Tolerance and Cadmium (Cd) Immobilization by Native Bacteria Isolated in Cocoa Soils with Increased Metal Content
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria Strains and Growth Conditions
2.2. Effect of Cadmium on Bacterial Growth
2.3. Cadmium Uptake Ability of Bacteria Strains
2.4. Transmission Electron Microscopy and Analyzer Energy Dispersive X-ray (TEM/EDX)
2.5. Fourier-Transform Infrared (FT-IR) Spectrum Analysis
2.6. Greenhouse Experiment
2.6.1. Location and Experimental Design
2.6.2. Pot Assays
2.6.3. Physical and Chemical Analysis of Soil Properties
2.6.4. Determination of Cd in Cacao Plant Parts
2.7. Statistical Analysis
3. Results
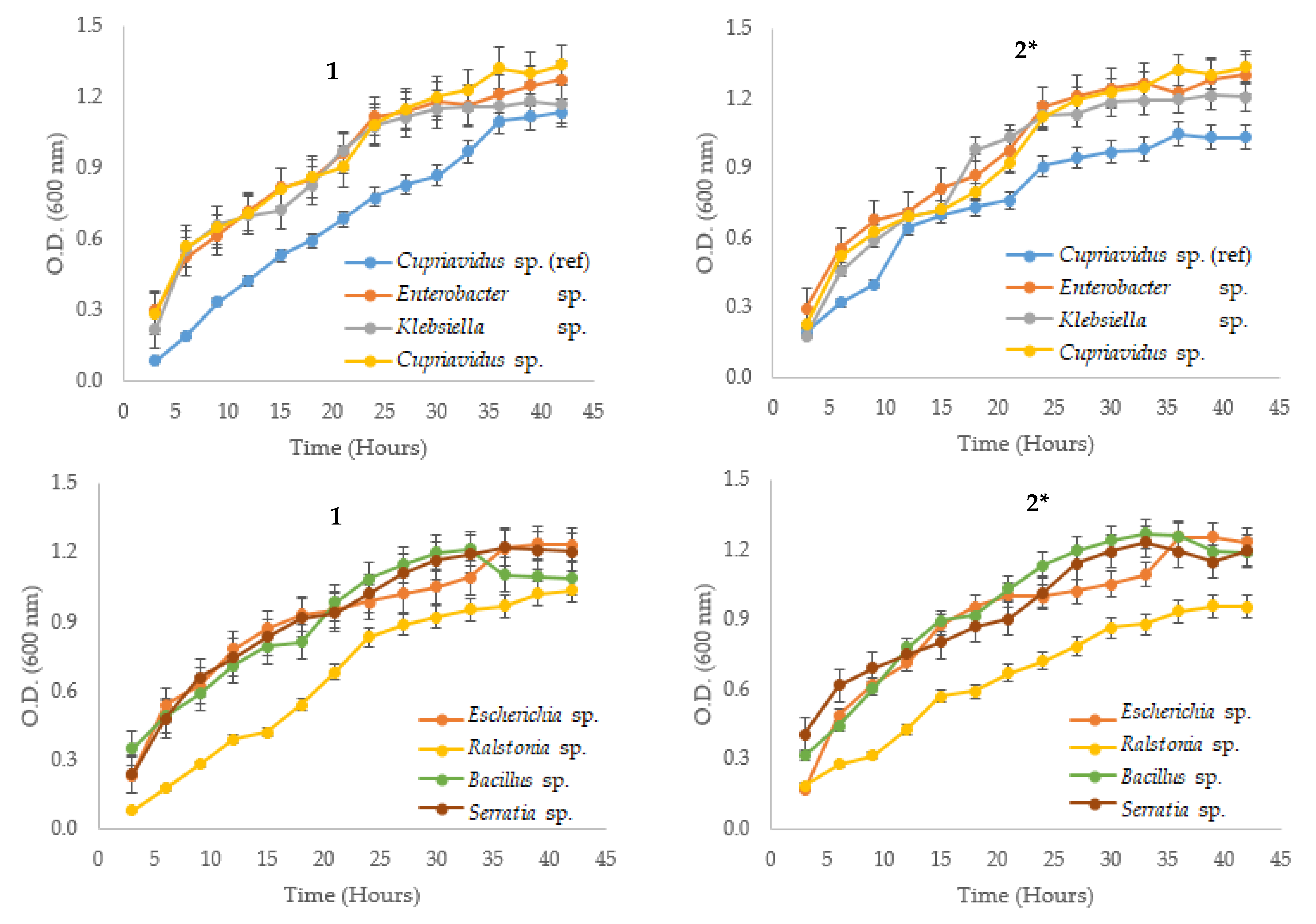
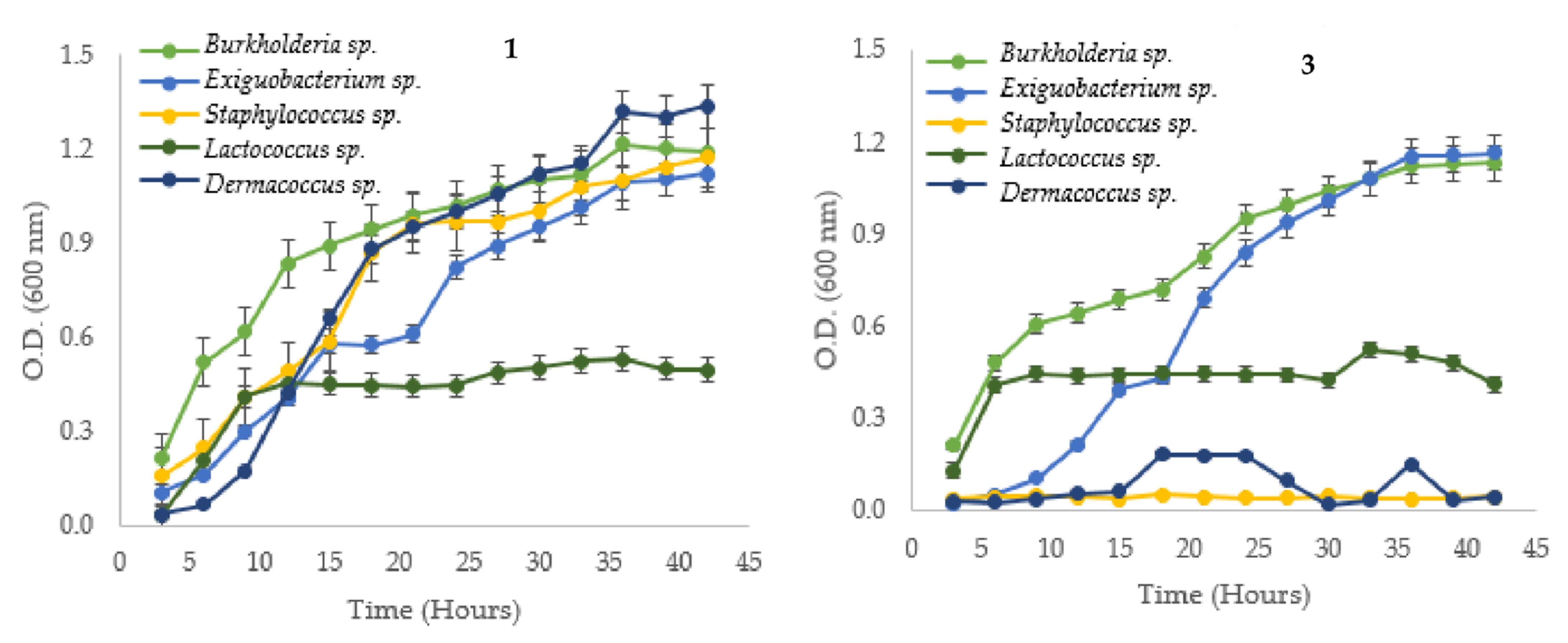
3.1. Bacterial Strain and Growth Curves Measurements
3.2. Cd Bioaccumulation in Native Bacteria Strains
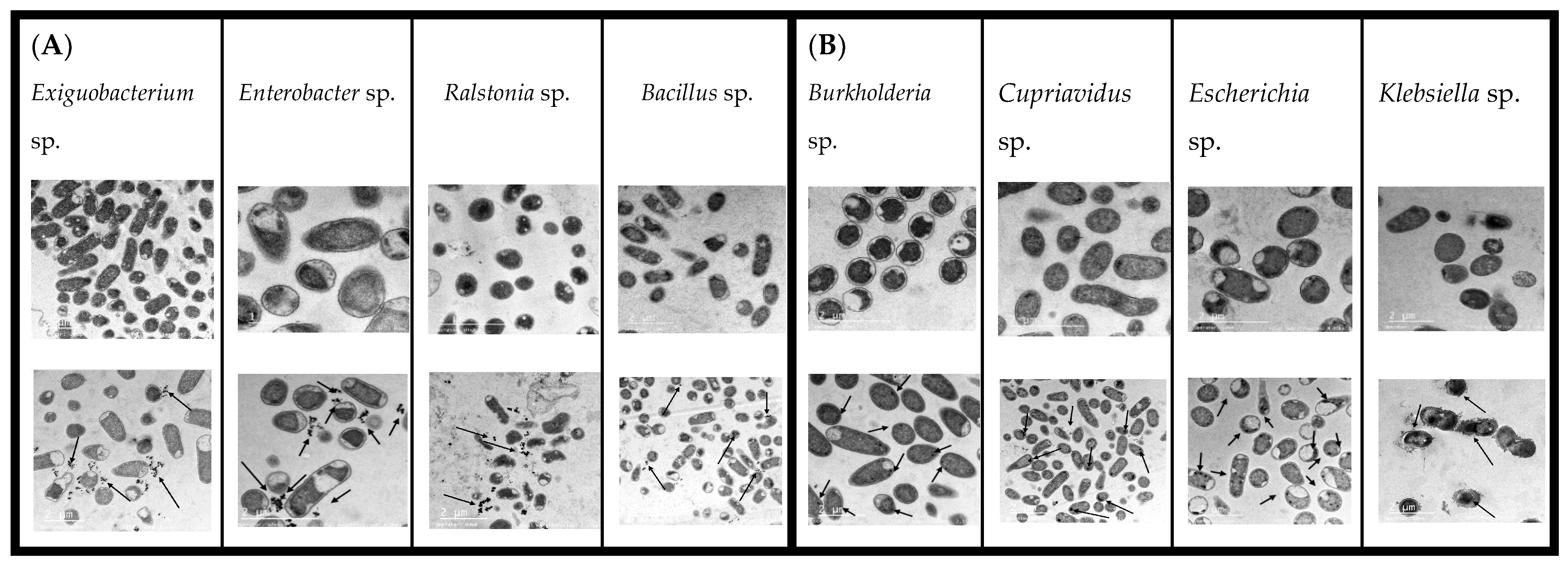
3.3. TEM, EDX, and FT-IR Analysis and Mechanisms of Cd Interaction Using Native Bacteria
3.3.1. TEM Results
3.3.2. EDX Spectra
3.3.3. FT-IR Spectra
3.4. Responses of Inoculation to Native Tolerant Cd Bacteria in Young Cacao Plants at Differents Cd Levels in the Soil
3.4.1. Soil Analysis and Cd Concentration Chemicals Used in the Study
3.4.2. Roots and Aerial Parts Cacao Biomass
3.4.3. Cd Accumulation in Cacao Plants, Bioconcentration, and Translocation Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zao, M.; Zhang, C.; Zeng, G.; Huang, D.; Xu, P.; Cheng, M. Growth. Growth, metabolism of Phanerochaete chrysosporium and route of lignin degradation in response to cadmium stress in solid-state fermentation. Chemosphere 2015, 138, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, R.; Banerjee, P. Mechanism of cadmium binding on the cell wall of an acidophilic bacterium. Bioresour. Technol. 2012, 108, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Siripornadulsil, S.; Siripornadulsil, W. Cadmium-tolerant bacteria reduce the uptake of cadmium in rice: Potential for microbial bioremediation. Ecotoxicol. Environ. Saf. 2013, 94, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Tossapol, L.; Sooksawat, N. Bioaccumulation and biosorption of Cd2+ and Zn2+ by bacteria isolated from a zinc mine in Thailand. Ecotoxicol. Environ. Saf. 2015, 122, 322–330. [Google Scholar] [CrossRef]
- Al-Saraj, M.; Abdel-Latif, M. Bioaccumulation of some hazardous metals by sol gel entrapped microorganisms. J. Non-Cryst. Solids 1999, 248, 137–140. [Google Scholar] [CrossRef]
- Wong, C.; Cobbett, C. HMA P-type ATPases are the major mechanism for root to shoot Cd translocation in Arabidopsis thaliana. New Phytol. 2009, 181, 71–78. [Google Scholar] [CrossRef]
- Wang, C.; Huang, Y. Burkholderia sp. Y4 inhibits cadmium accumulation in rice by increasing essential nutrient uptake and preferentially absorbing cadmium. Chemosphere 2020, 252, 126603. [Google Scholar] [CrossRef]
- Shi, Z.; Zhang, Z. Characterization of a high cadmium accumulating soil bacterium, Cupriavidus sp. WS2. Chemosphere 2020, 247, 125834. [Google Scholar] [CrossRef]
- Rani, A.; Souche, Y. Comparative assessment of in situ bioremediation potential of cadmium resistant acidophilic Pseudomonas putida 62BN and alkalophilic Pseudomonas monteilli 97AN strains on soybean. Int. Biodeterior. Biodegrad. 2009, 63, 62–66. [Google Scholar] [CrossRef]
- Engbersen, N.; Gramlich, A. Cadmium accumulation and allocation in different cacao cultivars. Sci. Total Environ. 2019, 678, 660–670. [Google Scholar] [CrossRef]
- Chavez, E.; He, Z. Concentration of cadmium in cacao beans and its relationship with soil cadmium in southern Ecuador. Sci. Total Environ. 2015, 533, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Bravo, D.; Pardo-Díaz, S. Cadmium and Cadmium-tolerant soil bacteria in cacao crops from northeastern Colombia. J. Appl. Microbiol. 2018, 124, 1175–1194. [Google Scholar] [CrossRef] [PubMed]
- Feria-Cáceres, P.; Penagos-Vélez, L. Theobroma cacao L. agricultural soils with natural low and high cadmium (Cd) in Santander (Colombia), contain a persistent shared bacterial composition shaped by multiple soil variables and bacterial isolates highly resistant to Cd concentrations. Curr. Res. Microb. Sci. 2021, 2, 100086. [Google Scholar] [CrossRef]
- Madigan, M.; Martinko, D. Brock Biology of Microorganisms, 13th ed.; Benjamin Cummings Editorial©: San Francisco, CA, USA, 2012. [Google Scholar]
- Huang, F.; Guo, C.-L.; Lu, G.-N.; Yi, X.-Y.; Zhu, L.-D.; Dang, Z. Bioaccumulation characterization of cadmium by growing Bacillus cereus RC-1 and its mechanism. Chemosphere 2014, 109, 134–142. [Google Scholar] [CrossRef]
- Ferreira, P.; Bomfeti, C. Cupriavidus necator strains: Zinc and cadmium tolerance and bioaccumulation. Sci. Agric. 2018, 75, 452–460. [Google Scholar] [CrossRef]
- Chen, Y.; Zhu, Q. How Serratia marcescens HB-4 absorbs Cadmium and its implication on phytoremediation. Ecol. Environ. Saf. 2019, 185, 109723. [Google Scholar] [CrossRef]
- Suarez, C.; Restrepo, J.; Quinchia, A. Colombian vegetal fibers as a reinforcement in polymeric matrix composites. Tecnura 2017, 21, 51. [Google Scholar] [CrossRef]
- Pereira de Araújo, R.; Furtado de Almeida, A. Photosynthetic, antioxidative, molecular and ultrastructural responses of young cacao plants to Cd toxicity in the soil. Ecotoxicol. Environ. Saf. 2017, 144, 148–157. [Google Scholar] [CrossRef]
- Kongor, J.; Hinneh, M. Factors influencing quality variation in cocoa (Theobroma cacao) bean flavour profile: A review. Food Reseach Int. 2016, 82, 44–52. [Google Scholar] [CrossRef]
- Padmavathiamma, P.; Loretta, L. Phytoremediation Technology: Hyper-accumulation Metals in Plants. Water Air Soil Pollut. 2007, 184, 105–126. [Google Scholar] [CrossRef]
- Yin, K.; Wang, Q. Microorganism remediation strategies towards heavy metals. Review. Chem. Eng. J. 2019, 360, 1553–1563. [Google Scholar] [CrossRef]
- Zoropogui, A.; Gambarelli, S. CzcE from Cupriavidus metallidurans CH34 is a copper-binding protein. Biochem. Biophys. Res. Commun. 2008, 365, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Banik, A.; Pandya, P. Characterization of halotolerant, pigmented, plant growth promoting bacteria of groundnut rhizosphere and its in-vitro evaluation of plant-microbe protocooperation to withstand salinity and metal stress. Sci. Total Environ. 2018, 630, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Shatrohan, L.; Sheel, R.; Ben, S.; Rajesh, K. Biosurfactant and exopolysaccharide-assisted rhizobacterial technique for the remediation of heavy metal contaminated soil: An advancement in metal phytoremediation technology. Environ. Technol. Innov. 2018, 10, 243–263. [Google Scholar] [CrossRef]
- Rajendran, P.; Muthukrishnan, J. Microbes in heavy metal remediation. Indian J. Exp. Biol. 2003, 41, 935–944. [Google Scholar]
- Xie, Y.; Li, H. Kinetic simulating of Cr (VI) removal by the waste Chlorella vulgaris biomass. J. Taiwan Inst. Chem. Eng. 2014, 45, 1773–1782. [Google Scholar] [CrossRef]
- Ozdemir, S.; Kilinc, E. Biosorption of Cd, Cu, Ni, Mn and Zn from aqueous solutions by thermophilic bacteria, Geobacillus toebii sub.sp. decanicus and Geobacillus thermoleovorans sub.sp. stromboliensis: Equilibrium, kinetic and thermodynamic studies. Chem. Eng. J. 2009, 152, 195–206. [Google Scholar] [CrossRef]
- Long, J.; Yu, M. Characterization of cadmium biosorption by inactive biomass of two cadmium-tolerant endophytic bacteria Microbacterium sp. D2-2 and Bacillus sp. C9-3. Ecotoxicology 2021, 30, 1419–1428. [Google Scholar] [CrossRef]
- Huang, F.; Dang, Z. Biosorption of Cd(II) by live and dead cells of Bacillus cereus RC-1 isolated from cadmium-contaminated soil. Colloids Surf. Biointerfaces 2013, 107, 11–18. [Google Scholar] [CrossRef]
- Mathew, B.; Biju, V. Accumulation of lead (Pb II) metal ions by Bacillus toyonensis SCE1 species, innate to industrial-area ground water and nanoparticle synthesis. Appl. Nanosci. 2019, 9, 49–66. [Google Scholar] [CrossRef]
- Kim, S.; Jin, M. Biosortion of cationic basic dye and cadmium by the novel biosorbent Bacillus catenulatus JB-022 strain. J. Biosci. Bioeng. 2015, 119, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Huang, Y. Two plant growth promoting bacterial Bacillus strains possess different mechanisms in adsortion and resistence to cadmium. Sci. Total Environ. 2020, 741, 140422. [Google Scholar] [CrossRef] [PubMed]
- Pugazhendhi, A.; Boovaragamoorthy, G. New insight into effective biosorption of lead from aqueos solution using Ralstonia solanacearum: Characterization and mechanism studies. J. Clean. Prod. 2018, 174, 1234–1239. [Google Scholar] [CrossRef]
- Park, Y.; Ko, J. Enhancement of bioremediation by Ralstonia sp. HM-1 in sediment polluted by Cd and Zn. Bioresour. Technol. 2008, 99, 7458–7463. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, P. Isolation, characterization and inoculation of Cd tolerant rice endophytes and their impacts on rice under contamined environment. Environ. Pollut. 2020, 260, 113990. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, Q. Biochemical and genetic basis of cadmium biosorption by Enterobacter ludwigii LY6, isolated from industrial contaminated soil. Environ. Pollut. 2020, 264, 114637. [Google Scholar] [CrossRef]
- Park, J.; Chon, H. Characterization of cadmium biosorption by Exiguobacterium sp. isolated from farmland soil near Cu-Pb-Zn mine. Environ. Sci. Pollut. Res. 2016, 23, 11814–11822. [Google Scholar] [CrossRef]
- Sheng, Y.; Wang, Y. Cadmium tolerant characteristic of a newly isolated Lactococcus lactis subsp. lactis. Environ. Toxicol. Pharmacol. 2016, 48, 183–190. [Google Scholar] [CrossRef]
- Valls, M.; Gonzalez-Duarte, R. Bioaccumulation of heavy metals with protein fusions of metallothionein to bacterial OMPs. Biochimie 1998, 80, 855–861. [Google Scholar] [CrossRef]
- Afzal, A.; Rasool, M. Assesment of heavy metal tolerance and biosorptive potencial of Klebsiella variicola isolated from industrial effluents. AMB Express 2017, 7, 184. [Google Scholar] [CrossRef]
- Shamin, S.; Rehman, A. Cadmium resistance and acumulation potencial of Klebsiella pneumoniae strain CBL-1 isolated from industrial wastewater. Pak. J. Zool. 2012, 44, 203–208. [Google Scholar]
- Holmes, J.; Richardson, D. Cadmium-specific formation of metal sulfide ‘Q-particles’ by Klebsiella pneumoniae. Microbiology 1997, 143, 2521–2530. [Google Scholar] [CrossRef] [PubMed]
- Sriram, M.; Gayathiri, S. Novel lipopeptide biosurfactant produced by hydrocarbon degrading and heavy metal tolerant bacterium Escherichia fergusonii KLU01 as a potential tool for bioremediation. Bioresour. Technol. 2011, 102, 9291–9295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Q. Bioaccumulation and distribution of cadmium by Burkholderia cepacia GYP1 under oligotrophic condition and mechanism analysis at proteome level. Ecotoxicol. Environ. Saf. 2019, 176, 162–169. [Google Scholar] [CrossRef]
- Li, J.; Liu, Y. Sorption mechanism and distribution of cadmium by different microbial species. J. Environ. Manag. 2019, 237, 552–559. [Google Scholar] [CrossRef]
- Huang, H.; Jia, Q. Screening strains for microbial technology of cadmium. Chemosphere 2020, 251, 126428. [Google Scholar] [CrossRef]
- BeĆ, K.; Grabska, J. Biomolecular and bioanalytical applications of infrared spectroscopy—A review. Anal. Chim. Acta 2020, 1133, 150–177. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, C. Cadmium adsorption mechanism of Serratia marcescens HB-4. CIE J. 2017, 68, 1574–1581. [Google Scholar] [CrossRef]
- Huang, J.; Liu, C. Identification of a novel heavy metal resistant Ralstonia strain and its growth response to cadmium exposure. J. Hazard. Mater. 2021, 416, 125942. [Google Scholar] [CrossRef]
- Sun, R.; Wang, L. Cadmium resistance mechanisms of a functional strain Enterobacter sp. DNB-S2, isolated from black soil in Northeast China. Environ. Pollut. 2020, 263, 114612. [Google Scholar] [CrossRef]
- Boza, E.J.; Motamayor, J.C.; Amores, F.M.; Cedeno-Amador, S.; Tondo, C.L.; Livingstone, D.S.; Gutiérrez, O.A. Genetic Characterization of the cacao (Theobroma cacao L.) clone ‘CCN 51′ and its impact and significance on global cacao improvement and production. J. Am. Soc. Hortic. Sci. 2014, 139, 219–229. [Google Scholar] [CrossRef]
- He, S.; He, Z. Soil biogeochemistry, plant physiology and phytoremediation of cadmium contaminated soils. Adv. Agron. 2015, 134, 135–225. [Google Scholar] [CrossRef]
- Qi, F.; Lamb, D. Cadmium solubility and bioavailability in soils amended with acidic and neutral biochar. Sci. Total Environ. 2018, 610, 1457–1466. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Dumat, C. Cadmium bioavailability, uptake, toxicity and detoxifcation in soilplant system. Rev. Environ. Contam. Toxicol. 2016, 241, 73–137. [Google Scholar] [CrossRef]
- Sauvé, S.; Norvell, W. Speciation and complexation of cadmium in extracted soil solutions. Environ. Sci. Technol. 2000, 34, 291–296. [Google Scholar] [CrossRef]
- Zug, K.; Huamaní Yupanqui, H. Cadmium Accumulation in Peruvian Cacao (Theobroma cacao L.) and Opportunities for Mitigation. Water Air Soil Pollut. 2019, 230, 72. [Google Scholar] [CrossRef]
- Liu, H.; Xie, Y. Efect of Serratia sp. K3 combined with organic materials on cadmium migration in soil-Vetiveria zizanioides L. system and bacterial community in contaminated soil. Chemosphere 2020, 242, 125164. [Google Scholar] [CrossRef]
- Li, Y.; Liu, K. Improvement of cadmium phytoremediation by Centella asiatica L. after soil inoculation with cadmium-resistant Enterobacter sp. FM-1. Chemosphere 2018, 202, 280–288. [Google Scholar] [CrossRef]
- Ehsan, M.; Santamaría-Delgado, K. Phytostabilization of cadmium contaminated soils by Lupinus uncinatus Schdl. Span. J. Agric. Res. 2009, 7, 390–397. [Google Scholar] [CrossRef]
- Verbruggen, N.; Hermans, C. Mechanisms to cope with arsenic or cadmium excess in plants. Curr. Opin. Plant Biol. 2009, 12, 364–372. [Google Scholar] [CrossRef]
- Antoniadis, V.; Levizou, E. Trace elements in the soil-plant interface: Phytoavailability, translocation, and phytoremediation–A review. Earth Sci. Rev. 2017, 171, 621–645. [Google Scholar] [CrossRef]
- Barraza, F.; Schreck, E. Beyond cadmium accumulation: Distribution of other trace elements in soils and cacao beans in Ecuador. Environ. Res. 2021, 192, 110241. [Google Scholar] [CrossRef] [PubMed]
- Arevalo-Hernandez, C.; Arevalo-Gardini, E. Growth and nutritional responses of wild and domesticated cacao genotypes to soil Cd stress. Sci. Total Environ. 2021, 763, 144021. [Google Scholar] [CrossRef] [PubMed]
- Oladipo, O.; Ezeokoli, O. Tolerance and growth kinetics of bacteria isolated from gold and gemstone mining sites in response to heavy metal concentrations. J. Environ. Manag. 2018, 212, 357–366. [Google Scholar] [CrossRef] [PubMed]

| Phylogenetic Affiliation (Related Reference Sequence) | Gram Stain | GenBank Accesión Number |
|---|---|---|
| Bacillus toyonensis | + | MN587894 |
| Burkholderia arboris | − | MN587896 |
| Cupriavidus necátor | − | MN587892 |
| Escherichia fergusonii | − | MN587901 |
| Exiguobacterium acetylicum | + | MN587893 |
| Ralstonia solanacearum | − | MN587895 |
| Serratia marcescens | − | MN587899 |
| Dermacoccus barathri | − | MN587890 |
| Enterobacter tabaco | − | MN587891 |
| Klebsiella variicola | − | MN587897 |
| Lactococcus lactis | + | MN587898 |
| Staphylococcus capitis | + | MN587900 |
| Native Strain | µ (h−1) | g (h) | K (Generation/h) | |||
|---|---|---|---|---|---|---|
| Without Cd | With Cd | Without Cd | With Cd | Without Cd | With Cd | |
| Cupriavidus metallidurans * | 0.093 | 0.058 | 7.492 | 11.907 | 0.133 | 0.084 |
| Serratia sp. (6-2) | 0.076 | 0.089 | 9.142 | 7.388 | 0.109 | 0.128 |
| Cupriavidus sp. (15-1) | 0.089 | 0.106 | 7.760 | 6.519 | 0.129 | 0.153 |
| Klebsiella sp. (18-4B) | 0.099 | 0.126 | 7.007 | 5.491 | 0.143 | 0.182 |
| Bacillus sp. (10-2) | 0.100 | 0.094 | 6.944 | 7.388 | 0.144 | 0.135 |
| Ralstonia sp. (16-1) | 0.123 | 0.065 | 5.639 | 10.728 | 0.177 | 0.093 |
| Escherichia sp. (4-2) | 0.066 | 0.074 | 10.548 | 9.365 | 0.095 | 0.107 |
| Enterobacter sp. (29-4B) | 0.111 | 0.104 | 6.249 | 6.657 | 0.160 | 0.150 |
| Exiguobacterium sp. (11-4A) | 0.115 | 0.144 | 6.005 | 4.806 | 0.167 | 0.208 |
| Burkholderia sp. (17-1) | 0.062 | 0.082 | 11.124 | 8.441 | 0.090 | 0.118 |
| Lactococcus sp. (22-4) | 0.240 | 0.392 | 2.885 | 1.769 | 0.347 | 0.565 |
| Bacteria | Initial (Cd) (mg/L) | Final pH Nutrient Broth | (Cd) Supernatant (mg/L) | (Cd) Captured (mg/L) | % Cd Capture | Bioaccumulation Factor (BF) |
|---|---|---|---|---|---|---|
| Cupriavidus metallidurans * | 6.36 ± 0.41 | 1.70 ± 0.11 | 1.61 ± 0.07 | 48.63 ± 0.49 | 0.43 ± 0.02 | |
| Serratia sp. (6-2) | 3.73 ± 0.31 | 6.25 ± 0.17 | 3.14 ± 0.09 | 0.28 ± 0.10 | 8.02 ± 2.73 | 0.07 ± 0.03 |
| Cupriavidus sp. (15-1) | 6.88 ± 0.30 | 2.38 ± 0.06 | 0.78 ± 0.14 | 24.61 ± 3.35 | 0.21 ± 0.04 | |
| Klebsiella sp. (18-4B) | 6.60 ± 0.38 | 0.69 ± 0.08 | 2.59 ± 0.40 | 78.86 ± 2.97 | 0.69 ± 0.11 | |
| Bacillus sp. (10-2) | 6.34 ± 0.23 | 2.20 ± 0.09 | 0.87 ± 0.19 | 28.21 ± 5.37 | 0.23 ± 0.05 | |
| Ralstonia sp. (16-1) | 6.62 ± 0.25 | 1.41 ± 0.19 | 2.17 ± 0.21 | 60.59 ± 5.59 | 0.58 ± 0.06 | |
| Escherichia sp. (4-2) | 6.22 ± 0.42 | 1.50 ± 0.21 | 1.78 ± 0.39 | 53.93 ± 9.09 | 0.48 ± 0.11 | |
| Enterobacter sp. (29-4B) | 6.73 ± 0.44 | 1.56 ± 0.05 | 1.39 ± 0.04 | 47.16 ± 1.40 | 0.37 ± 0.01 | |
| Exiguobacterium sp. (11-4A) | 3.22 ± 0.27 | 5.14 ± 0.38 | 2.92 ± 0.03 | 0.90 ± 0.09 | 28.24 ± 2.0 | 0.24 ± 0.02 |
| Burkholderia sp. (17-1) | 6.06 ± 0.20 | 2.50 ± 0.05 | 0.68 ± 0.02 | 21.33 ± 2.21 | 0.18 ± 0.01 | |
| Lactococcus sp. (22-4) | 5.16 ± 0.19 | 1.73 ± 0.04 | 1.51 ± 0.18 | 46.38 ± 3.45 | 0.40 ± 0.05 |
| Wavenumber (cm−1) | Assignment | Vibration Types |
|---|---|---|
| ~3275 | Amide A | Stretch N-H proteins |
| 2958/2873 | Methyl CH3 | Stretch C-H asymmetric lipids/carbohydrates/proteins |
| 2923/2852 | Methyl CH2 | Stretch C-H asymmetric lipids/carbohydrates/proteins |
| ~1636 | Amide I | Stretch C=O proteins |
| 1529 | Amide II | Flexion N-H proteins |
| 1467 | Methyl CH2 | Flexion C-H proteins/lipids |
| 1455 | Methyl CH3 | Flexion C-H proteins/lipids |
| 1389 | Carbonyl | Stretch C=O asymmetric fatty acids and amino acids |
| ~1230/~1060 | Phosphate PO2− | Stretch P=O asymmetric phospholipids, nucleic acids |
| ~966 | Phosphate PO2− | Stretch P=O symmetric phospholipids, nucleic acids |
| ~914 | Ketones | C-O ring vibrations of nucleic acids “sugars” |
| ~860 | C-H group | Tri-substituted bending C-H |
| Cd Level | Treatment | Fresh Weight (g) | Dry Weight (g) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Roots | Stems | Aerial Parts | Roots | Stems | Aerial Parts | ||||||||
| T1 a | T2 b | T1 a | T2 b | T1 a | T2 b | T1 a | T2 b | T1 a | T2 b | T1 a | T2 b | ||
| Low | Control | 5.55 ± 1.91 | 6.58 ± 2.10 | 7.4 ± 4.2 | 8.0 ± 1.2 | 8.3 ± 1.8 | 10.5 ± 0.2 | 0.74 ± 0.47 | 1.17 ± 0.15 | 1.37 ± 0.55 | 2.31 ± 0.29 | 1.89 ± 0.58 | 3.52 ± 0.49 |
| Bacteria 1 | 3.56 ± 0.09 | 6.13 ± 2.56 | 4.6 ± 0.1 | 8.5 ± 2.7 | 6.9 ± 0.9 | 12.0 ± 4.9 | 0.42 ± 0.06 | 1.14 ± 0.13 | 1.29 ± 0.63 | 2.29 ± 0.54 | 1.57 ± 0.82 | 3.49 ± 1.52 | |
| Bacteria 2 | 3.39 ± 0.08 | 5.70 ± 1.73 | 5.4 ± 0.9 | 8.4 ± 0.8 | 7.7 ± 0.5 | 12.7 ± 1.9 | 0.58 ± 0.19 | 1.13 ± 0.23 | 1.58 ± 0.47 | 2.18 ± 0.27 | 1.85 ± 0.74 | 3.95 ± 0.96 | |
| Bacteria 3 | 3.43 ± 0.52 | 7.34 ± 2.49 | 5.1 ± 1.5 | 9.5 ± 2.1 | 7.7 ± 0.6 | 14.0 ± 2.3 | 0.44 ± 0.11 | 1.35 ± 0.10 | 1.31 ± 0.43 | 2.50 ± 0.88 | 1.78 ± 0.60 | 4.17 ± 0.84 | |
| High | Control | 3.83 ± 1.22 | 4.81 ± 2.03 | 4.9 ± 1.7 | 8.6 ± 1.1 | 6.6 ± 0.5 | 10.4 ± 0.5 | 0.44 ± 0.05 | 1.01 ± 0.36 | 1.19 ± 0.34 | 2.47 ± 0.29 | 1.57 ± 0.65 | 3.25 ± 0.51 |
| Bacteria 1 | 2.53 ± 1.17 | 3.97 ± 0.70 | 4.2 ± 0.8 | 7.0 ± 1.3 | 5.4 ± 0.7 | 9.5 ± 1.8 | 0.40 ± 0.02 | 0.86 ± 0.18 | 1.14 ± 0.41 | 2.04 ± 0.29 | 1.33 ± 0.78 | 3.25 ± 0.58 | |
| Bacteria 2 | 3.18 ± 0.49 | 4.28 ± 2.36 | 5.0 ± 0.5 | 6.2 ± 2.8 | 5.9 ± 1.0 | 8.3 ± 3.6 | 0.48 ± 0.26 | 0.87 ± 0.14 | 1.05 ± 0.25 | 1.81 ± 0.52 | 1.58 ± 0.95 | 2.81 ± 1.07 | |
| Bacteria 3 | 3.13 ± 1.36 | 3.42 ± 1.28 | 4.2 ± 1.9 | 6.1 ± 0.5 | 5.2 ± 1.4 | 7.8 ± 1.9 | 0.50 ± 0.07 | 0.79 ± 0.41 | 1.04 ± 0.30 | 1.92 ± 0.58 | 1.22 ± 0.76 | 2.77 ± 0.87 | |
| ANOVA (Type II) | |||||||||||||
| Treatment | 8.82 | 1.91 | 6.46 | 5.81 | 1.53 | 0.09 | 0.22 | 0.16 | |||||
| Time | 12.56 | 64.09 | 18.31 | 18.77 | 10.91 | 2.81 *** | 10.7 *** | 38.81 *** | |||||
| Cd level | 3.10 | 32.25* | 19.59 | 21.14 | 59.92* | 0.51 ** | 0.90 | 3.75 * | |||||
| Treatment: Cd | 1.66 | 5.55 | 4.68 | 0.85 | 29.35 | 0.02 | 0.38 | 1.09 | |||||
| Time | Factors Treatment | Bioconcentration (BCF) | Translocatión (TF) | ||
|---|---|---|---|---|---|
| Low | High | Low | High | ||
| T1 | Control | 4.92 ± 4.17 (0.39–11.72) | 2.54 ± 0.07 (0.97–4.70) | 1.13 ± 1.54 (0.25–13.72) | 4.30 ± 2.54 (0.97–4.70) |
| Klebsiella sp. (18-4B) | 3.12 ± 0.70 (0.79–6.27) | 1.51 ± 0.07 (0.77–2.16) | 0.76 ± 0.68 (0.36–3.29) | 8.24 ± 6.61 (0.77–2.16) | |
| Exiguobacterium sp. (11-4A) | 2.75 ± 1.91 (0.93–7.28) | 2.39 ± 1.06 (1.40–3.57) | 0.24 ± 0.17 (1.65–7.44) | 5.18 ± 3.03 (1.40–3.57) | |
| Enterobacter sp. (29-4B) | 3.56 ± 1.64 (0.82–6.75) | 2.51 ± 1.36 (0.82–5.16) | 0.95 ± 1.20 (0.31–4.62) | 3.77 ± 1.31 (0.82–5.16) | |
| T2 | Control | 4.60 ± 1.49 (0.18–18.57) | 3.36 ± 0.90 (0.82–10.39) | 5.82 ± 10.83 (0.19–18.57) | 7.69 ± 10.47 (0.50–13.75) |
| Klebsiella sp. (18-4B) | 2.28 ± 0.59 (0.15–7.88) | 2.27 ± 0.56 (0.51–5.54) | 5.32 ± 7.80 (0.43–7.88) | 7.76 ± 9.47 (0.084–15.12) | |
| Exiguobacterium sp. (11-4A) | 4.64 ± 3.19 (0.45–8.01) | 2.53 ± 0.28 (0.74–4.94) | 5.82 ± 10.83 (0.45–33.84) | 6.39 ± 6.70 (0.69–13.49) | |
| Enterobacter sp. (29-4B) | 3.53 ± 3.99 (0.31–17.73) | 2.48 ± 0.77 (0.71–3.66) | 6.64 ± 11.87 (0.31–17.73) | 4.37 ± 1,62 (0.73–7.22) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feria-Cáceres, P.F.; Penagos-Velez, L.; Moreno-Herrera, C.X. Tolerance and Cadmium (Cd) Immobilization by Native Bacteria Isolated in Cocoa Soils with Increased Metal Content. Microbiol. Res. 2022, 13, 556-573. https://doi.org/10.3390/microbiolres13030039
Feria-Cáceres PF, Penagos-Velez L, Moreno-Herrera CX. Tolerance and Cadmium (Cd) Immobilization by Native Bacteria Isolated in Cocoa Soils with Increased Metal Content. Microbiology Research. 2022; 13(3):556-573. https://doi.org/10.3390/microbiolres13030039
Chicago/Turabian StyleFeria-Cáceres, Pedro F., Lucas Penagos-Velez, and Claudia X. Moreno-Herrera. 2022. "Tolerance and Cadmium (Cd) Immobilization by Native Bacteria Isolated in Cocoa Soils with Increased Metal Content" Microbiology Research 13, no. 3: 556-573. https://doi.org/10.3390/microbiolres13030039
APA StyleFeria-Cáceres, P. F., Penagos-Velez, L., & Moreno-Herrera, C. X. (2022). Tolerance and Cadmium (Cd) Immobilization by Native Bacteria Isolated in Cocoa Soils with Increased Metal Content. Microbiology Research, 13(3), 556-573. https://doi.org/10.3390/microbiolres13030039






