Bioinformatics Analysis Identifies a Small ORF in the Genome of Fish Nidoviruses of Genus Oncotshavirus Predicted to Encode a Novel Integral Protein
Abstract
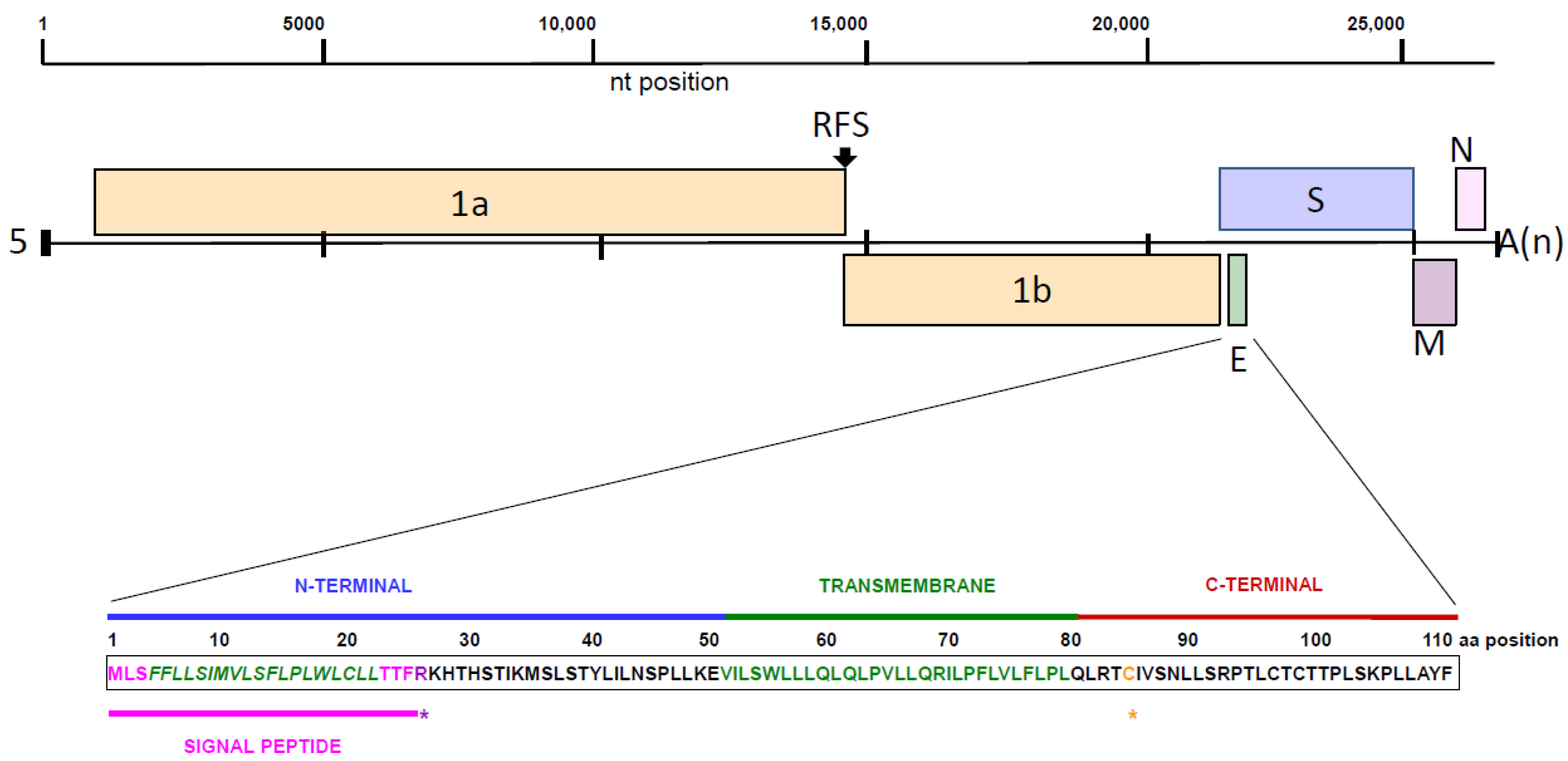
:1. Introduction
2. Materials and Methods
3. Results and Discussion




Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- International Committee on Taxonomy of Viruses. ICTV Master Species List 2019 v2. Checklist Dataset. 2019. Available online: https://www.gbif.org/dataset/e01b0cbb-a10a-420c-b5f3-a3b20cc266ad (accessed on 3 July 2020).
- Sola, I.; Almazan, F.; Zuñiga, S.; Enjuanes, L. Continuous and Discontinuous RNA Synthesis in Coronaviruses. Annu. Rev. Virol. 2015, 2, 265–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boon, J.A.D.; Snijder, E.J.; Chirnside, E.D.; A de Vries, A.; Horzinek, M.C.; Spaan, W.J. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 1991, 65, 2910–2920. [Google Scholar] [CrossRef] [Green Version]
- Saberi, A.; Gulyaeva, A.A.; Brubacher, J.L.; Newmark, P.A.; Gorbalenya, A.E. A planarian nidovirus expands the limits of RNA genome size. PLoS Pathog. 2018, 14, e1007314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worldometer. Coronavirus Update (Live). Available online: https://www.worldometers.info/coronavirus/ (accessed on 1 October 2021).
- Bukhari, K.; Mulley, G.; Gulyaeva, A.A.; Zhao, L.; Shu, G.; Jiang, J.; Neuman, B.W. Description and initial characterization of metatranscriptomic nidovirus-like genomes from the proposed new family Abyssoviridae, and from a sister group to the Coronavirinae, the proposed genus Alphaletovirus. Virology 2018, 524, 160–171. [Google Scholar] [CrossRef] [PubMed]
- GenBank Database. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KY130432 (accessed on 9 July 2021).
- McKibbon, A. Analysis of Structural Proteins of Atlantic Salmon Bafinivirus—A Coronavirus of Fish. Master’s Thesis, University of Prince Edward Island, Charlottetown, PE, Canada, 21 May 2020. [Google Scholar]
- Schütze, H. Coronaviruses in aquatic organisms. In Aquaculture Virology; Kibenge, F.S.B., Godoy, M.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 323–332. [Google Scholar]
- Cano, I.; Stone, D.; Savage, J.; Wood, G.; Mulhearn, B.; Gray, J.; Stinton, N.; Ross, S.; Bonar, M.; Taylor, N.G.H.; et al. Isolation of a Chinook Salmon Bafinivirus (CSBV) in Imported Goldfish Carassius auratus L. in the United Kingdom and Evaluation of Its Virulence in Resident Fish Species. Viruses 2020, 12, 578. [Google Scholar] [CrossRef]
- Zhang, X.; Shen, W.; Xu, C.; Wang, Y.; Xu, H.; Liu, X.; Wei, Y. Discovery of a novel Piscanivirus in yellow catfish (Pelteobagrus fulvidraco) in China. Infect. Genet. Evol. 2019, 74, 103924. [Google Scholar] [CrossRef]
- De Groot, R.J.; Baker, S.C.; Baric, R.; Enjuanes, L.; Gorbalenya, A.E.; Holmes, K.V.; Perlman, S.; Poon, L.; Rottier, P.J.M.; Talbot, P.J.; et al. Family Coronaviridae. In Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2012; pp. 806–828. [Google Scholar]
- Fischer, F.; Stegen, C.F.; Masters, P.S.; Samsonoff, W.A. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 1998, 72, 7885–7894. [Google Scholar] [CrossRef] [Green Version]
- MacLachlan, N.J.; Dubovi, E.J. Chapter 24, Coronaviridae. In Fenner’s Veterinary Virology, 5th ed.; MacLachlan, N.J., Dubovi, E.J., Eds.; Academic Press: Amsterdam, The Netherlands, 2017; pp. 393–413. [Google Scholar]
- GenBank Database. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KJ681496 (accessed on 9 July 2021).
- Shi, M.; Lin, X.-D.; Chen, X.; Tian, J.-H.; Chen, L.-J.; Li, K.; Wang, W.; Eden, J.-S.; Shen, J.-J.; Liu, L.; et al. The evolutionary history of vertebrate RNA viruses. Nat. Cell Biol. 2018, 556, 197–202. [Google Scholar] [CrossRef]
- Chen, X.-Y.; Zhou, Y.; Chen, X.; Zhang, J.; Zeng, X.-D.; Ji, F.; Xu, L.-M. Isolation and genetic analysis of a nidovirus from crucian carp (Carassius auratus). Arch. Virol. 2019, 164, 1651–1654. [Google Scholar] [CrossRef]
- GenBank Database. Available online: https://www.ncbi.nlm.nih.gov/nuccore/MT424676 (accessed on 9 July 2021).
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Ostell, J.; Pruitt, K.D.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2019, 48, D84–D86. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Higgins, D.; Sharp, P. CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene 1988, 73, 237–244. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeepTMHMM. Available online: https://biolib.com/DTU/DeepTMHMM (accessed on 9 July 2021).
- Tsirigos, K.; Peters, C.; Shu, N.; Käll, L.; Elofsson, A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015, 43, W401–W407. [Google Scholar] [CrossRef] [PubMed]
- Käll, L. Prediction of Transmembrane Topology and Signal Peptide Given a Protein’s Amino Acid Sequence. Methods Mol. Biol. 2010, 673, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Lao, D.M.; Arai, M.; Ikeda, M.; Shimizu, T. The presence of signal peptide significantly affects transmembrane topology prediction. Bioinformatics 2002, 18, 1562–1566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- SignalP-5.0 Signal Peptide and Cleavage Sites in Gram+, Gram-and Eukaryotic Amino Acid Sequences. Available online: https://services.healthtech.dtu.dk/service.php?SignalP-5.0. (accessed on 9 July 2021).
- Gupta, R.; Brunak, S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002, 7, 310–322. [Google Scholar]
- Ning, W.; Jiang, P.; Guo, Y.; Wang, C.; Tan, X.; Zhang, W.; Peng, D.; Xue, Y. GPS-Palm: A deep learning-based graphic presentation system for the prediction of S-palmitoylation sites in proteins. Brief. Bioinform. 2021, 22, 1836–1847. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Stothard, P. The Sequence Manipulation Suite: JavaScript programs for analyzing and for-matting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. Available online: https://www.bioinformatics.org/sms2/ (accessed on 9 July 2021). [CrossRef] [PubMed] [Green Version]
- Brierley, I.; Boursnell, M.E.; Binns, M.M.; Bilimoria, B.; Blok, V.C.; Brown, T.D.; Inglis, S.C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987, 6, 3779–3785. [Google Scholar] [CrossRef] [PubMed]
- Brierley, I.; Digard, P.; Inglis, S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: Requirement for an RNA pseudoknot. Cell 1989, 57, 537–547. [Google Scholar] [CrossRef]
- Brierley, I. Ribosomal frameshifting on viral RNAs. J. Gen. Virol. 1995, 76, 1885–1892. [Google Scholar] [CrossRef]
- Lenard, J. Viral membranes. In Encyclopedia of Virology, 3rd ed.; Mahy, B.W.J., Van Re-genmortel, M.H.V., Eds.; Academic Press: Cambridge, MA, USA, 2008; pp. 308–314. [Google Scholar]
- Kim, D.; Lee, J.-Y.; Yang, J.-S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e10. [Google Scholar] [CrossRef]
- Pasternak, A.; Spaan, W.J.M.; Snijder, E. Nidovirus transcription: How to make sense…? J. Gen. Virol. 2006, 87, 1403–1421. [Google Scholar] [CrossRef]
- Stewart, H.; Brown, K.; Dinan, A.M.; Irigoyen, N.; Snijder, E.J.; Firth, A.E. Transcriptional and Translational Landscape of Equine Torovirus. J. Virol. 2018, 92, e00589-18. [Google Scholar] [CrossRef] [Green Version]
- Duart†, G.; García-Murria†, M.J.; Grau†, B.; Acosta-Cáceres, J.M.; Martínez-Gil, L.; Mingarro, I. SARS-CoV-2 envelope protein topology in eukaryotic membranes. Open Biol. 2020, 10, 200209. [Google Scholar] [CrossRef]
- Nieto-Torres, J.L.; DeDiego, M.L.; Álvarez, E.; Jiménez-Guardeño, J.M.; Regla-Nava, J.A.; Llorente, M.; Kremer, L.; Shuo, S.; Enjuanes, L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology 2011, 415, 69–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fung, T.S.; Liu, D.X. Post-translational modifications of coronavirus proteins: Roles and function. Futur. Virol. 2018, 13, 405–430. [Google Scholar] [CrossRef] [Green Version]
- Ruch, T.R.; Machamer, C.E. The Coronavirus E Protein: Assembly and Beyond. Viruses 2012, 4, 363–382. [Google Scholar] [CrossRef] [Green Version]
- Raamsman, M.J.B.; Locker, J.K.; de Hooge, A.; de Vries, A.A.F.; Griffiths, G.; Vennema, H.; Rottier, P.J.M. Characterization of the Coronavirus Mouse Hepatitis Virus Strain A59 Small Membrane Protein E. J. Virol. 2000, 74, 2333–2342. [Google Scholar] [CrossRef] [Green Version]
- Goder, V.; Spiess, M. Topogenesis of membrane proteins: Determinants and dynamics. FEBS Lett. 2001, 504, 87–93. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. Available online: http://www.expasy.org/tools/peptidecutter/ (accessed on 9 July 2021).
- Kapp, K.; Schrempf, S.; Lemberg, M.K.; Dobberstein, B. Chapter 1, Post-Targeting Functions of Signal Peptides. In Protein Transport into the Endoplasmic Reticulum; Zimmermann, R., Ed.; Landes Bioscience: Austin, TX, USA, 2009. Available online: https://www.ncbi.nlm.nih.gov/books/NBK6322/ (accessed on 9 July 2021).
- Mandala, V.S.; McKay, M.J.; Shcherbakov, A.A.; Dregni, A.J.; Kolocouris, A.; Hong, M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 2020, 27, 1202–1208. [Google Scholar] [CrossRef]
- Yuan, Q.; Liao, Y.; Torres, J.; Tam, J.P.; Liu, D. Biochemical evidence for the presence of mixed membrane topologies of the severe acute respiratory syndrome coronavirus envelope protein expressed in mammalian cells. FEBS Lett. 2006, 580, 3192–3200. [Google Scholar] [CrossRef] [Green Version]
- Corse, E.; Machamer, C.E. The Cytoplasmic Tail of Infectious Bronchitis Virus E Protein Directs Golgi Targeting. J. Virol. 2002, 76, 1273–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Yuan, Q.; Torres, J.; Tam, J.P.; Liu, D. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology 2006, 349, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Lopez, L.A.; Riffle, A.J.; Pike, S.L.; Gardner, D.; Hogue, B.G. Importance of Conserved Cysteine Residues in the Coronavirus Envelope Protein. J. Virol. 2008, 82, 3000–3010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breitinger, U.; Farag, N.; Ali, N.K.; Breitinger, H.-G. Patch-clamp study of Hepatitis C p7 channels reveals genotype-specific sensitivity to inhibitors. Biophys. J. 2016, 110, 2419–2429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farag, N.S.; Breitinger, U.; El-Azizi, M.; Breitinger, H.-G. The p7 viroporin of hepatitis C virus contributes to liver inflammation by stimulating production of Interleukin-1β. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 712–720. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Zheng, B.J.; Xu, K.; Schwarz, W.; Du, L.; Wong, C.K.L.; Chen, J.; Duan, S.; Deubel, V.; Sun, B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. USA. 2006, 103, 12540–12545. [Google Scholar] [CrossRef] [Green Version]
- Stöver, B.C.; Müller, K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform. 2010, 11, 7. Available online: http://www.biomedcentral.com/1471-2105/11/7 (accessed on 3 October 2021). [CrossRef] [PubMed] [Green Version]
| Virus Isolate | GenBank Accession No. | Genome Length (nt) | % Sequence Identity | Host Fish Species | Reference |
|---|---|---|---|---|---|
| CSBV NIDO | KJ681496 | 27,004 | - | Chinook salmon, Oncorhynchus tshawytscha | [15] |
| ASBV VT01292015-09 | KY130432 | 26,496 | 99.43% | Atlantic salmon, Salmo salar | [7] |
| CSBV WHQSR4345 | MG600027 | 26,466 | 97.40% | Lesser spiny eel, Macrognathus aculeatus | [16] |
| CSBV HB93 | MH171482 | 25,971 | 97.21% | Crucian carp, Carassius auratus | [17] |
| YCBV Shaoxing | MH822145 | 26,985 | 96.24% | Yellow catfish, Pelteobagrus fulvidraco | [11] |
| CSBV Cefas-W054 | MT123520 | 25,969 | 97.99% | Goldfish, Carassius auratus | [10] |
| PFO-1 ZJLH18531 | MT424676 | 26,996 | 96.14% | Yellow catfish, Tachysurus fulvidraco | [18] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kibenge, F.; McKibbon, A.; Kibenge, M.; Wang, Y. Bioinformatics Analysis Identifies a Small ORF in the Genome of Fish Nidoviruses of Genus Oncotshavirus Predicted to Encode a Novel Integral Protein. Microbiol. Res. 2021, 12, 753-764. https://doi.org/10.3390/microbiolres12040055
Kibenge F, McKibbon A, Kibenge M, Wang Y. Bioinformatics Analysis Identifies a Small ORF in the Genome of Fish Nidoviruses of Genus Oncotshavirus Predicted to Encode a Novel Integral Protein. Microbiology Research. 2021; 12(4):753-764. https://doi.org/10.3390/microbiolres12040055
Chicago/Turabian StyleKibenge, Frederick, Ashley McKibbon, Molly Kibenge, and Yingwei Wang. 2021. "Bioinformatics Analysis Identifies a Small ORF in the Genome of Fish Nidoviruses of Genus Oncotshavirus Predicted to Encode a Novel Integral Protein" Microbiology Research 12, no. 4: 753-764. https://doi.org/10.3390/microbiolres12040055
APA StyleKibenge, F., McKibbon, A., Kibenge, M., & Wang, Y. (2021). Bioinformatics Analysis Identifies a Small ORF in the Genome of Fish Nidoviruses of Genus Oncotshavirus Predicted to Encode a Novel Integral Protein. Microbiology Research, 12(4), 753-764. https://doi.org/10.3390/microbiolres12040055







