Rhizopus oryzae Inulinase Production and Characterization with Application in Chicory Root Saccharification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cultures and Media
2.2. Culture Screening for Enzyme Production
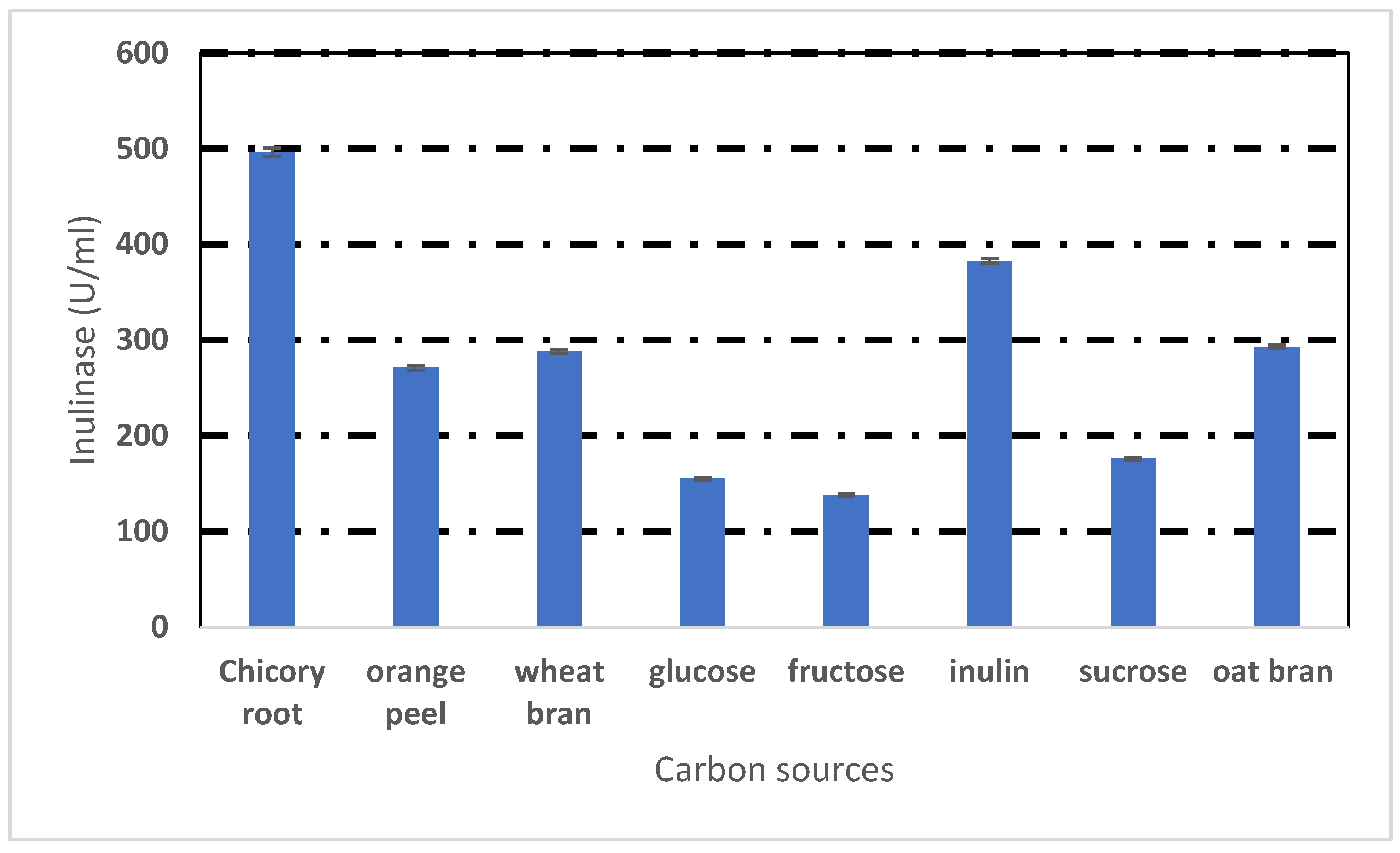
2.3. Effect of Different Carbon Sources
2.4. Multifactorial Experiments for Optimizing Inulinase Production
2.5. Screening of Variables Influencing the Production of Inulinase Enzyme
2.6. Optimization of Significant Variables Using Box–Behnken Design
2.7. Analytical Methods
2.8. Purification of Inulinase
2.9. Characterization of Inulinase
2.10. Fermentation of Chicory Root Using Produced Inulinase Enzyme
2.11. Multifactorial Experiments for Fermentation of Chicory Root Using Produced Inulinase Enzyme
2.11.1. Screening of Factors Affecting Inulinase Production
2.11.2. Optimization of Significant Variables Using Box–Behnken Design
3. Results and Discussion
3.1. Screening for Inulinase Producing Strains
3.2. Effect of Different Carbon Sources on Inulinase Production by Rhizopus oryzae NRRL 3563
3.3. Multifactorial Designs for Optimizing Inulinase Production
3.3.1. Screening of Significant Variables Using the Plackett–Burman Design
3.3.2. Optimization of Medium Components Using Box–Behnken Design
3.4. Model Validation
3.5. Comparison of Optimization of Inulinase Production by Rhizopus oryzaei NRRL 3563 with other Fungi
3.6. Purification of Inulinase Enzyme
3.7. Characterization of Partially Purified Inulinase Enzyme
3.7.1. Molecular Determination
3.7.2. Effect of Temperature on Inulinase Activity and Stability
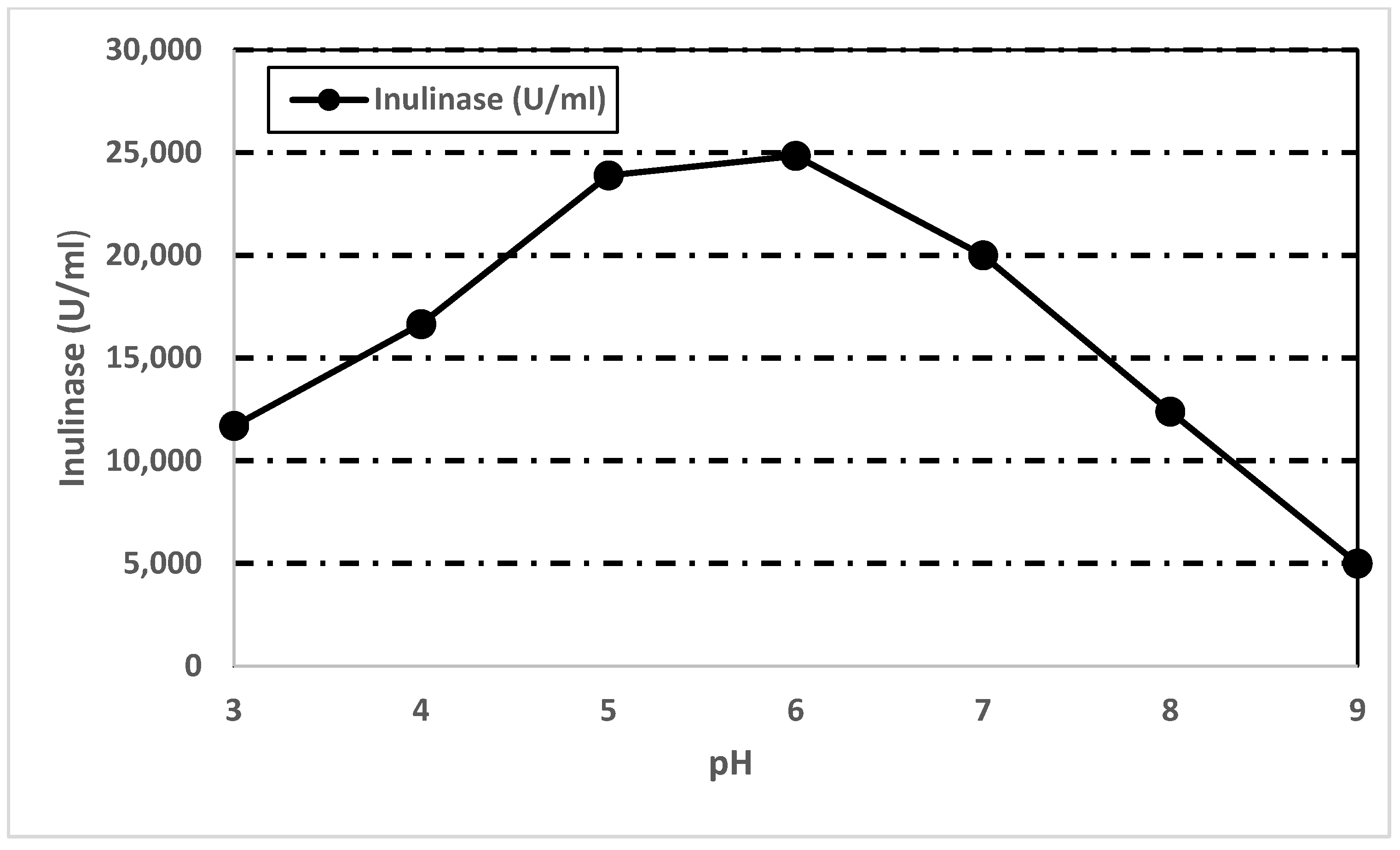
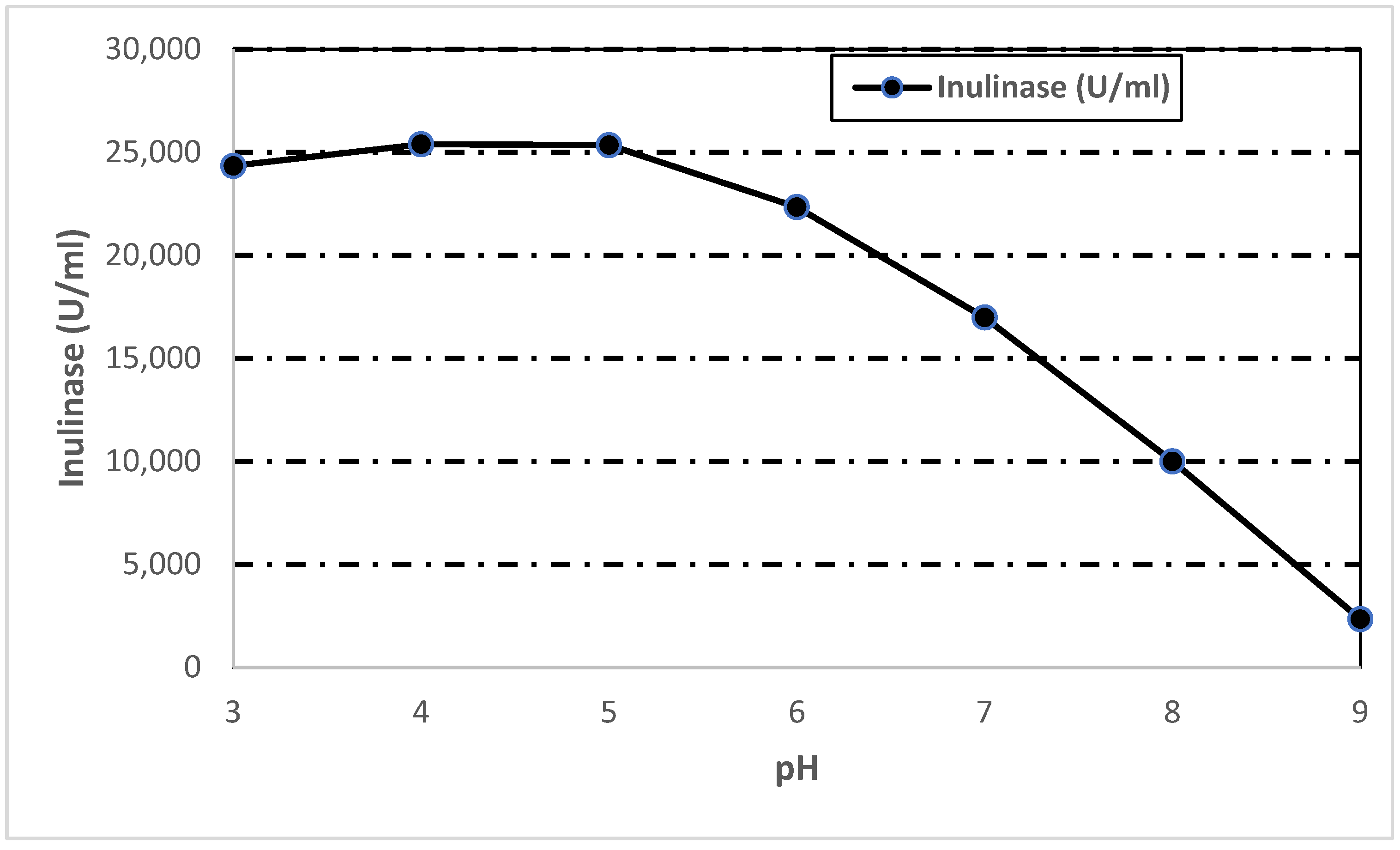
3.7.3. Effect of pH on Inulinase Activity and Stability
3.7.4. Substrate Specificity
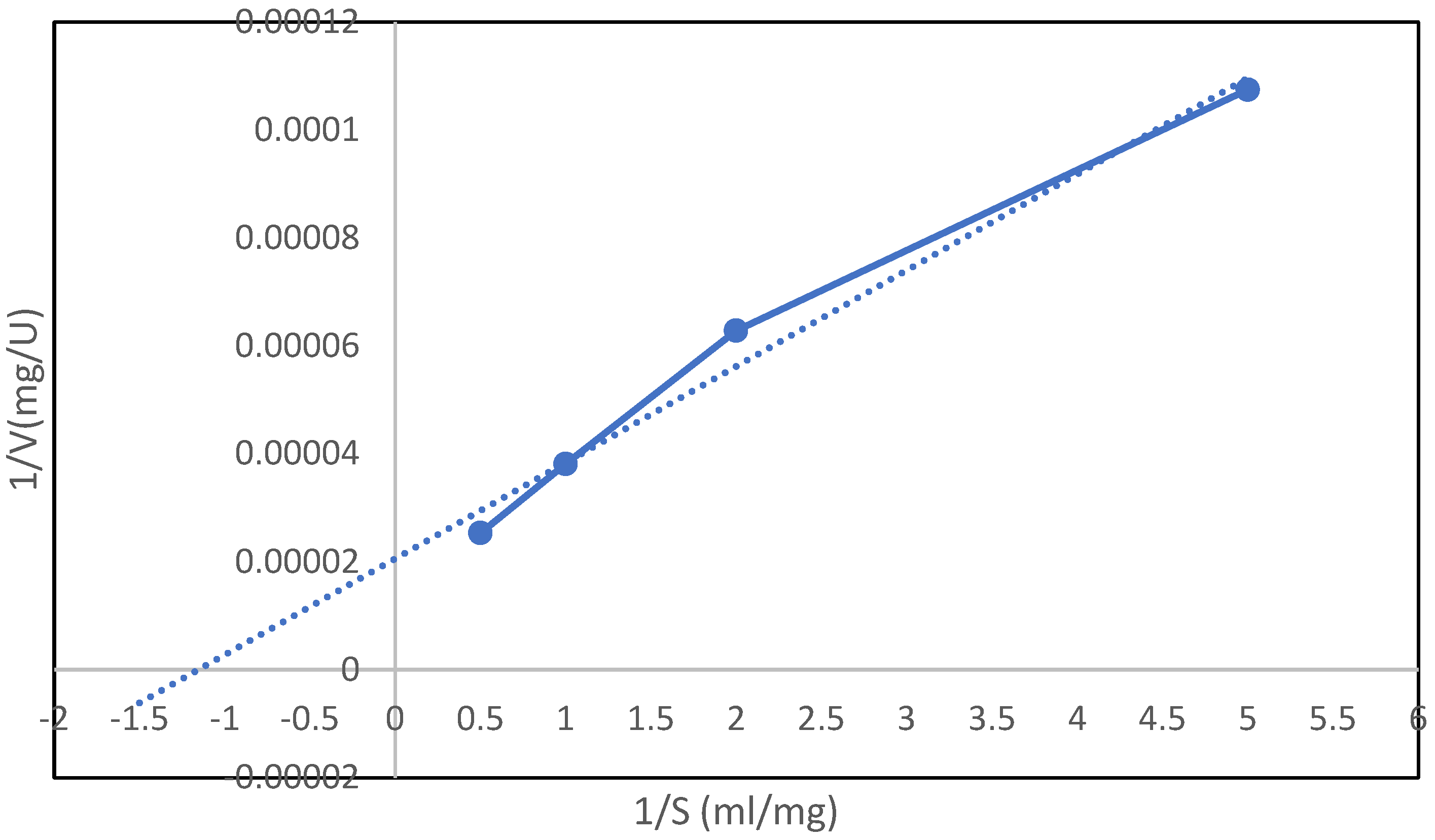
3.7.5. Determination of Kinetic Parameters of Inulinase
3.8. Multifactorial Experiments for Fermentation of Chicory Root Produced Using Inulinase Enzymes
3.8.1. Screening of Factors Affecting Fermentation of Chicory Root
3.8.2. Optimization of Medium Components Using Box–Behnken Design
3.9. Model Validation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kango, N.; Jain, S.C. Production and Properties of Microbial Inulinases: Recent Advances. Food Biotechnol. 2011, 25, 165–212. [Google Scholar] [CrossRef]
- Singh, R.S.; Chauhan, K. Inulinase production from a new inulinase producer, Penicillium oxalicum BGPUP-4. Biocatal. Agric. Biotechnol. 2017, 9, 1–10. [Google Scholar] [CrossRef]
- Das, D.; Selvaraj, R.; Bhat, M.R. Optimization of inulinase production by a newly isolated strain Aspergillus flavus var. flavus by solid state fermentation of Saccharum arundinaceum. Biocatal. Agric. Biotechnol. 2019, 22, 101363. [Google Scholar] [CrossRef]
- Yang, Z.; Hu, J.; Zhao, M. Isolation and quantitative determination of inulin-type oligosaccharides in roots of Morinda officinalis. Carbohydr. Polym. 2011, 83, 1997–2004. [Google Scholar] [CrossRef]
- Saber, W.I.A.; El-Naggar, N.E. Optimization of fermentation conditions for the biosynthesis of inulinase by the new source; Aspergillus tamarii and hydrolysis of some inulin containing agro-wastes. Biotechnology 2009, 8, 425–433. [Google Scholar] [CrossRef]
- Das, D.; Bhat, R.; Selvaraj, R. Review of inulinase production using solid-state fermentation. Ann. Microbiol. 2019, 69, 201–209. [Google Scholar] [CrossRef]
- Gencheva, P.; Dobrev, G.; Delchev, N.; Hristov, J.; Ivanova, V. Jerusalem artichoke and pea hulls-based substrates as raw material for ethanol production by Saccharomyces cerevisiae. Int. Rev. Chem. Eng. 2012, 4, 84–90. [Google Scholar]
- Ilgın, M.; Germec, M.; Turhan, I. Inulinase production and mathematical modeling from carob extract by using Aspergillus niger. Biotech. Prog. 2020, 36, e2919. [Google Scholar] [CrossRef]
- Nath, S.; Kango, N. Recent Developments in Industrial Mycozymes: A Current Appraisa. Mycology 2022, 13, 81–105. [Google Scholar] [CrossRef]
- Mohamed, S.A.; Salah, H.A.; Moharam, M.E.; Foda, M.S.; Fahmy, A.S. Characterization of two thermostable inulinases from Rhizopus oligosporus NRRL 2710. Genet. Eng. Biotechnol. 2015, 13, 65–69. [Google Scholar] [CrossRef] [Green Version]
- Yazdanpanah, L.; Mohamadi, N. Antifungal activity of clove essential oil from Syzygium aromaticum on Paecilomyces variotii agent of pistachio dieback. J. Biodivers. Environ. Sci. 2014, 4, 42–45. [Google Scholar]
- Moreno-Gavíra, A.; Huertas, V.; Diánez, F.; Santos, M.; Sánchez-Montesinos, B. Paecilomyces and its importance in the biological control of agricultural pests and diseases. Plants 2020, 9, 1746. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Chi, Z.; Liu, G.L.; Wang, F.; Madzak, C.; Chi, Z.M. Inulin hydrolysis and citric acid production from inulin using the surface-engineered Yarrowia lipolytica displaying inulinase. Metab. Eng. 2010, 12, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Trivedia, S.; Divechab, J.; Shaha, A. Optimization of inulinase productionby a newly isolated Aspergillus tubingensis CR16 using low-cost substrates. Car-Bohydrate Polym. 2012, 90, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Alma, E.; Cruz-Guerrero, J.; Olvera, L.; Garcıá-Garibay, M.; Gómez-Ruiz, L. Inulinase-hyperproducing strains of Kluyveromyces sp. isolated from aguamiel (Agave sap) and pulqueWorld. J. Microbiol. Biotechnol. 2006, 22, 115–117. [Google Scholar]
- Onilude, A.A.; Fadaunsi, I.F.; Garuba, E.O. Inulinase production by Saccharomyces sp. in solid state fermentation using wheat bran as substrate. Annal. Microbiol. 2012, 62, 843–848. [Google Scholar] [CrossRef]
- Chellapandi, P.; Jani, H.M. Production of endoglucanase by the native strains of Streptomyces isolates in submerged fermentation. Braz. J. Microbiol. 2008, 39, 122–127. [Google Scholar] [CrossRef] [Green Version]
- Plackett, R.L.; Burman, J.P. The design of optimum multifactorial experiments. Biometrika 1946, 33, 305–325. [Google Scholar] [CrossRef]
- Box, G.E.P.; Behnken, D.W. Some new three level designs for the study of quantitative variables. Technometrics 1960, 2, 455–475. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Bradford, M.M. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structura l proteins during the assembly of the head of bacteriop hage. Nature 1970, T4, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Lineweaver, H.; Burk, D. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 1934, 56, 658–666. [Google Scholar] [CrossRef]
- Lan, T.Q.; Wei, D.; Yang, S.-T.; Liu, X. Enhanced cellulase production by Trichoderma viride in a rotating fibrous bed bioreactor. Bioresour. Technol. 2013, 133, 175–182. [Google Scholar] [CrossRef]
- Scott, T.A.; Melvin, E.H. Determination of Dextran with Anthrone. Anal. Chem. 1953, 25, 1656–1661. [Google Scholar] [CrossRef]
- Su, X.; Schmitz, G.; Zhang, M.; Roderick, I.; Mackie, R.I.; Cann, K.I. Heterologous Gene Expression in Filamentous Fungi. Adv. Appl. Microbiol. 2012, 81, 1–61. [Google Scholar]
- Yazici, S.O.; Sahin, S.; Biyik, H.H.; Geroglu, Y.; Ozmen, I. Optimization of fermentation parameters for high-activity inulinase production and purification from Rhizopus oryzae by Plackett–Burman and Box–Behnken. J. Food Sci. Technol. 2021, 58, 739–751. [Google Scholar] [CrossRef]
- Gupta, A.; Sharma, A.; Pathak, R.; Kumar, A.; Sharma, S. Solid State Fermentation of Non-Edible Oil Seed Cakes for Production of Proteases and Cellulases and Degradation of Anti-Nutritional Factors. J. Food Biotechnol. Res. 2018, 2, 2–7. [Google Scholar]
- Huitrón, C.; Pérez, R.; Gutiérrez, L.; Lappe, P.; Petrosyan, P.; Villegas, J.; Aguilar, C.; Rocha-Zavaleta, L.; Blancas, A. Bioconversion of Agave tequilana fructans by exo-inulinases from indigenous Aspergillus niger CH-A-2010 enhances ethanol production from raw Agave tequilana juice. J. Ind. Microbiol. Biotechnol. 2013, 40, 123–132. [Google Scholar] [CrossRef]
- Trivedi, S.; Divecha, J.; Shah, T.; Shah, A. Rapid and efficient bioconversion of chicory inulin to fructose by immobilized thermostable inulinase from Aspergillus tubingensis CR16. Bioresour. Bioprocess. 2015, 2, 32. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.S.; Singh, R.P. Production of fructooligosaccharides from inulin by endoinulinases and their prebiotic potential. Food Technol. Biotechnol. 2010, 48, 435–450. [Google Scholar]
- Reeta, R.S. Beta-glucosidase from Aspergillus niger NII 08121: Molecular characterization and applications in bioethanol production. Ph.D. Thesis, National Institute for Interdisciplinary Science & Technology, Thiruvananthapuram, India, 2011. [Google Scholar]
- Abdella, A.; Mazeed, T.E.; Yang, S.T.; El-Baz, A.F. Production of β-Glucosidase by Aspergillus niger on Wheat Bran and Glycerol in Submerged Culture: Factorial Experimental Design and Process Optimization. Curr. Biotechnol. 2014, 3, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Garuba, E.O.; Onilude, A.A. Exo-Inulinase Production by a catabolite repression-resistant mutant Thermophilic Aspergillus tamarii -U4 in solid state fermentation. Biotechnol. J. Inter. 2020, 24, 21–31. [Google Scholar] [CrossRef]
- Cazetta, M.L.; Contiero, J. Influence of nitrogen source and sucrose concentrationon inulinase production by Kluyveromyces marxianusin fed-batch fermentation. Afr. J. Biotechnol. 2011, 10, 12012–12017. [Google Scholar]
- Akinyele, B.J.; Adetuyi, F.C. Effect of agrowastes, pH and temperature variation on the growth of Volvariella volvacea. Afr. J. Biotechnol. 2005, 4, 1390–1395. [Google Scholar]
- Skowronek, M.; Fiedurek, J. Optimisation of Inulinase Production by Aspergillus niger Using Simplex and Classical Method. Food Technol. Biotechnol. 2004, 42, 141–146. [Google Scholar]
- Abou-Taleb, K.A.; Amin, S.A.; Ahmed, A.I. Production of exo-inulinase from Aspergillus niger and Candida oleophila for degradation of chicory root inulin and ethanol production. Middle East J. Agric. Res. 2019, 8, 855–867. [Google Scholar]
- Jain, S.C.; Jain, P.C.; Kango, N. Production of inulinase from kluyveromyces marxianus using dahlia tuber extract. Braz. J. Microbiol. 2012, 2012, 62–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapala, J.; Bustos-Jaimes, I.; Manzanares, P.; Bárzana, E.; Montiel, C.A. Purification and characterization of an inulinase produced by a Kluyveromyces marxianus strain isolated from blue agave bagasse. Protein Expr. Purif. 2020, 176, 105718. [Google Scholar] [CrossRef] [PubMed]
- Laowklom, N.; Chantanaphan, R.; Pinphanichakarn, P. Production, Purification and Characterization of Inulinase from a Newly Isolated Streptomyces sp. CP01. Nat. Resour. 2012, 3, 137–144. [Google Scholar]
- Ramapriya, R.; Thirumurugan, A.; Sathishkumar, T.; Manimaran, D.R. Partial purification and characterization of exoinulinase produced from Bacillus sp. J. Genet. Eng. Biotechnol. 2018, 16, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Fawzi, E.M. Comparative study of two purified inulinases from thermophile thielavia terrestris NRRL 8126 and mesophile Aspergillus foetidus NRRL 337 grown on Cichorium Intybus L. Braz. J. Microbiol. 2011, 42, 633–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Germec, M.; Turhan, I. Thermostability of Aspergillus niger inulinase from sugar beet molasses in the submerged fermentation and determination of its kinetic and thermodynamic parameters. Biomass Convers. Biorefinery 2022, 12, 3219–3227. [Google Scholar] [CrossRef]
- Cho, Y.J.; Yun, J.W. Purification and characterization of an endoinulinase from Xanthomonas oryzae no. 5. Process Biochem. 2002, 37, 1325–1331. [Google Scholar] [CrossRef]
- Naidoo, K.; Kumar, A.; Sharma, V.; Permaul, K.; Singh, S. Purifi cation and Characterization of an Endoinulinase from Xanthomonas campestris pv. phaseoli KM 24 Mutant. Food Technol. Biotechnol. 2015, 53, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, D.S.; Junior, G.V.; Fernandes, P.; Ribeiro, G.C.A.; Lima, D.M.; Góes-Neto, A.; Oliveira, R.Q.; Figueiredo-Ribeiro, R.d.C.L.; de Assis, S. Production, characterization and application of inulinase from fungal endophyte CCMB 328. An. Acad. Bras. Ciências 2012, 84, 443–453. [Google Scholar] [CrossRef] [Green Version]
- AEl-souod, S.M.A.; Mohamed, T.M.; Ali, E.M.; El-badry, M.O.; El-keiy, M.M. Partial purification of extracellular exo-inulinase from Ulocladium atrum. J. Genet. Eng. Biotechnol. 2014, 12, 15–20. [Google Scholar] [CrossRef]
- Garuba, O.E.; Onilude, A.A.; Oyinlola, A.K. Isolation and molecular characterization of inulinase- producing thermophilic fungi from waste dumpsite located at apete-awotan in ibadan, south-western Nigeria. Innov. Rom. Food Biotechnol. 2015, 16, 31–36. [Google Scholar]
- Abdella, A.; Ramadan, S.; Hamouda, R.A.; Saddiq, A.A.; Alhazmi, N.M.; Al-Saman, M.A. Paecilomyces variotii xylanase production, purification and characterization with antioxidant xylo-oligosaccharides production. Sci. Rep. 2021, 11, 16468. [Google Scholar] [CrossRef]
- Flores-Gallegos, A.C.; Contreras-Esquivel, J.C.; Morlett-Chávez, J.A.; Aguilar, C.N.; Rodríguez-Herrera, R. Comparative study of fungal strains for thermostable inulinase production. J. Biosci. Bioeng. 2015, 119, 421–426. [Google Scholar] [CrossRef]
- Torabizadeha, H.; Mahmoud, A. Inulin hydrolysis by inulinase immobilized covalently on magnetic nanoparticles prepared with wheat gluten hydrolysates. Biotechnol. Rep. 2018, 17, 97–103. [Google Scholar] [CrossRef]
- Chen, H.Q.; Chen, X.M.; Li, Y.; Wanga, J.; Jin, Z.Y. Purification and characterisation of exo- and endo-inulinase from Aspergillus ficuum JNSP5-06. Food Chem. 2009, 115, 1206–1212. [Google Scholar] [CrossRef]
- Corrado, I.; Cascelli, N.; Ntasi, G.; Birolo, L.; Sannia, G.; Pezzella, C. Optimization of Inulin Hydrolysis by Penicillium lanosocoeruleum Inulinases and Efficient Conversion into Polyhydroxyalkanoates. Front. Bioeng. Biotechnol. 2021, 9, 108. [Google Scholar] [CrossRef]
- Barclaya, T.; Ginic-Markovica, M.; Johnstona, M.R.; Cooper, P.; Petrovsky, N. Analysis of the hydrolysis of inulin using real time 1H NMR spectroscopy. Carbohydr. Res. 2012, 352, 117–125. [Google Scholar] [CrossRef] [Green Version]
- Popoola-Akinola, O.O.; Raji, T.J.; Olawoye, B. Lignocellulose, dietary fibre, inulin and their potential application in food. Heliyon 2022, 8, e10459. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.-F.; Chi, Z.; Liu, G.-L.; Jiang, H.; Hu, Z.; Ch, Z.-M. Inulinase hyperproduction by Kluyveromyces marxianus through codon optimization, selection of the promoter, and high-cell-density fermentation for efficient inulin hydrolysis. Ann. Microbiol. 2019, 69, 647–657. [Google Scholar] [CrossRef]
- Abdella, A.; El-Baz, A.F.; Ibrahim, I.A.; Mahrous, E.E.; Yang, S.T. Biotransformation of soy flour isoflavones by Aspergillus niger NRRL 3122 β-glucosidase enzyme. Nat. Prod. Res. 2018, 32, 2382–2391. [Google Scholar] [CrossRef] [PubMed]
- Baston, O.; Neagu, C.; Bahrim, G. Establishing the Optimum Conditions for Inulin Hydrolysisby Using Commercial Inulinase. Rev. Chim. 2013, 64, 649–653. [Google Scholar]



| Factor | Low Level (−1) | High Level (+1) |
|---|---|---|
| chicory root | 10 g/L | 20 g/L |
| K2HPO4 | 0.5 | 2 |
| MgSO·7H2O | 0.3 | 1 |
| NaNO3 | 0.5 | 3 |
| KCI | 0.3 | 1 |
| FeSO4·7H2O | 0.05 | 0.5 |
| (NH4) H2PO4 | 1 | 4 |
| Temperature | 25 | 35 |
| pH | 5.5 | 7.5 |
| Factor | Symbol | Low Level (−1) | High Level (+1) |
|---|---|---|---|
| pH | X1 | 5 | 8 |
| Temp (°C) | X2 | 40 | 60 |
| Time (h) | X3 | 1 | 5 |
| Substrate conc. % (w/v) | X4 | 0.5 | 3 |
| Enzyme conc. (U/mL) | X5 | 10,000 | 20,000 |
| Agitation (rpm) | X6 | 100 | 200 |
| Strain | Incubation Period (Day) | Cell Dry Weight (g/L) | pH | Protein (mg/mL) | Inulinase (IU/mL) |
|---|---|---|---|---|---|
| Aspergillus niger NRRL 3122 | 2 | 3.4 | 6.42 | 0.02 | 95 ± 3.6 |
| 4 | 5.14 | 6.74 | 0.05 | 110 ± 7.1 | |
| 6 | 5.16 | 6.48 | 0.07 | 135 ± 9.3 | |
| 8 | 4.94 | 6.63 | 0.07 | 130 ± 8.5 | |
| Aspergillus oryzae NRRL 2217 | 2 | 3.1 | 8.36 | 0.017 | 75 ± 3.2 |
| 4 | 5.52 | 8.48 | 0.02 | 90 ± 4.7 | |
| 6 | 5.91 | 8.07 | 0.025 | 110± 5.3 | |
| 8 | 4.83 | 8.70 | 0.024 | 108 ± 6.1 | |
| Pacilomyces variotti NRRL 1115 | 2 | 5.3 | 7.31 | 0.01 | 43 ± 1.2 |
| 4 | 7.46 | 7.16 | 0.03 | 72 ± 2.3 | |
| 6 | 7.98 | 6.88 | 0.045 | 92 ± 3.1 | |
| 8 | 7.26 | 8.24 | 0.042 | 89 ± 2.7 | |
| Rhizopus oryzae NRRL 3563 | 2 | 3.8 | 2.5 | 0.07 | 160 ± 10 |
| 4 | 5.21 | 3.71 | 0.09 | 250 ± 12 | |
| 6 | 5.82 | 4.5 | 0.103 | 380 ± 15 | |
| 8 | 5.4 | 5 | 0.101 | 370 ± 11 |
| Pattern | Chicory Root (g/L) | K2HPO4 (g/L) | NaNO3 (g/L) | MgSO4 (g/L) | KCL (g/L) | FeSO4 (g/L) | NH4H2PO4 (g/L) | Temp °C | pH | Inulinase (U/mL) |
|---|---|---|---|---|---|---|---|---|---|---|
| −+−−+−+++ | 20 | 2 | 0.5 | 0.3 | 1 | 0.05 | 1 | 25 | 7.5 | 139.16 |
| +++++++++ | 10 | 2 | 0.5 | 1 | 0.3 | 0.5 | 1 | 35 | 7.5 | 328.33 |
| −+++−−−+− | 10 | 2 | 3 | 0.3 | 0.3 | 0.05 | 4 | 35 | 5.5 | 657.16 |
| −−−+−−+−+ | 20 | 0.5 | 3 | 1 | 0.3 | 0.05 | 4 | 25 | 7.5 | 197.20 |
| +−+++−−−+ | 10 | 0.5 | 0.5 | 1 | 1 | 0.5 | 4 | 25 | 5.5 | 279.33 |
| −+−+++−−− | 20 | 2 | 0.5 | 0.3 | 0.3 | 0.5 | 4 | 25 | 5.5 | 131.46 |
| −−+−+++−− | 10 | 0.5 | 3 | 0.3 | 0.3 | 0.5 | 1 | 25 | 7.5 | 542.3 |
| −−+−−+−++ | 20 | 0.5 | 3 | 0.3 | 1 | 0.5 | 1 | 35 | 5.5 | 279.36. |
| +−−+−+++− | 20 | 2 | 3 | 1 | 1 | 0.5 | 4 | 35 | 7.5 | 262.93 |
| +−−−+−−+− | 10 | 2 | 3 | 1 | 1 | 0.05 | 1 | 25 | 5.5 | 438.22 |
| +++−−−+−− | 20 | 0.5 | 0.5 | 1 | 0.3 | 0.05 | 1 | 35 | 5.5 | 147.9 |
| ++−−−+−−+ | 10 | 0.5 | 0.5 | 0.3 | 1 | 0.05 | 4 | 35 | 7.5 | 262.93 |
| Factor | Estimate | Std Error | t Ratio | p-Value |
|---|---|---|---|---|
| Chicory root | −31.08 | 9.481 | −4.33 | 0.0022 ** |
| NaNO3 | 122.33 | 37.92 | 3.49 | 0.032 ** |
| KCL | −138.8 | 135.44 | −3.51 | 0.0343 ** |
| FeSO4 | −12.35 | 11.67 | 0.43 | 0.7231 |
| MgSO4 | 35.32 | 39.23 | 0.82 | 0.4325 |
| K2HPO4 | 40.55 | 63.20 | 0.80 | 0.4823 |
| NH4H2PO4 | 8.23 | 5.43 | 0.32 | 0.8231 |
| Temp | 6.54 | 9.481 | 0.68 | 0.5471 |
| pH | −30.32 | 47.406 | −0.64 | 0.5668 |
| Pattern | Chicory Root (g/L) | NaNO3 (g/L) | KCL | Experimental Inulinase (U/mL) | Predicted Inulinase (U/mL) |
|---|---|---|---|---|---|
| 18,440−− | 7.5 | 3 | 0.3 | 860.16 | 850.14 |
| +0+ | 10 | 4 | 0.3 | 1020.02 | 1010 |
| +0− | 10 | 4 | 0.1 | 2140.6 | 1060.22 |
| 0 | 7.5 | 4 | 0.2 | 1070.3 | 890.225 |
| +−0 | 10 | 3 | 0.2 | 990.37 | 980.38 |
| 0+− | 7.5 | 5 | 0.3 | 845.36 | 840.01 |
| ++0 | 10 | 5 | 0.2 | 1085.11 | 1075.02 |
| −0+ | 5 | 4 | 0.1 | 650.07 | 640.41 |
| 0−+ | 7.5 | 3 | 0.1 | 875.03 | 865.15 |
| 0 | 7.5 | 4 | 0.2 | 885.33 | 875.44 |
| 0++ | 7.5 | 5 | 0.1 | 925.3 | 915.37 |
| −−0 | 5 | 3 | 0.2 | 635.27 | 630.14 |
| 0 | 7.5 | 4 | 0.2 | 845.36 | 845.36 |
| −+0 | 5 | 5 | 0.2 | 570.2 | 560.43 |
| −0− | 5 | 4 | 0.3 | 575.27 | 765.49 |
| Factor | Estimate | Std Error | t Ratio | p-Value |
|---|---|---|---|---|
| Intercept | 1582.6667 | 18.99898 | 83.30 | <0.0001 ** |
| Chicory root | 390.375 | 11.63445 | 33.55 | <0.0001 ** |
| NaNO3 | −49.25 | 11.63445 | −4.23 | 0.0082 ** |
| KCL | 15.125 | 11.63445 | 1.30 | 0.2503 |
| Chicory root × NaNO3 | 71.25 | 16.4536 | 4.33 | 0.0075 ** |
| Chicory root × KCL | 9 | 16.4536 | 0.55 | 0.6079 |
| NaNO3 × KCL | −29.5 | 16.4536 | −1.79 | 0.1330 |
| Chicory root × Chicory root | −94.45833 | 17.12545 | −5.52 | 0.0027 ** |
| Microorganism | Substrate | Inulinase (U/mL) | Productivity (U/mL/Day) | References |
|---|---|---|---|---|
| Rhizopus oryzae NRRL 3563 | 1% chicory root | 1085 | 180 | This study |
| Aspergillus tamarii AR-IN9 | 1% Dahlia tuber | 71.97 | 24 | [5] |
| Aspergillus niger 13/36 | 1% sucrose | 80 | 20 | [37] |
| Candida oleophila | 1% chicory root | 46.38 | 11.59 | [38] |
| Penicillium oxalicum | 2% inulin | 11.06 | 2.5 | [2] |
| Kluyveromyces marxianus | 3% chicory root | 129.21 | 32.3 | [39] |
| Volumetric Activity U/mL | Total Activity (U) | Protein Conc (mg/mL) | Total Protein (mg) | Specific Activity (U/mg) | Purification Folds | Recovery % | |
|---|---|---|---|---|---|---|---|
| Crude enzyme | 1085 | 217,000 | 0.18 | 36 | 6027.7 | 1 | 100 |
| 70% Ammonium sulphate/dialysis | 8685 | 173,300 | 0.623 | 12.46 | 13,940 | 2.3 | 80 |
| Ultra-filtration | 26,065 | 130,325 | 1.187 | 5.935 | 21,950 | 3.63 | 60 |
| Substrate | Relative Activity (%) |
|---|---|
| Inulin | 100 |
| Sucrose | 30 |
| Cellobiose | 0 |
| Soluble starch | 0 |
| Pattern | pH | Temperature (°C) | Substrate Conc. % (w/v) | Time (h) | Enzyme (U/mL) | Agitation (RPM) | Saccharification Yield % |
|---|---|---|---|---|---|---|---|
| −−+−−+ | 5 | 40 | 0.5 | 5 | 10,000 | 200 | 5.83 |
| ++++++ | 8 | 60 | 3 | 5 | 20,000 | 200 | 10.22 |
| −+−−+− | 5 | 60 | 0.5 | 1 | 20,000 | 100 | 27.21 |
| −+−+++ | 5 | 60 | 3 | 1 | 20,000 | 200 | 21.6 |
| +−−+−+ | 8 | 40 | 3 | 1 | 10,000 | 200 | 5.1 |
| −+++−− | 5 | 60 | 3 | 5 | 10,000 | 100 | 18.86 |
| +−+++− | 8 | 40 | 3 | 5 | 20,000 | 100 | 5.61 |
| ++−−−+ | 8 | 60 | 0.5 | 1 | 10,000 | 200 | 11.6 |
| +++−−− | 8 | 60 | 0.5 | 5 | 10,000 | 100 | 10.8 |
| −−−+−− | 5 | 40 | 3 | 1 | 10,000 | 100 | 10.22 |
| +−−−+− | 8 | 40 | 0.5 | 1 | 20,000 | 100 | 6.04 |
| −−+−++ | 5 | 40 | 3 | 5 | 20,000 | 200 | 7.2 |
| Factor | Estimate | Std Error | t Ratio | p-Value |
|---|---|---|---|---|
| Enzyme conc. (U/mL) (10,000, 20,000) | 1.639 | 0.92 | 2.92 | 0.0332 ** |
| Time (h) (1, 5) | 6.545 | 0.934 | 7.49 | 0.0007 ** |
| Substrate conc. % (w/v) (0.5, 3) | 0.3033 | 1.679 | 0.18 | 0.8637 |
| Temperature (°C) (40, 60) | 4.768 | 0.5609 | 5.10 | 0.0038 ** |
| pH (5, 8) | −1.751 | 0.9348 | −1.87 | 0.1199 |
| Agitation (rpm) | −0.03 | 0.0186 | −2.15 | 0.0840 |
| Pattern | Temp (°C) | Enzyme Conc. (U/mL) | Time (h) | Exp | Pred |
|---|---|---|---|---|---|
| 0 | 80 | 30,000 | 9 | 20.39 | 21.63 |
| −0− | 60 | 30,000 | 5 | 44.67 | 43.71 |
| −−0 | 80 | 40,000 | 7 | 24.42 | 23.32 |
| +−0 | 70 | 30,000 | 7 | 48.86 | 44.26 |
| 0+− | 80 | 30,000 | 5 | 34.9 | 34.56 |
| 0−− | 60 | 30,000 | 9 | 33.5 | 33.66 |
| ++0 | 70 | 40,000 | 5 | 34.9 | 36.25 |
| +0− | 70 | 30,000 | 7 | 41.71 | 44.26 |
| 0 | 80 | 20,000 | 7 | 57.94 | 58.2 |
| 0 | 70 | 40,000 | 9 | 27.91 | 28.22 |
| −0+ | 70 | 30,000 | 7 | 41.87 | 44.82 |
| 0−+ | 60 | 20,000 | 7 | 41.87 | 42.92 |
| 0++ | 60 | 40,000 | 7 | 58.07 | 57.51 |
| −+0 | 70 | 20,000 | 9 | 34.9 | 33.71 |
| +0+ | 70 | 20,000 | 5 | 48.72 | 48.72 |
| Factor | Estimate | Std Error | t Ratio | p-Value |
|---|---|---|---|---|
| Intercept | −392.73 | 108.27 | −3.62 | 0.0017 ** |
| Temp (°C) | 6.99 | 2.24 | 3.15 | 0.0262 ** |
| Time (h) | 32.64 | 8.76 | 3.73 | 0.0196 ** |
| Enzyme conc. (U/mL) | 62.3 | 18.06 | 3.46 | 0.0173 ** |
| Temp (°C) × Enzyme conc. (U/mL) | −5.309 | 0.157 | −8.6 | 0.032 ** |
| Enzyme conc. (U/mL) × Time(h) | 0.968 | 0.793 | 1.22 | 0.2782 |
| Temp (°C) × Time(h) | −0.038 | 0.075 | −0.49 | 0.662 |
| Temp (°C) × Temp (°C) | −0.0148 | 0.012 | −1.19 | 0.292 |
| Time (h) × Time(h) | −2.642447 | 0.425 | −6.21 | 0.042 ** |
| Enzyme conc. (U/mL) × Enzyme conc. (U/mL) | −0.03887 | 1.661 | 1.32 | 0.2442 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdella, A.; Al-Saman, M.; Abou-Elazm, F.I.; El-Far, S.W. Rhizopus oryzae Inulinase Production and Characterization with Application in Chicory Root Saccharification. Microbiol. Res. 2023, 14, 297-315. https://doi.org/10.3390/microbiolres14010024
Abdella A, Al-Saman M, Abou-Elazm FI, El-Far SW. Rhizopus oryzae Inulinase Production and Characterization with Application in Chicory Root Saccharification. Microbiology Research. 2023; 14(1):297-315. https://doi.org/10.3390/microbiolres14010024
Chicago/Turabian StyleAbdella, Asmaa, Mahmoud Al-Saman, Fatma I. Abou-Elazm, and Shaymaa Wagdy El-Far. 2023. "Rhizopus oryzae Inulinase Production and Characterization with Application in Chicory Root Saccharification" Microbiology Research 14, no. 1: 297-315. https://doi.org/10.3390/microbiolres14010024
APA StyleAbdella, A., Al-Saman, M., Abou-Elazm, F. I., & El-Far, S. W. (2023). Rhizopus oryzae Inulinase Production and Characterization with Application in Chicory Root Saccharification. Microbiology Research, 14(1), 297-315. https://doi.org/10.3390/microbiolres14010024






