CrossRef]
- Inoue, D.; Hiroshima, N.; Ishizawa, H.; Dohra, H.; Ike, M. Complete Genome Sequences of Two Predatory Bacterial Strains, Bacteriovorax sp. HI3 and Myxococcus sp. MH1, Isolated from a Freshwater Pond. Microbiol. Resour. Announc. 2022, 11, e0114622. [Google Scholar] [CrossRef]
- Pasternak, Z.; Pietrokovski, S.; Rotem, O.; Gophna, U.; Lurie-Weinberger, M.N.; Jurkevitch, E. By Their Genes Ye Shall Know Them: Genomic Signatures of Predatory Bacteria. ISME J. 2013, 7, 756–769. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.E.; Cullen, N.; DeGiorgis, J.A.; Martinez, K.J.; Mellone, J.; Oser, M.; Wang, J.; Zhang, Y. Variation in Genome Content and Predatory Phenotypes between Bdellovibrio sp. NC01 Isolated from Soil and B. bacteriovorus Type Strain HD100. Microbiology 2019, 165, 1315–1330. [Google Scholar] [CrossRef]
- Bratanis, E.; Andersson, T.; Lood, R.; Bukowska-Faniband, E. Biotechnological Potential of Bdellovibrio and like Organisms and Their Secreted Enzymes. Front. Microbiol. 2020, 11, 662. [Google Scholar] [CrossRef]
- Dwidar, M.; Monnappa, A.K.; Mitchell, R.J. The Dual Probiotic and Antibiotic Nature of Bdellovibrio bacteriovorus. BMB Rep. 2012, 45, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Martínez, V.; Herencias, C.; Jurkevitch, E.; Prieto, M.A. Engineering a Predatory Bacterium as a Proficient Killer Agent for Intracellular Bio-Products Recovery: The Case of the Polyhydroxyalkanoates. Sci. Rep. 2016, 6, 24381. [Google Scholar] [CrossRef]
- Martínez, V.; Jurkevitch, E.; García, J.L.; Prieto, M.A. Reward for Bdellovibrio bacteriovorus for Preying on a Polyhydroxyalkanoate Producer. Environ. Microbiol. 2013, 15, 1204–1215. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Tan, C.H.; Constancias, F.; Kohli, G.S.; Cohen, Y.; Rice, S.A. Predation by Bdellovibrio bacteriovorus Significantly Reduces Viability and Alters the Microbial Community Composition of Activated Sludge Flocs and Granules. FEMS Microbiol. Ecol. 2017, 93, fix020. [Google Scholar] [CrossRef]
- Yan, C.; Zhan, M.; Xv, K.; Zhang, S.; Liang, T.; Yu, R. Sludge Dewaterability Enhancement under Low Temperature Condition with Cold-Tolerant Bdellovibrio sp. CLL13. Sci. Total Environ. 2022, 820, 153269. [Google Scholar] [CrossRef]
- Choi, S.Y.; Im, H.; Mitchell, R.J. Violacein and Bacterial Predation: Promising Alternatives for Priority Multidrug Resistant Human Pathogens. Future Microbiol. 2017, 12, 835–838. [Google Scholar] [CrossRef] [PubMed]
- Pérez, J.; Contreras-Moreno, F.J.; Marcos-Torres, F.J.; Moraleda-Muñoz, A.; Muñoz-Dorado, J. The Antibiotic Crisis: How Bacterial Predators Can Help. Comput. Struct. Biotechnol. J. 2020, 18, 2547–2555. [Google Scholar] [CrossRef]
- DePas, W.H.; Syed, A.K.; Sifuentes, M.; Lee, J.S.; Warshaw, D.; Saggar, V.; Csankovszki, G.; Boles, B.R.; Chapman, M.R. Biofilm Formation Protects Escherichia coli against Killing by Caenorhabditis elegans and Myxococcus xanthus. Appl. Environ. Microbiol. 2014, 80, 7079–7087. [Google Scholar] [CrossRef]
- Nair, R.R.; Vasse, M.; Wielgoss, S.; Sun, L.; Yu, Y.-T.N.; Velicer, G.J. Bacterial Predator-Prey Coevolution Accelerates Genome Evolution and Selects on Virulence-Associated Prey Defences. Nat. Commun. 2019, 10, 4301. [Google Scholar] [CrossRef]
- Livingstone, P.G.; Millard, A.D.; Swain, M.T.; Whitworth, D.E. Transcriptional Changes When Myxococcus xanthus Preys on Escherichia coli Suggest Myxobacterial Predators Are Constitutively Toxic but Regulate Their Feeding. Microb. Genom. 2018, 4, e000152. [Google Scholar] [CrossRef]
- Zhang, N.; Li, T.; Pan, H.; Wang, Y.; Li, Q.; Luan, J.; He, X.; Shi, W.; Li, Y.; Wang, C.; et al. Genetic Components of Escherichia coli Involved in Its Complex Prey-Predator Interaction with Myxococcus xanthus. Front. Microbiol. 2023, 14, 1304874. [Google Scholar] [CrossRef] [PubMed]
- Sert, D.; Mercan, E.; Kara, Ü. Butter Production from Ozone-Treated Cream: Effects on Characteristics of Physicochemical, Microbiological, Thermal and Oxidative Stability. LWT 2020, 131, 109722. [Google Scholar] [CrossRef]
- Baggio, A.; Marino, M.; Innocente, N.; Celotto, M.; Maifreni, M. Antimicrobial Effect of Oxidative Technologies in Food Processing: An Overview. Eur. Food Res. Technol. 2020, 246, 669–692. [Google Scholar] [CrossRef]
- Collineau, L.; Chapman, B.; Bao, X.; Sivapathasundaram, B.; Carson, C.A.; Fazil, A.; Reid-Smith, R.J.; Smith, B.A. A Farm-to-Fork Quantitative Risk Assessment Model for Salmonella Heidelberg Resistant to Third-Generation Cephalosporins in Broiler Chickens in Canada. Int. J. Food Microbiol. 2020, 330, 108559. [Google Scholar] [CrossRef]
- Rothrock, M.J.; Guard, J.Y.; Oladeinde, A. Salmonella Diversity along the Farm-to-Fork Continuum of Pastured Poultry Flocks in the Southeastern United States. Front. Anim. Sci. 2021, 2, 761930. [Google Scholar] [CrossRef]
- Dourou, D.; Beauchamp, C.S.; Yoon, Y.; Geornaras, I.; Belk, K.E.; Smith, G.C.; Nychas, G.-J.E.; Sofos, J.N. Attachment and Biofilm Formation by Escherichia coli O157:H7 at Different Temperatures, on Various Food-Contact Surfaces Encountered in Beef Processing. Int. J. Food Microbiol. 2011, 149, 262–268. [Google Scholar] [CrossRef]
- Silagyi, K.; Kim, S.-H.; Lo, Y.M.; Wei, C. Production of Biofilm and Quorum Sensing by Escherichia coli O157:H7 and Its Transfer from Contact Surfaces to Meat, Poultry, Ready-to-Eat Deli, and Produce Products. Food Microbiol. 2009, 26, 514–519. [Google Scholar] [CrossRef]
- Wang, H.; Ding, S.; Dong, Y.; Ye, K.; Xu, X.; Zhou, G. Biofilm Formation of Salmonella Serotypes in Simulated Meat Processing Environments and Its Relationship to Cell Characteristics. J. Food Prot. 2013, 76, 1784–1789. [Google Scholar] [CrossRef] [PubMed]
- Dashiff, A.; Junka, R.A.; Libera, M.; Kadouri, D.E. Predation of Human Pathogens by the Predatory Bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J. Appl. Microbiol. 2011, 110, 431–444. [Google Scholar] [CrossRef]
- Dashiff, A.; Kadouri, D.E. Predation of Oral Pathogens by Bdellovibrio bacteriovorus 109J. Mol. Oral Microbiol. 2011, 26, 19–34. [Google Scholar] [CrossRef]
- Im, H.; Dwidar, M.; Mitchell, R.J. Bdellovibrio bacteriovorus HD100, a Predator of Gram-Negative Bacteria, Benefits Energetically from Staphylococcus aureus Biofilms without Predation. ISME J. 2018, 12, 2090–2095. [Google Scholar] [CrossRef] [PubMed]
- Inoue, D.; Hiroshima, N.; Nakamura, S.; Ishizawa, H.; Ike, M. Characterization of Two Novel Predatory Bacteria, Bacteriovorax Stolpii HI3 and Myxococcus sp. MH1, Isolated from a Freshwater Pond: Prey Range, and Predatory Dynamics and Efficiency. Microorganisms 2022, 10, 1816. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wuertz, S. Bacteria and Archaea on Earth and Their Abundance in Biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef]
- Galié, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the Food Industry: Health Aspects and Control Methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef] [PubMed]
- Kadouri, D.; O’Toole, G.A. Susceptibility of Biofilms to Bdellovibrio bacteriovorus Attack. Appl. Environ. Microbiol. 2005, 71, 4044–4051. [Google Scholar] [CrossRef] [PubMed]
- Kadouri, D.; Venzon, N.C.; O’Toole, G.A. Vulnerability of Pathogenic Biofilms to Micavibrio aeruginosavorus. Appl. Environ. Microbiol. 2007, 73, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Joseph, B.; Otta, S.K.; Karunasagar, I.; Karunasagar, I. Biofilm Formation by Salmonella spp. on Food Contact Surfaces and Their Sensitivity to Sanitizers. Int. J. Food Microbiol. 2001, 64, 367–372. [Google Scholar] [CrossRef]
- Merino, L.; Procura, F.; Trejo, F.M.; Bueno, D.J.; Golowczyc, M.A. Biofilm Formation by Salmonella sp. in the Poultry Industry: Detection, Control and Eradication Strategies. Food Res. Int. Ott. Ont 2019, 119, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Tang, C.; Tran, M.; Kadouri, D.E. Effect of Predatory Bacteria on Human Cell Lines. PLoS ONE 2016, 11, e0161242. [Google Scholar] [CrossRef]
- Monnappa, A.K.; Bari, W.; Choi, S.Y.; Mitchell, R.J. Investigating the Responses of Human Epithelial Cells to Predatory Bacteria. Sci. Rep. 2016, 6, 33485. [Google Scholar] [CrossRef]
- Cho, G.; Kwon, J.; Soh, S.M.; Jang, H.; Mitchell, R.J. Sensitivity of Predatory Bacteria to Different Surfactants and Their Application to Check Bacterial Predation. Appl. Microbiol. Biotechnol. 2019, 103, 8169–8178. [Google Scholar] [CrossRef] [PubMed]
- Niemira, B.A.; Fan, X. Fruits and vegetables|Advances in Processing Technologies to Preserve and Enhance the Safety of Fresh and Fresh-Cut Fruits and Vegetables. In Encyclopedia of Food Microbiology; Elsevier: Amsterdam, The Netherlands, 2014; pp. 983–991. ISBN 978-0-12-384733-1. [Google Scholar]
- Olanya, O.M.; Niemira, B.A.; Cassidy, J.M.; Boyd, G.; Uknalis, J. Pathogen Reduction by Predatory Bacteria and Survival of Bdellovibrio bacteriovorus and Escherichia coli on Produce and Buffer Treated with Low-Dose Gamma Radiation. LWT 2020, 130, 109630. [Google Scholar] [CrossRef]
- Li, Y.; Qiu, F.; Yan, H.; Wan, X.; Wang, M.; Ren, K.; Xu, Q.; Lv, L.; Yin, C.; Liu, X.; et al. Increasing the Autotrophic Growth of Chlorella USTB-01 via the Control of Bacterial Contamination by Bdellovibrio USTB-06. J. Appl. Microbiol. 2018, 124, 1131–1138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Guo, L.; Cui, Z.; Ju, F. Exploiting Predatory Bacteria as Biocontrol Agents across Ecosystems. Trends Microbiol. 2024, 32, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Scherff, R.H. Control of Bacterial Blight of Soybean by Bdellovibrio bacteriovorus. Phytopathology 1973, 63, 400. [Google Scholar] [CrossRef]
- Youdkes, D.; Helman, Y.; Burdman, S.; Matan, O.; Jurkevitch, E. Potential Control of Potato Soft Rot Disease by the Obligate Predators Bdellovibrio and like Organisms. Appl. Environ. Microbiol. 2020, 86, e02543-19. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Xu, X.; Gao, R.; Li, Y.; Li, A.; Yao, Q.; Zhu, H. Myxococcus xanthus R31 Suppresses Tomato Bacterial Wilt by Inhibiting the Pathogen Ralstonia solanacearum with Secreted Proteins. Front. Microbiol. 2021, 12, 801091. [Google Scholar] [CrossRef] [PubMed]
- Najnine, F.; Cao, Q.; Zhao, Y.; Cai, J. Antibacterial Activities of Bdellovibrio and like Organisms in Aquaculture. In The Ecology of Predation at the Microscale; Jurkevitch, E., Mitchell, R.J., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 89–126. ISBN 978-3-030-45598-9. [Google Scholar]
- Lyu, J.; Yang, L.; Zhang, L.; Ye, B.; Wang, L. Antibiotics in Soil and Water in China—A Systematic Review and Source Analysis. Environ. Pollut. 2020, 266, 115147. [Google Scholar] [CrossRef] [PubMed]
- Richards, G.P.; Watson, M.A.; Williams, H.N.; Jones, J.L. Predator-Prey Interactions between Halobacteriovorax and Pathogenic Vibrio parahaemolyticus Strains: Geographical Considerations and Influence of Vibrio Hemolysins. Microbiol. Spectr. 2023, 11, e0235323. [Google Scholar] [CrossRef]
- Ottaviani, D.; Pieralisi, S.; Chierichetti, S.; Rocchegiani, E.; Hattab, J.; Mosca, F.; Tiscar, P.G.; Leoni, F.; Angelico, G. Vibrio parahaemolyticus Control in Mussels by a Halobacteriovorax Isolated from the Adriatic Sea, Italy. Food Microbiol. 2020, 92, 103600. [Google Scholar] [CrossRef]
- Ooi, M.C.; Goulden, E.F.; Smith, G.G.; Bridle, A.R. Predatory Bacteria in the Haemolymph of the Cultured Spiny Lobster Panulirus ornatus. Microbiology 2021, 167, 001113. [Google Scholar] [CrossRef] [PubMed]
- Ju, F.; Xia, Y.; Guo, F.; Wang, Z.; Zhang, T. Taxonomic Relatedness Shapes Bacterial Assembly in Activated Sludge of Globally Distributed Wastewater Treatment Plants. Environ. Microbiol. 2014, 16, 2421–2432. [Google Scholar] [CrossRef] [PubMed]
- Mookherjee, A.; Jurkevitch, E. Interactions between Bdellovibrio and like Organisms and Bacteria in Biofilms: Beyond Predator-Prey Dynamics. Environ. Microbiol. 2022, 24, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Waso, M.; Khan, S.; Singh, A.; McMichael, S.; Ahmed, W.; Fernández-Ibáñez, P.; Byrne, J.A.; Khan, W. Predatory Bacteria in Combination with Solar Disinfection and Solar Photocatalysis for the Treatment of Rainwater. Water Res. 2020, 169, 115281. [Google Scholar] [CrossRef] [PubMed]
- Atterbury, R.J.; Tyson, J. Predatory Bacteria as Living Antibiotics—Where Are We Now? Microbiology 2021, 167. [Google Scholar] [CrossRef] [PubMed]
- Schwudke, D.; Linscheid, M.; Strauch, E.; Appel, B.; Zahringer, U.; Moll, H.; Muller, M.; Brecker, L.; Gronow, S.; Lindner, B. The Obligate Predatory Bdellovibrio bacteriovorus Possesses a Neutral Lipid A Containing Alpha-D-Mannoses That Replace Phosphate Residues: Similarities and Differences between the Lipid as and the Lipopolysaccharides of the Wild Type Strain B. bacteriovorus HD100 and Its Host-Independent Derivative HI100. J. Biol. Chem. 2003, 278, 27502–27512. [Google Scholar] [CrossRef] [PubMed]
- Shanks, R.M.Q.; Davra, V.R.; Romanowski, E.G.; Brothers, K.M.; Stella, N.A.; Godboley, D.; Kadouri, D.E. An Eye to a Kill: Using Predatory Bacteria to Control Gram-Negative Pathogens Associated with Ocular Infections. PLoS ONE 2013, 8, e66723. [Google Scholar] [CrossRef] [PubMed]
- Raghunathan, D.; Radford, P.M.; Gell, C.; Negus, D.; Moore, C.; Till, R.; Tighe, P.J.; Wheatley, S.P.; Martinez-Pomares, L.; Sockett, R.E.; et al. Engulfment, Persistence and Fate of Bdellovibrio bacteriovorus Predators inside Human Phagocytic Cells Informs Their Future Therapeutic Potential. Sci. Rep. 2019, 9, 4293. [Google Scholar] [CrossRef]
- Seidler, R.J.; Starr, M.P. Structure of the Flagellum of Bdellovibrio bacteriovorus. J. Bacteriol. 1968, 95, 1952–1955. [Google Scholar] [CrossRef]
- Findlay, J.S.; Flick-Smith, H.C.; Keyser, E.; Cooper, I.A.; Williamson, E.D.; Oyston, P.C.F. Predatory Bacteria Can Protect SKH-1 Mice from a Lethal Plague Challenge. Sci. Rep. 2019, 9, 7225. [Google Scholar] [CrossRef]
- Iebba, V.; Totino, V.; Santangelo, F.; Gagliardi, A.; Ciotoli, L.; Virga, A.; Ambrosi, C.; Pompili, M.; De Biase, R.V.; Selan, L.; et al. Bdellovibrio bacteriovorus Directly Attacks Pseudomonas aeruginosa and Staphylococcus aureus Cystic Fibrosis Isolates. Front. Microbiol. 2014, 5, 280. [Google Scholar] [CrossRef] [PubMed]
- Kahraman Vatansever, S.; Tekintas, Y.; Cilli, F.F.; Hosgor-Limoncu, M. Effect of Predator Bacteria Bdellovibrio bacteriovorus on Clinical Pathogens and Biofilms. Indian J. Microbiol. 2023, 63, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Patini, R.; Cattani, P.; Marchetti, S.; Isola, G.; Quaranta, G.; Gallenzi, P. Evaluation of Predation Capability of Periodontopathogens Bacteria by Bdellovibrio bacteriovorus HD100. An in Vitro Study. Mater. Basel Switz. 2019, 12, 2008. [Google Scholar] [CrossRef] [PubMed]
- Dharani, S.; Kim, D.H.; Shanks, R.M.Q.; Doi, Y.; Kadouri, D.E. Susceptibility of Colistin-Resistant Pathogens to Predatory Bacteria. Res. Microbiol. 2018, 169, 52–55. [Google Scholar] [CrossRef] [PubMed]
- Kadouri, D.E.; To, K.; Shanks, R.M.Q.; Doi, Y. Predatory Bacteria: A Potential Ally against Multidrug-Resistant Gram-Negative Pathogens. PLoS ONE 2013, 8, e63397. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.; Negus, D.; Raghunathan, D.; Radford, P.; Moore, C.; Clark, G.; Diggle, M.; Tyson, J.; Twycross, J.; Sockett, R.E. Measuring and Modelling the Response of Klebsiella pneumoniae KPC Prey to Bdellovibrio bacteriovorus Predation, in Human Serum and Defined Buffer. Sci. Rep. 2017, 7, 8329. [Google Scholar] [CrossRef]
- Sun, Y.; Ye, J.; Hou, Y.; Chen, H.; Cao, J.; Zhou, T. Predation Efficacy of Bdellovibrio bacteriovorus on Multidrug-Resistant Clinical Pathogens and Their Corresponding Biofilms. Jpn. J. Infect. Dis. 2017, 70, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Im, H.; Choi, S.Y.; Son, S.; Mitchell, R.J. Combined Application of Bacterial Predation and Violacein to Kill Polymicrobial Pathogenic Communities. Sci. Rep. 2017, 7, 14415. [Google Scholar] [CrossRef] [PubMed]
- Westergaard, J.M.; Kramer, T.T. Bdellovibrio and the Intestinal Flora of Vertebrates. Appl. Environ. Microbiol. 1977, 34, 506–511. [Google Scholar] [CrossRef]
- Atterbury, R.J.; Hobley, L.; Till, R.; Lambert, C.; Capeness, M.J.; Lerner, T.R.; Fenton, A.K.; Barrow, P.; Sockett, R.E. Effects of Orally Administered Bdellovibrio bacteriovorus on the Well-Being and Salmonella Colonization of Young Chicks. Appl. Environ. Microbiol. 2011, 77, 5794–5803. [Google Scholar] [CrossRef]
- Romanowski, E.G.; Stella, N.A.; Brothers, K.M.; Yates, K.A.; Funderburgh, M.L.; Funderburgh, J.L.; Gupta, S.; Dharani, S.; Kadouri, D.E.; Shanks, R.M.Q. Predatory Bacteria Are Nontoxic to the Rabbit Ocular Surface. Sci. Rep. 2016, 6, 30987. [Google Scholar] [CrossRef] [PubMed]
- Shatzkes, K.; Singleton, E.; Tang, C.; Zuena, M.; Shukla, S.; Gupta, S.; Dharani, S.; Onyile, O.; Rinaggio, J.; Connell, N.D.; et al. Predatory Bacteria Attenuate Klebsiella pneumoniae Burden in Rat Lungs. mBio 2016, 7, e01847-16. [Google Scholar] [CrossRef] [PubMed]
- Shatzkes, K.; Singleton, E.; Tang, C.; Zuena, M.; Shukla, S.; Gupta, S.; Dharani, S.; Rinaggio, J.; Kadouri, D.E.; Connell, N.D. Examining the Efficacy of Intravenous Administration of Predatory Bacteria in Rats. Sci. Rep. 2017, 7, 1864. [Google Scholar] [CrossRef] [PubMed]
- Shatzkes, K.; Tang, C.; Singleton, E.; Shukla, S.; Zuena, M.; Gupta, S.; Dharani, S.; Rinaggio, J.; Connell, N.D.; Kadouri, D.E. Effect of Predatory Bacteria on the Gut Bacterial Microbiota in Rats. Sci. Rep. 2017, 7, 43483. [Google Scholar] [CrossRef] [PubMed]
- Shatzkes, K.; Chae, R.; Tang, C.; Ramirez, G.C.; Mukherjee, S.; Tsenova, L.; Connell, N.D.; Kadouri, D.E. Examining the Safety of Respiratory and Intravenous Inoculation of Bdellovibrio bacteriovorus and Micavibrio aeruginosavorus in a Mouse Model. Sci. Rep. 2015, 5, 12899. [Google Scholar] [CrossRef] [PubMed]
- Willis, A.R.; Moore, C.; Mazon-Moya, M.; Krokowski, S.; Lambert, C.; Till, R.; Mostowy, S.; Sockett, R.E. Injections of Predatory Bacteria Work Alongside Host Immune Cells to Treat Shigella Infection in Zebrafish Larvae. Curr. Biol. CB 2016, 26, 3343–3351. [Google Scholar] [CrossRef] [PubMed]
- Torraca, V.; Mostowy, S. Zebrafish Infection: From Pathogenesis to Cell Biology. Trends Cell Biol. 2018, 28, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Gomes, M.C.; Mostowy, S. The Case for Modeling Human Infection in Zebrafish. Trends Microbiol. 2020, 28, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The Zebrafish Reference Genome Sequence and Its Relationship to the Human Genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef]
- Meijer, A.H.; Spaink, H.P. Host-Pathogen Interactions Made Transparent with the Zebrafish Model. Curr. Drug Targets 2011, 12, 1000–1017. [Google Scholar] [CrossRef]
- Russo, R.; Kolesnikova, I.; Kim, T.; Gupta, S.; Pericleous, A.; Kadouri, D.E.; Connell, N.D. Susceptibility of Virulent Yersinia pestis Bacteria to Predator Bacteria in the Lungs of Mice. Microorganisms 2018, 7, 2. [Google Scholar] [CrossRef]
- Boileau, M.J.; Mani, R.; Breshears, M.A.; Gilmour, M.; Taylor, J.D.; Clinkenbeard, K.D. Efficacy of Bdellovibrio bacteriovorus 109J for the Treatment of Dairy Calves with Experimentally Induced Infectious Bovine Keratoconjunctivitis. Am. J. Vet. Res. 2016, 77, 1017–1028. [Google Scholar] [CrossRef]
- Boileau, M.J.; Clinkenbeard, K.D.; Iandolo, J.J. Assessment of Bdellovibrio bacteriovorus 109J Killing of Moraxella bovis in an in Vitro Model of Infectious Bovine Keratoconjunctivitis. Can. J. Vet. Res. Rev. Can. Rech. Vet. 2011, 75, 285–291. [Google Scholar]
- Romanowski, E.G.; Brothers, K.M.; Calvario, R.C.; Stella, N.A.; Kim, T.; Elsayed, M.; Kadouri, D.E.; Shanks, R.M.Q. Predatory Bacteria Prevent the Proliferation of Intraocular Serratia marcescens and Fluoroquinolone-Resistant Pseudomonas aeruginosa. Microbiology 2024, 170, 001433. [Google Scholar] [CrossRef]
- Rendulic, S.; Jagtap, P.; Rosinus, A.; Eppinger, M.; Baar, C.; Lanz, C.; Keller, H.; Lambert, C.; Evans, K.J.; Goesmann, A.; et al. A Predator Unmasked: Life Cycle of Bdellovibrio bacteriovorus from a Genomic Perspective. Science 2004, 303, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Monnappa, A.K.; Dwidar, M.; Seo, J.K.; Hur, J.-H.; Mitchell, R.J. Bdellovibrio bacteriovorus Inhibits Staphylococcus aureus Biofilm Formation and Invasion into Human Epithelial Cells. Sci. Rep. 2014, 4, 3811. [Google Scholar] [CrossRef] [PubMed]
- Waso, M.; Reyneke, B.; Havenga, B.; Khan, S.; Khan, W. Insights into Bdellovibrio spp. Mechanisms of Action and Potential Applications. World J. Microbiol. Biotechnol. 2021, 37, 85. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, H.; Jiang, H.; Yao, Q.; Zhu, H. Characteristics of a Lipase ArEstA with Lytic Activity against Drug-Resistant Pathogen from a Novel Myxobacterium, Archangium Lipolyticum sp. Nov. Front. Microbiol. 2023, 14, 1320827. [Google Scholar] [CrossRef]
- Hobley, L.; Summers, J.K.; Till, R.; Milner, D.S.; Atterbury, R.J.; Stroud, A.; Capeness, M.J.; Gray, S.; Leidenroth, A.; Lambert, C.; et al. Dual Predation by Bacteriophage and Bdellovibrio bacteriovorus Can Eradicate Escherichia coli Prey in Situations Where Single Predation Cannot. J. Bacteriol. 2020, 202, e00629-19. [Google Scholar] [CrossRef]
- Marine, E.; Milner, D.S.; Lambert, C.; Sockett, R.E.; Pos, K.M. A Novel Method to Determine Antibiotic Sensitivity in Bdellovibrio bacteriovorus Reveals a DHFR-Dependent Natural Trimethoprim Resistance. Sci. Rep. 2020, 10, 5315. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alexakis, K.; Baliou, S.; Ioannou, P. Predatory Bacteria in the Treatment of Infectious Diseases and Beyond. Infect. Dis. Rep. 2024, 16, 684-698. https://doi.org/10.3390/idr16040052
Alexakis K, Baliou S, Ioannou P. Predatory Bacteria in the Treatment of Infectious Diseases and Beyond. Infectious Disease Reports. 2024; 16(4):684-698. https://doi.org/10.3390/idr16040052
Chicago/Turabian StyleAlexakis, Konstantinos, Stella Baliou, and Petros Ioannou. 2024. "Predatory Bacteria in the Treatment of Infectious Diseases and Beyond" Infectious Disease Reports 16, no. 4: 684-698. https://doi.org/10.3390/idr16040052




