Homologs of Phycobilisome Abundance Regulator PsoR Are Widespread across Cyanobacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Analysis and Protein Modeling
2.2. Sequence Homology and Phylogeny
2.3. Promoter Analyses
2.4. Predicted Protein–Protein Interactions
3. Results
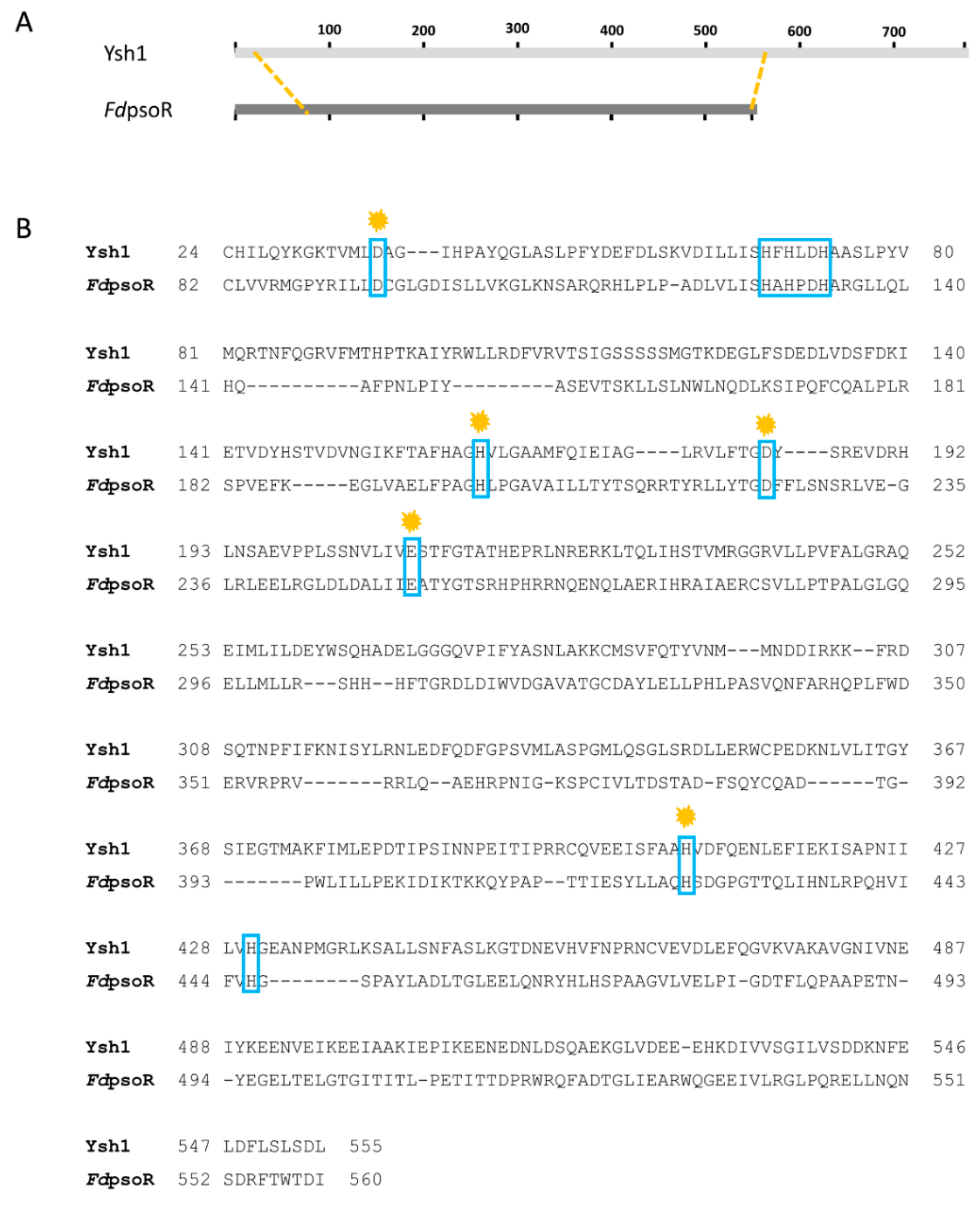
3.1. PsoR Is a Putative β-CASP Domain-Containing Ribonuclease
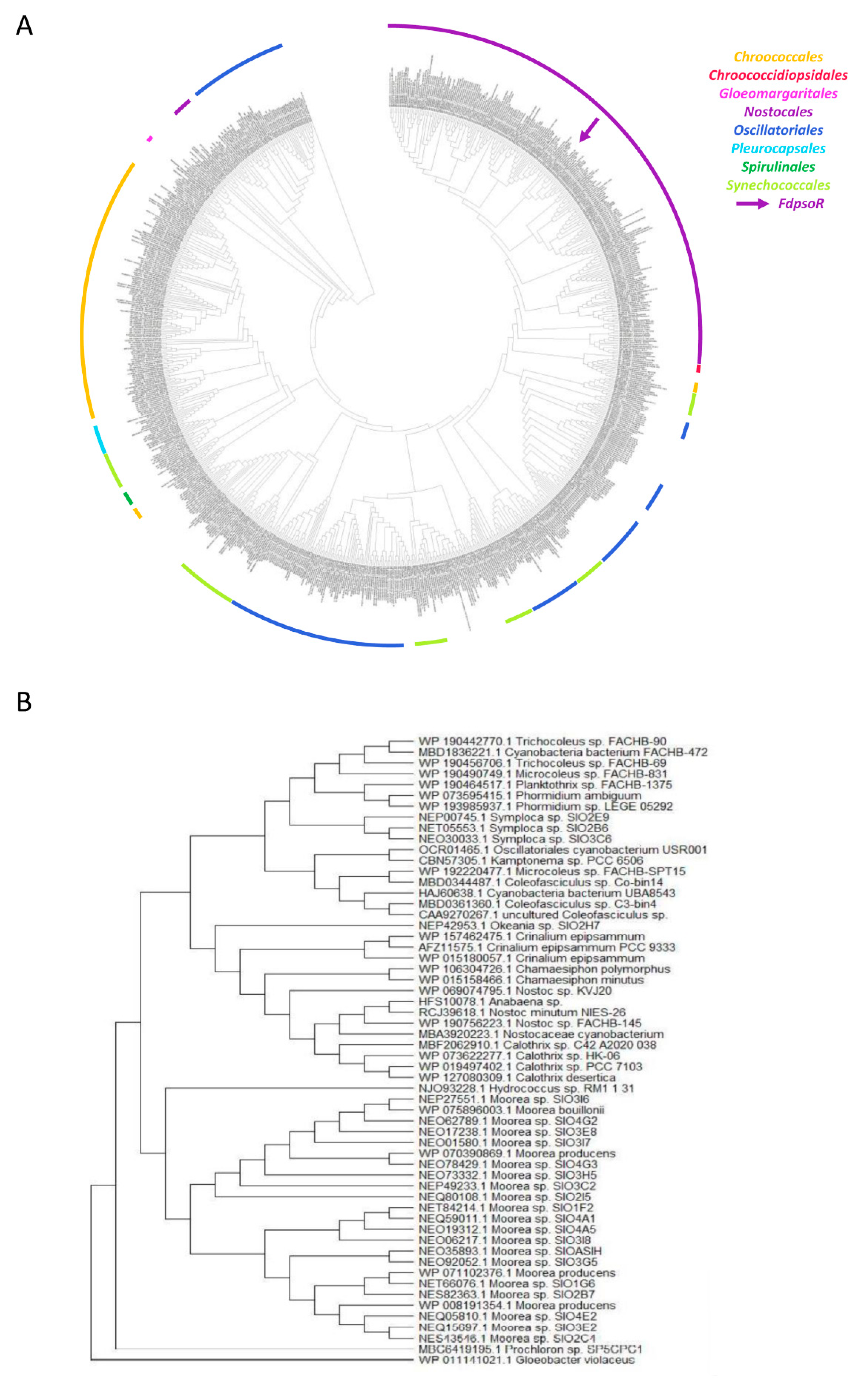
3.2. PsoR Is Widespread throughout the Cyanobacteria Phylum
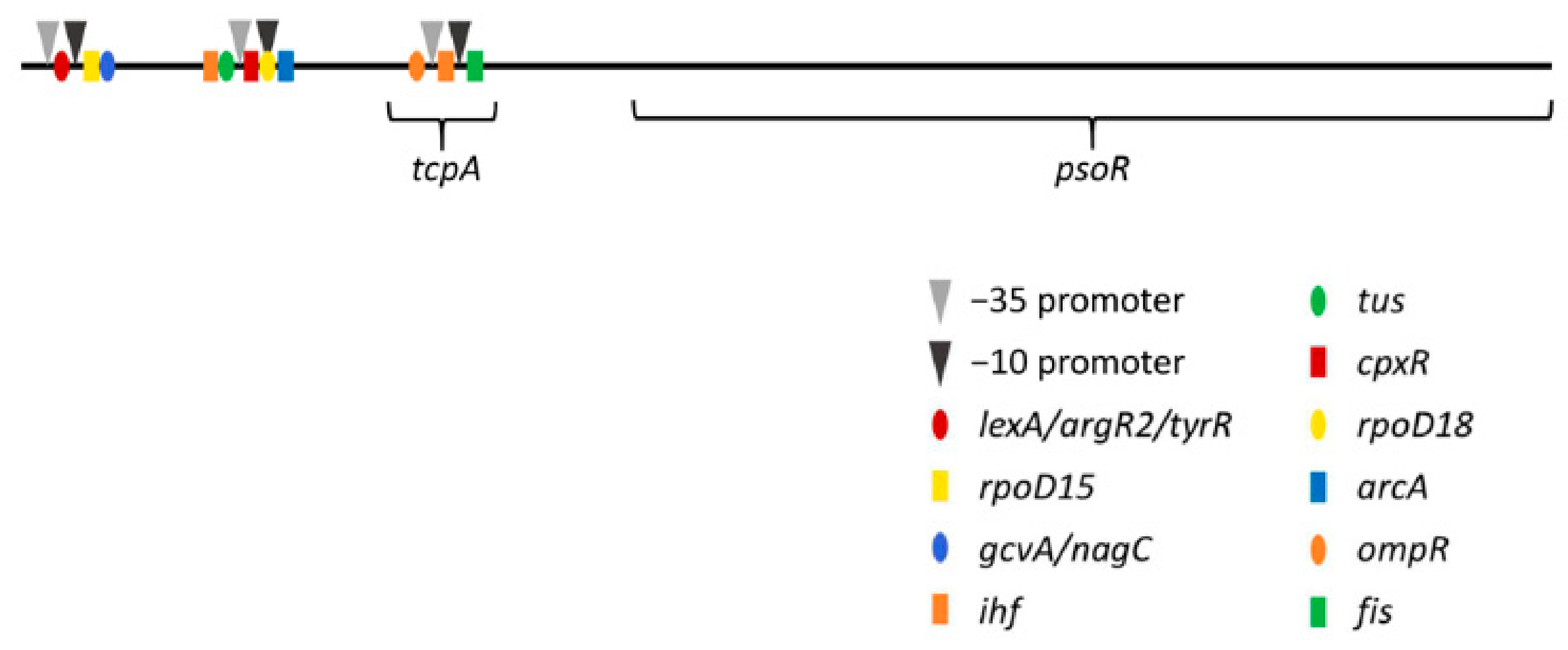
3.3. Promoter Analysis for psoR in Fremyella Diplosiphon
3.4. Protein–Protein Interactions of FdPsoR and Synechocystis PsoR Homologs
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zwirglmaier, K.; Jardillier, L.; Ostrowski, M.; Mazard, S.; Garczarek, L.; Vaulot, D.; Not, F.; Massana, R.; Ulloa, O.; Scanlan, D.J. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ. Microbiol. 2008, 10, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Zehr, J.P. Nitrogen fixation by marine cyanobacteria. Trends Microbiol. 2011, 19, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Santos-Merino, M.; Singh, A.K.; Ducat, D.C. New applications of synthetic biology tools for cyanobacterial metabolic engineering. Front. Bioeng. Biotechnol. 2019, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Hays, S.G.; Ducat, D.C. Engineering cyanobacteria as photosynthetic feedstock factories. Photosyn. Res. 2015, 123, 285–295. [Google Scholar] [CrossRef]
- Jungblut, A.D.; Hawes, I.; Mountfort, D.; Hitzfeld, B.; Dietrich, D.R.; Burns, B.P.; Neilan, B.A. Diversity within cyanobacterial mat communities in variable salinity meltwater ponds of McMurdo Ice Shelf, Antarctica. Environ. Microbiol. 2005, 7, 519–529. [Google Scholar] [CrossRef]
- Martinez, J.N.; Nishihara, A.; Lichtenberg, M.; Trampe, E.; Kawai, S.; Tank, M.; Kühl, M.; Hanada, S.; Thiel, V. Vertical distribution and diversity of phototrophic bacteria within a hot spring microbial mat (Nakabusa Hot Springs, Japan). Microbes Environ. 2019, 34, 374–387. [Google Scholar] [CrossRef]
- Samolov, E.; Baumann, K.; Büdel, B.; Jung, P.; Leinweber, P.; Mikhailyuk, T.; Karsten, U.; Glaser, K. Biodiversity of algae and cyanobacteria in biological soil crusts collected along a climatic gradient in Chile using an integrative approach. Microorganisms 2020, 8, 1047. [Google Scholar] [CrossRef]
- Montgomery, B.L. Mechanisms and fitness implications of photomorphogenesis during chromatic acclimation in cyanobacteria. J. Exp. Bot. 2016, 67, 4079–4090. [Google Scholar] [CrossRef]
- Ho, M.-Y.; Soulier, N.T.; Canniffe, D.P.; Shen, G.; Bryant, D.A. Light regulation of pigment and photosystem biosynthesis in cyanobacteria. Curr. Opin. Plant Biol. 2017, 37, 24–33. [Google Scholar] [CrossRef]
- Sanfilippo, J.E.; Garczarek, L.; Partensky, F.; Kehoe, D.M. Chromatic acclimation in cyanobacteria: A diverse and widespread process for optimizing photosynthesis. Annu. Rev. Microbiol. 2019, 73, 407–433. [Google Scholar] [CrossRef]
- Gutu, A.; Kehoe, D.M. Emerging perspectives on the mechanisms, regulation, and distribution of light color acclimation in cyanobacteria. Mol. Plant 2012, 5, 1–13. [Google Scholar] [CrossRef]
- Adir, N.; Dines, M.; Klartag, M.; McGregor, A.; Melamed-Frank, M. Assembly and disassembly of phycobilisomes. In Complex Intracellular Structures in Prokaryotes; Shively, J.M., Ed.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 47–77. [Google Scholar] [CrossRef]
- Zhao, K.H.; Su, P.; Bohm, S.; Song, B.; Zhou, M.; Bubenzer, C.; Scheer, H. Reconstitution of phycobilisome core-membrane linker, LCM, by autocatalytic chromophore binding to ApcE. Biochim. Biophys. Acta 2005, 1706, 81–87. [Google Scholar] [CrossRef]
- Bennett, A.; Bogorad, L. Complementary chromatic adaptation in a filamentous blue-green alga. J. Cell Biol. 1973, 58, 419–435. [Google Scholar] [CrossRef]
- Singh, A.K.; McIntyre, L.M.; Sherman, L.A. Microarray analysis of the genome-wide response to iron deficiency and iron reconstitution in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2003, 132, 1825–1839. [Google Scholar] [CrossRef]
- Tamary, E.; Kiss, V.; Nevo, R.; Adam, Z.; Bernát, G.; Rexroth, S.; Rögner, M.; Reich, Z. Structural and functional alterations of cyanobacterial phycobilisomes induced by high-light stress. Biochim. Biophys. Acta 2012, 1817, 319–327. [Google Scholar] [CrossRef]
- Collier, J.L.; Grossman, A.R. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 1994, 13, 1039–1047. [Google Scholar] [CrossRef]
- Nagarajan, A.; Zhou, M.; Nguyen, A.Y.; Liberton, M.; Kedia, K.; Shi, T.; Piehowski, P.; Shukla, A.; Fillmore, T.L.; Nicora, C.; et al. Proteomic insights into phycobilisome degradation, a selective and tightly controlled process in the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Biomolecules 2019, 9, 374. [Google Scholar] [CrossRef]
- Six, C.; Joubin, L.; Partensky, F.; Holtzendorff, J.; Garczarek, L. UV-induced phycobilisome dismantling in the marine picocyanobacterium Synechococcus sp. WH8102. Photosynth. Res. 2007, 92, 75–86. [Google Scholar] [CrossRef]
- Chenu, A.; Keren, N.; Paltiel, Y.; Nevo, R.; Reich, Z.; Cao, J. Light adaptation in phycobilisome antennas: Influence on the rod length and structural arrangement. J. Phys. Chem. B 2017, 121, 9196–9202. [Google Scholar] [CrossRef]
- Kolodny, Y.; Zer, H.; Propper, M.; Yochelis, S.; Paltiel, Y.; Keren, N. Marine cyanobacteria tune energy transfer efficiency in their light-harvesting antennae by modifying pigment coupling. FEBS J. 2021, 288, 980–994. [Google Scholar] [CrossRef]
- Montgomery, B.L. Seeing new light: Recent insights into the occurrence and regulation of chromatic acclimation in cyanobacteria. Curr. Opin. Plant Biol. 2017, 37, 18–23. [Google Scholar] [CrossRef]
- Tandeau de Marsac, N. Occurrence and nature of chromatic adaptation in cyanobacteria. J. Bacteriol. 1977, 130, 82–91. [Google Scholar] [CrossRef]
- Everroad, C.; Six, C.; Partensky, F.; Thomas, J.-C.; Holtzendorff, J.; Wood, A.M. Biochemical bases of type IV chromatic adaptation in marine Synechococcus spp. J. Bacteriol. 2006, 188, 3345–3356. [Google Scholar] [CrossRef]
- Gan, F.; Shen, G.; Bryant, D.A. Occurrence of far-red light photoacclimation (FaRLiP) in diverse cyanobacteria. Life 2014, 5, 4. [Google Scholar] [CrossRef]
- Zhao, C.; Gan, F.; Shen, G.; Bryant, D.A. RfpA, RfpB, and RfpC are the master control elements of Far-Red Light Photoacclimation (FaRLiP). Front. Microbiol. 2015, 6, 1303. [Google Scholar] [CrossRef]
- Kehoe, D.M.; Grossman, A.R. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 1996, 273, 1409–1412. [Google Scholar] [CrossRef]
- Kehoe, D.M.; Grossman, A.R. New classes of mutants in complementary chromatic adaptation provide evidence for a novel four-step phosphorelay system. J. Bacteriol. 1997, 179, 3914–3921. [Google Scholar] [CrossRef]
- Cobley, J.G.; Miranda, R.D. Mutations affecting chromatic adaptation in the cyanobacterium Fremyella diplosiphon. J. Bacteriol. 1983, 153, 1486–1492. [Google Scholar] [CrossRef]
- Terauchi, K.; Montgomery, B.L.; Grossman, A.R.; Lagarias, J.C.; Kehoe, D.M. RcaE is a complementary chromatic adaptation photoreceptor required for green and red light responsiveness. Mol. Microbiol. 2004, 51, 567–577. [Google Scholar] [CrossRef]
- Li, L.; Alvey, R.M.; Bezy, R.P.; Kehoe, D.M. Inverse transcriptional activities during complementary chromatic adaptation are controlled by the response regulator RcaC binding to red and green light-responsive promoters. Mol. Microbiol. 2008, 68, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Kehoe, D.M. In vivo analysis of the roles of conserved aspartate and histidine residues within a complex response regulator. Mol. Microbiol. 2005, 55, 1538–1552. [Google Scholar] [CrossRef]
- Bezy, R.P.; Kehoe, D.M. Functional characterization of a cyanobacterial OmpR/PhoB class transcription factor binding site controlling light color responses. J. Bacteriol. 2010, 192, 5923–5933. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alvey, R.M.; Bezy, R.P.; Frankenberg-Dinkel, N.; Kehoe, D.M. A light regulated OmpR-class promoter element co-ordinates light-harvesting protein and chromophore biosynthetic enzyme gene expression. Mol. Microbiol. 2007, 64, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Bezy, R.P.; Wiltbank, L.; Kehoe, D.M. Light-dependent attenuation of phycoerythrin gene expression reveals convergent evolution of green light sensing in cyanobacteria. Proc. Natl. Acad. Sci. USA 2011, 108, 18542–18547. [Google Scholar] [CrossRef]
- Cobley, J.; Khan, S.; Ahmad, H.; Bailey, S. The gene, tcpA, required for photoregulation of phycobilisome abundance and for heterotrophic growth in Fremyella diplosiphon, is a useful phylogenetic marker specific for the phylum, Cyanobacteria. In Proceedings of the 7th European Workshop on Molecular Biology of Cyanobacteria, České Budějovice, Czech Republic, 31 August–4 September 2008; p. 101. [Google Scholar]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Solovyev, V.; Salamov, A. Automatic annotation of microbial genomes and metagenomic sequences. In Metagenomics and Its Applications in Agriculture; Li, R.W., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2011; pp. 61–78. [Google Scholar]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Daiyasu, H.; Osaka, K.; Ishino, Y.; Toh, H. Expansion of the zinc metallo-hydrolase family of the beta-lactamase fold. FEBS Lett. 2001, 503, 1–6. [Google Scholar] [CrossRef]
- Callebaut, I.; Moshous, D.; Mornon, J.P.; de Villartay, J.P. Metallo-beta-lactamase fold within nucleic acids processing enzymes: The beta-CASP family. Nucleic Acids Res. 2002, 30, 3592–3601. [Google Scholar] [CrossRef] [PubMed]
- Dominski, Z.; Carpousis, A.J.; Clouet-d’Orval, B. Emergence of the β-CASP ribonucleases: Highly conserved and ubiquitous metallo-enzymes involved in messenger RNA maturation and degradation. Biochim. Biophys. Acta 2013, 1829, 532–551. [Google Scholar] [CrossRef] [PubMed]
- Mandel, C.R.; Kaneko, S.; Zhang, H.; Gebauer, D.; Vethantham, V.; Manley, J.L.; Tong, L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 2006, 444, 953–956. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.P.; Chechik, M.; Byrne, R.T.; Waterman, D.G.; Ng, C.L.; Dodson, E.J.; Koonin, E.V.; Antson, A.A.; Smits, C. Structure and activity of a novel archaeal β-CASP protein with N-terminal KH domains. Structure 2011, 19, 622–632. [Google Scholar] [CrossRef]
- Mathy, N.; Hébert, A.; Mervelet, P.; Bénard, L.; Dorléans, A.; Li de la Sierra-Gallay, I.; Noirot, P.; Putzer, H.; Condon, C. Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behaviour. Mol. Microbiol. 2010, 75, 489–498. [Google Scholar] [CrossRef]
- Phung, D.K.; Rinaldi, D.; Langendijk-Genevaux, P.S.; Quentin, Y.; Carpousis, A.J.; Clouet-d’Orval, B. Archaeal β-CASP ribonucleases of the aCPSF1 family are orthologs of the eukaryal CPSF-73 factor. Nucleic Acids Res. 2013, 41, 1091–1103. [Google Scholar] [CrossRef]
- States, D.J.; Gish, W. Combined use of sequence similarity and codon bias for coding region identification. J. Comput. Biol. 1994, 1, 39–50. [Google Scholar] [CrossRef]
- Ryan, K.; Calvo, O.; Manley, J.L. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 2004, 10, 565–573. [Google Scholar] [CrossRef]
- Krogmann, D.W.; Pérez-Gómez, B.; Gutiérrez-Cirlos, E.B.; Chagolla-López, A.; González de la Vara, L.; Gómez-Lojero, C. The presence of multidomain linkers determines the bundle-shape structure of the phycobilisome of the cyanobacterium Gloeobacter violaceus PCC 7421. Photosynth. Res. 2007, 93, 27–43. [Google Scholar] [CrossRef]
- Guglielmi, G.; Cohen-Bazire, G.; Bryant, D.A. The structure of Gloeobacter violaceus and its phycobilisomes. Arch. Microbiol. 1981, 129, 181–189. [Google Scholar] [CrossRef]
- Shih, P.M.; Wu, D.; Latifi, A.; Axen, S.D.; Fewer, D.P.; Talla, E.; Calteau, A.; Cai, F.; Tandeau de Marsac, N.; Rippka, R.; et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 1053–1058. [Google Scholar] [CrossRef]
- Monteiro, L.M.O.; Sanches-Medeiros, A.; Westmann, C.A.; Silva-Rocha, R. Unraveling the complex interplay of Fis and IHF through synthetic promoter engineering. Front. Bioeng. Biotechnol. 2020, 8, 510. [Google Scholar] [CrossRef]
- Takashima, K.; Nagao, S.; Kizawa, A.; Suzuki, T.; Dohmae, N.; Hihara, Y. The role of transcriptional repressor activity of LexA in salt-stress responses of the cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 2020, 10, 17393. [Google Scholar] [CrossRef]
- Nukui, M.; Mello, L.V.; Littlejohn, J.E.; Setlow, B.; Setlow, P.; Kim, K.; Leighton, T.; Jedrzejas, M.J. Structure and molecular mechanism of Bacillus anthracis cofactor-independent phosphoglycerate mutase: A crucial enzyme for spores and growing cells of Bacillus species. Biophys. J. 2007, 92, 977–988. [Google Scholar] [CrossRef]
- Connelly, J.C.; Kirkham, L.A.; Leach, D.R. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl. Acad. Sci. USA 1998, 95, 7969–7974. [Google Scholar] [CrossRef]
- Patel, P.H.; Suzuki, M.; Adman, E.; Shinkai, A.; Loeb, L.A. Prokaryotic DNA polymerase I: Evolution, structure, and “base flipping” mechanism for nucleotide selection. J. Mol. Biol. 2001, 308, 823–837. [Google Scholar] [CrossRef]
- Condon, C.; Gilet, L. The metallo-β-lactamase family of ribonucleases. In Ribonucleases; Nicholson, A.W., Ed.; Springer: Berlin, Germany, 2011; pp. 245–267. [Google Scholar] [CrossRef]
- Yokono, M.; Akimoto, S.; Koyama, K.; Tsuchiya, T.; Mimuro, M. Energy transfer processes in Gloeobacter violaceus PCC 7421 that possesses phycobilisomes with a unique morphology. Biochim. Biophys. Acta 2008, 1777, 55–65. [Google Scholar] [CrossRef][Green Version]
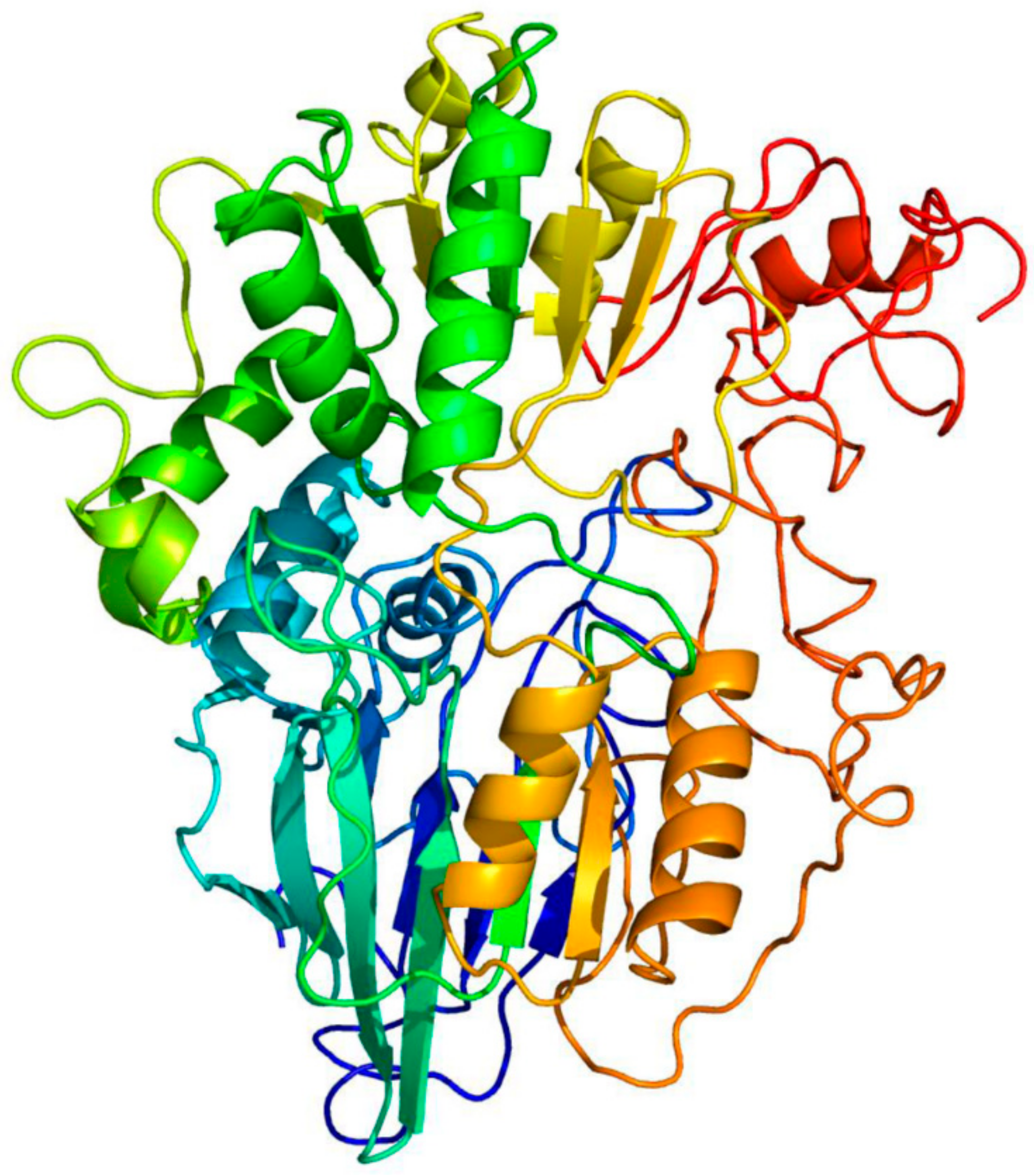


| PDB ID | Organism | Description | Alignment Coverage | Confidence | Percent ID |
|---|---|---|---|---|---|
| c2ycbA_ | Methanothermobacter thermautotrophicus str. Delta H | Hydrolase; archaeal β-CASP protein with n-terminal kh domains | 15–430 | 100.0 | 22% |
| c2xr1B_ | Methanosarcina mazei | Hydrolase; dimeric archaeal cleavage and polyadenylation specificity factor with n-terminal kh domains (kh-cpsf) | 15–430 | 100.0 | 18% |
| c2xr1A_ | Methanosarcina mazei | Hydrolase; dimeric archaeal cleavage and polyadenylation specificity factor with n-terminal kh domains (kh-cpsf) | 18–430 | 100.0 | 16% |
| c3af5A_ | Pyrococcus horikoshii OT3 | Hydrolase; archaeal cpsf subunit | 15–430 | 100.0 | 20% |
| d2i7xa1 | Saccharomyces cerevisiae | Family: β-CASP RNA-metabolizing hydrolases | 19–430 | 100.0 | 14% |
| c2i7xA_ | Saccharomyces cerevisiae | RNA-binding protein, protein binding | 19–430 | 100.0 | 14% |
| d2dkfa1 | Thermus thermophilus | Family: β-CASP RNA-metabolizing hydrolases | 20–430 | 100.0 | 23% |
| c5habB_ | Methanolobus psychrophilus R15 | Hydrolase: mpy-rnase j (mutant h84a), an archaeal rnase j | 15–430 | 100.0 | 16% |
| c5a0tA_ | Streptomyces coelicolor A3(2) | Hydrolase: catalysis and 5′ end sensing by ribonuclease rnase j of the metallo-beta-lactamase family | 16–431 | 100.0 | 17% |
| d2i7ta1 | Human (Homo sapiens); Saccharomyces cerevisiae | Family: beta-CASP RNA-metabolizing hydrolases | 18–431 | 100.0 | 24% |
| Β-CASP Motif | Ysh1 Residue | FdPsoR Residue |
|---|---|---|
| Motif 1 | D37 | D47 |
| Motif 2 | H73 | H85 |
| Motif 3 | H163 | H156 |
| Motif 4 | D184 | D175 |
| Motif A | E209 | E203 |
| Motif B | H408 | H376 |
| Motif C | H430 | H398 |
| Promoter | −10 Promoter | −35 Promoter | Oligonucleotides from Known TF Binding Sites | ||||
|---|---|---|---|---|---|---|---|
| bp | Sequence | bp | Sequence | bp | Name | Sequence | bp |
| 84 | CGATATACT | 69 | TTGTAT | 45 | lexA | TATATAAA | 55 |
| argR2 | ATATAAAT | 56 | |||||
| lexA | ATAAATAA | 58 | |||||
| tyrR | TAAATAAA | 59 | |||||
| rpoD15 | TAAGGTTA | 83 | |||||
| gcvA | TTATATTT | 92 | |||||
| nagC | ATATTTTA | 94 | |||||
| 402 | AAATATATT | 387 | TTACTA | 364 | ihf | AAATAAAA | 343 |
| tus | CATTAGTA | 351 | |||||
| cpxR | TAAAAAGA | 376 | |||||
| rpoD18 | AAATATAT | 387 | |||||
| arcA | TTAATTAA | 404 | |||||
| 818 | TTTTATGAT | 803 | TTGTTT | 779 | ompR | TCATATTT | 759 |
| ihf | ACAAAAAA | 791 | |||||
| fis | ACAATTAT | 816 | |||||
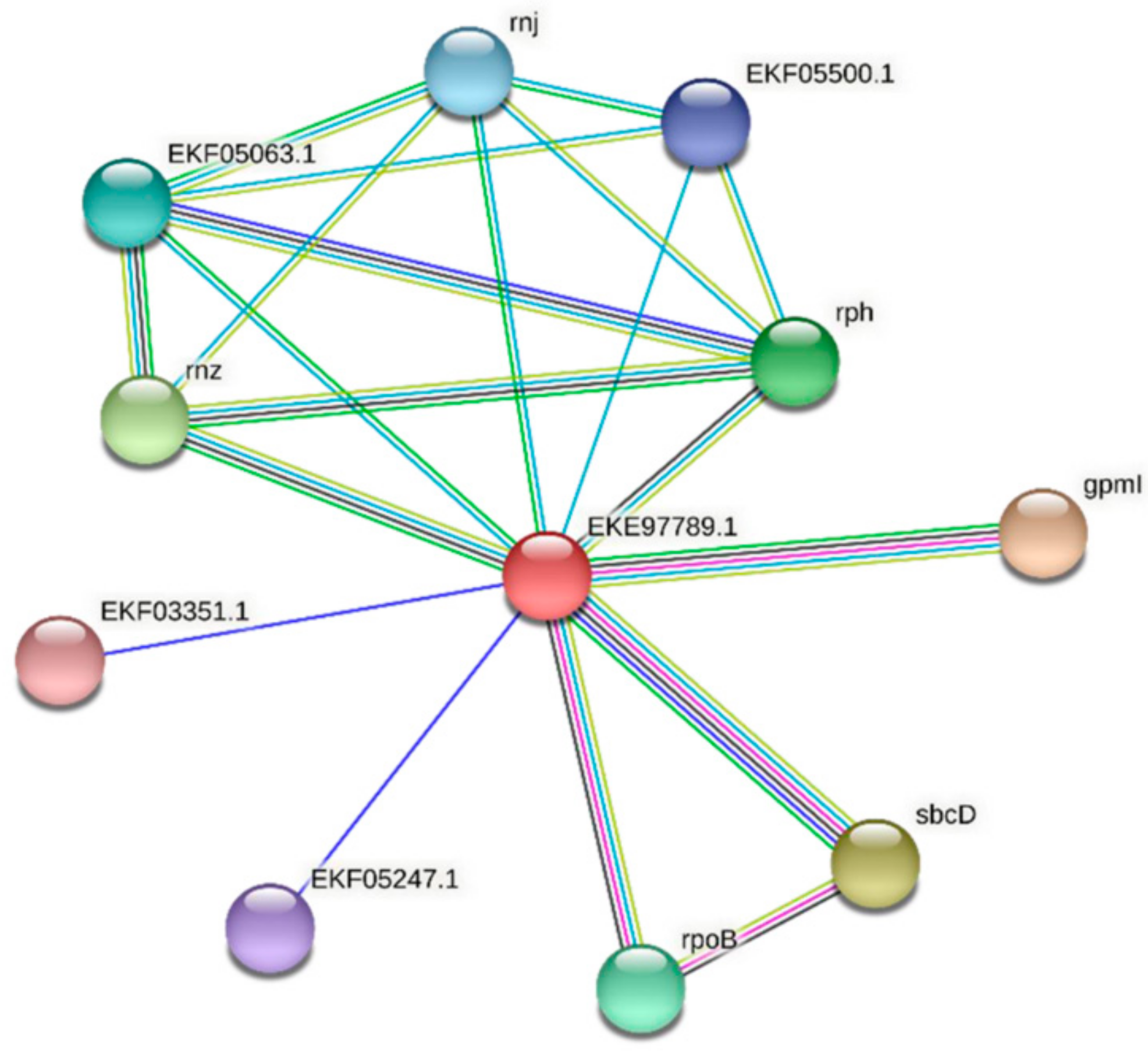
| Predicted Interaction Partner | Description | Confidence Score |
|---|---|---|
| GpmI | Catalyzes the interconversion of 2-phosphoglycerate and 3-phosphoglycerate. | 0.871 |
| SbcC/D | SbcD cleaves DNA hairpin structures. Subunit D. The complex acts as a 3′->5′ double-strand exonuclease that can open hairpins. It also has 5′ single-stranded endonuclease activity. | 0.749 |
| Rnz | Zinc phosphodiesterase displays some tRNA 3′-processing endonuclease activity. It is probably involved in tRNA maturation by removing a 3′-trailer from precursor tRNA. | 0.727 |
| Rph | Phosphorolytic 3′–5′ exoribonuclease plays an important role in tRNA 3′-end maturation. | 0.689 |
| RpoB | DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. | 0.688 |
| EKF05063.1 | Ribonuclease E/G | 0.680 |
| Rnj | An RNase has a 5′–3′ exonuclease and possibly endonuclease activity. Involved in maturation of rRNA, and in some organisms, mRNA maturation and/or decay. | 0.678 |
| EKF05500.1 | Ribonuclease T(2) family protein | 0.666 |
| EKF05247.1 | Uncharacterized protein | 0.656 |
| EKF03351.1 | Phycocyanin-associated protein | 0.654 |
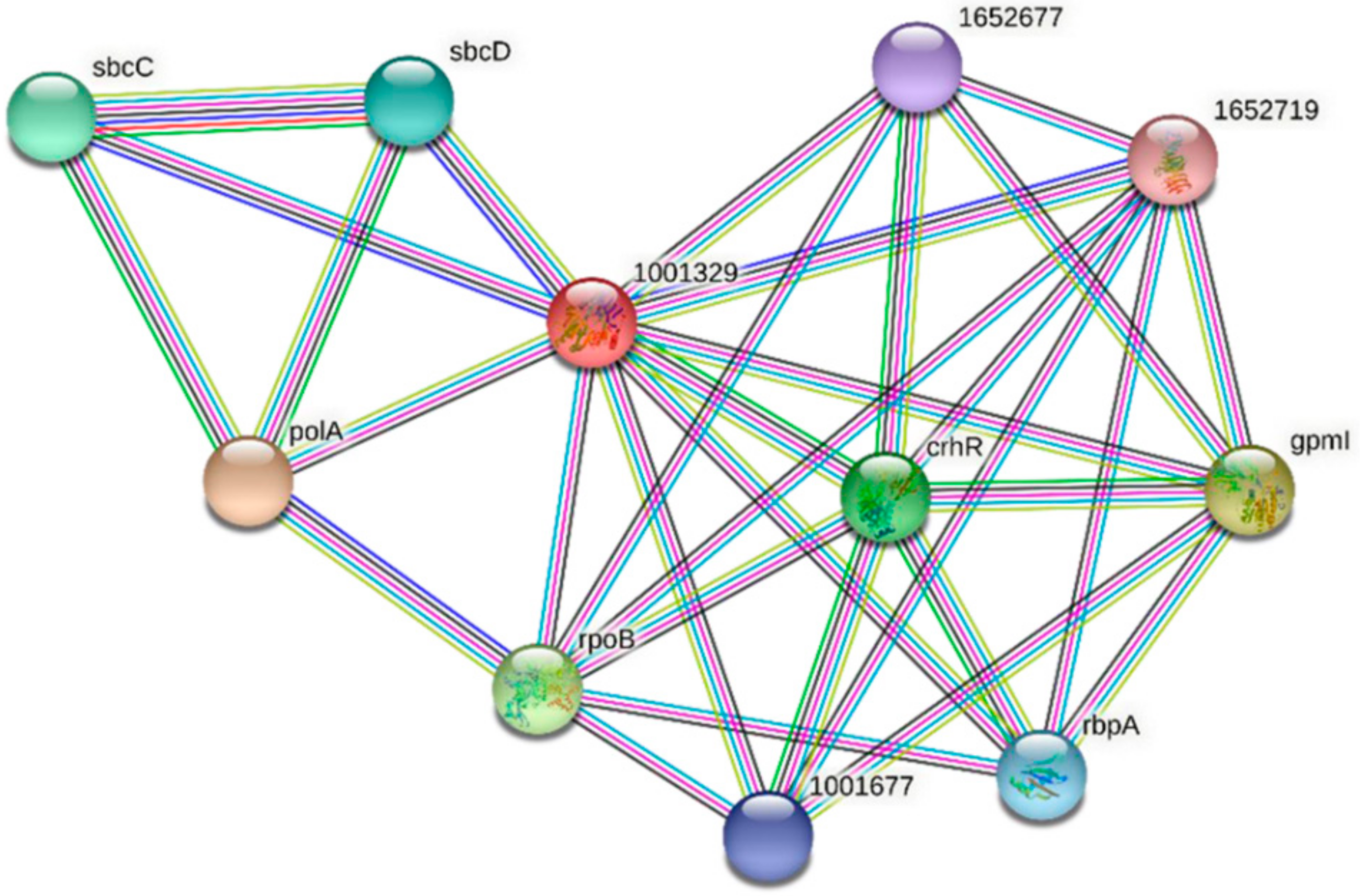
| Predicted Interaction Partner | Description | Confidence Score |
|---|---|---|
| PolA | DNA polymerase I. In addition to polymerase activity, this DNA polymerase exhibits 3′–5′ and 5′–3′ exonuclease activity. | 0.937 |
| GpmI | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase. Catalyzes the interconversion of 2-phosphoglycerate and 3-phosphoglycerate. | 0.900 |
| RpoB | DNA-directed RNA polymerase subunit beta. DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. | 0.736 |
| CrhR | RNA helicase CrhR. An ATP-dependent bidirectional RNA helicase with RNA-dependent ATPase activity. | 0.700 |
| SbcC | Nuclease SbcCD subunit C. SbcCD cleaves DNA hairpin structures. | 0.691 |
| RbpA | Putative RNA-binding protein | 0.674 |
| 1001677 | Slr0193; RNA-binding protein | 0.674 |
| 1652677 | Ssr1480; RNA-binding protein | 0.674 |
| 1652719 | sSlr1410; uncharacterized WD repeat-containing protein | 0.658 |
| Predicted Interaction Partner | Description | Confidence Score |
|---|---|---|
| PolA | DNA polymerase I. In addition to polymerase activity, this DNA polymerase exhibits 5′–3′ exonuclease activity. | 0.932 |
| Gpm | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase. Catalyzes the interconversion of 2-phosphoglycerate and 3-phosphoglycerate. | 0.870 |
| SbcD | Nuclease SbcCD subunit D. SbcCD cleaves DNA hairpin structures. | 0.757 |
| ACA99530.1 | Uncharacterized protein | 0.751 |
| SbcC | Nuclease SbcCD subunit C. SbcCD cleaves DNA hairpin structures. | 0.740 |
| ACA98931.1 | FHA-domain protein | 0.728 |
| ACA98998.1 | SH3b domain-containing protein | 0.721 |
| ACA98040.1 | WD-repeat protein | 0.718 |
| ACA99688.1 | TPR-repeat containing protein | 0.711 |
| ACA99716.1 | Serine/threonine kinase | 0.681 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Layer, A.; Montgomery, B.L. Homologs of Phycobilisome Abundance Regulator PsoR Are Widespread across Cyanobacteria. Microbiol. Res. 2022, 13, 167-182. https://doi.org/10.3390/microbiolres13020014
Layer A, Montgomery BL. Homologs of Phycobilisome Abundance Regulator PsoR Are Widespread across Cyanobacteria. Microbiology Research. 2022; 13(2):167-182. https://doi.org/10.3390/microbiolres13020014
Chicago/Turabian StyleLayer, Alicia, and Beronda L. Montgomery. 2022. "Homologs of Phycobilisome Abundance Regulator PsoR Are Widespread across Cyanobacteria" Microbiology Research 13, no. 2: 167-182. https://doi.org/10.3390/microbiolres13020014
APA StyleLayer, A., & Montgomery, B. L. (2022). Homologs of Phycobilisome Abundance Regulator PsoR Are Widespread across Cyanobacteria. Microbiology Research, 13(2), 167-182. https://doi.org/10.3390/microbiolres13020014






