Construction of Recombinant Escherichia coli with a High L-Phenylalanine Production Yield from Glucose
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Gene Deletion
2.3. Plasmids Construction
2.4. Growth Conditions
2.5. Analytical Methods
3. Results and Discussion
3.1. Engineering a Base E. coli Strain for L-Phenylalanine Production
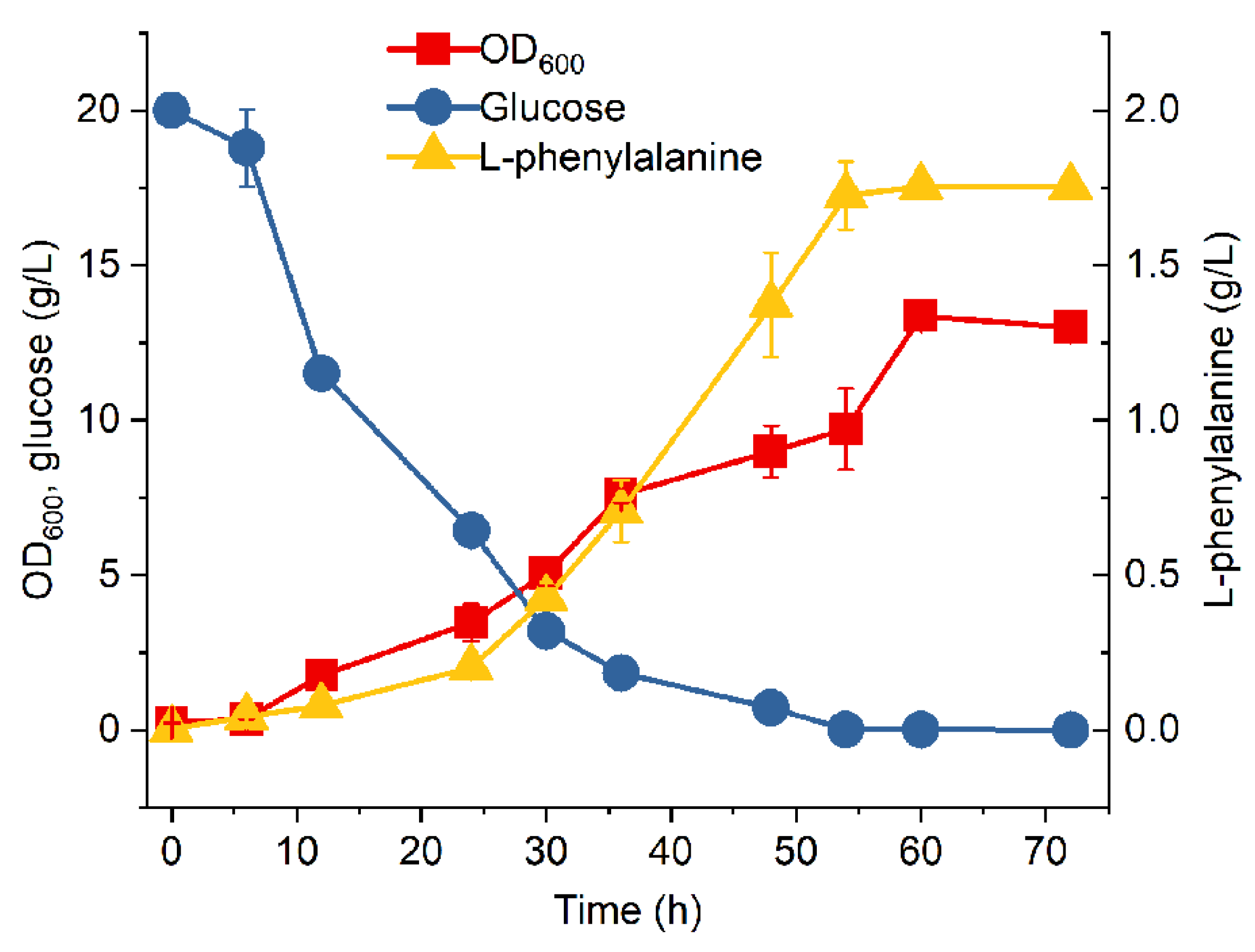
3.2. Introduction of Glf to Improve Glucose Consumption of MPH-2
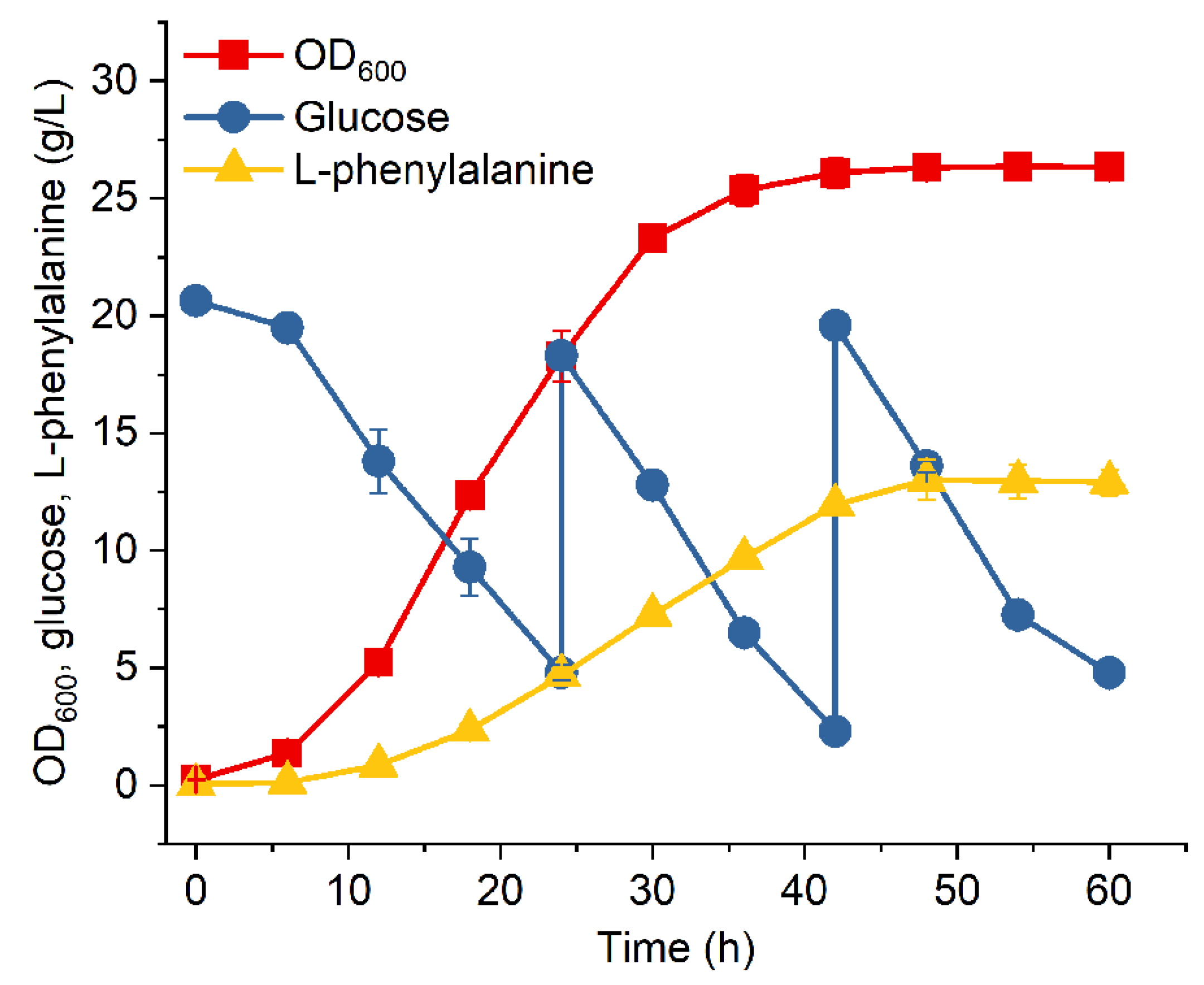
3.3. Fed-Batch Fermentation of MPH-3
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, H.; Liao, X.; Wang, T.; Du, G.; Chen, J. Enhanced L-phenylalanine biosynthesis by co-expression of pheA(fbr) and aroF(wt). Bioresour. Technol. 2010, 101, 4151–4156. [Google Scholar] [CrossRef] [PubMed]
- Huccetogullari, D.; Luo, Z.W.; Lee, S.Y. Metabolic engineering of microorganisms for production of aromatic compounds. Microb. Cell. Fact. 2019, 18, 41. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, N.; Matsutani, M.; Matsushita, K.; Yakushi, T. Stepwise metabolic engineering of Corynebacterium glutamicum for the production of phenylalanine. J. Gen. Appl. Microbiol. 2023, 69, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Niu, H.; Li, Q.; Gu, P. Metabolic engineering for the production of L-phenylalanine in Escherichia coli. 3 Biotech 2019, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Liao, X.; Liu, L.; Wang, T.; Du, G.; Chen, J. Enhanced L-phenylalanine production by recombinant Escherichia coli BR-42 (pAP-B03) resistant to bacteriophage BP-1 via a two-stage feeding approach. J. Ind. Microbiol. Biotechnol. 2011, 38, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Umbarger, H.E. Amino acid biosynthesis and its regulation. Annu. Rev. Biochem. 1978, 47, 532–606. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M. Towards bacterial strains overproducing L-tryptophan and other aromatics by metabolic engineering. Appl. Microbiol. Biotechnol. 2006, 69, 615–626. [Google Scholar] [CrossRef]
- Wallace, B.J.; Pittard, J. Regulator gene controlling enzymes concerned in tyrosine biosynthesis in Escherichia coli. J. Bacteriol. 1969, 97, 1234–1241. [Google Scholar] [CrossRef]
- Rihtar, E.; Zgur Bertok, D.; Podlesek, Z. The Uropathogenic specific protein gene usp from Escherichia coli and Salmonella bongori is a Novel Member of the TyrR and H-NS Regulons. Microorganisms 2020, 8, 330. [Google Scholar] [CrossRef]
- Lawley, B.; Fujita, N.; Ishihama, A.; Pittard, A.J. The TyrR protein of Escherichia coli is a class I transcription activator. J. Bacteriol. 1995, 177, 238–241. [Google Scholar] [CrossRef]
- Gunsalus, R.P.; Yanofsky, C. Nucleotide sequence and expression of Escherichia coli trpR, the structural gene for the trp aporepressor. Proc. Natl. Acad. Sci. USA 1980, 77, 7117–7121. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.; Martinez, J.A.; Flores, N.; Escalante, A.; Gosset, G.; Bolivar, F. Engineering Escherichia coli to overproduce aromatic amino acids and derived compounds. Microb. Cell. Fact. 2014, 13, 126. [Google Scholar] [CrossRef] [PubMed]
- Gavini, N.; Davidson, B.E. Regulation of pheA expression by the pheR product in Escherichia coli is mediated through attenuation of transcription. J. Biol. Chem. 1991, 266, 7750–7753. [Google Scholar] [CrossRef] [PubMed]
- Postma, P.W.; Lengeler, J.W. Phosphoenolpyruvate:carbohydrate phosphotransferase system of bacteria. Microbiol. Rev. 1985, 49, 232–269. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Niu, H.; Huang, Z.; Li, Q.; Gu, P. Construction of a switchable synthetic Escherichia coli for aromatic amino acids by a tunable switch. J. Ind. Microbiol. Biotechnol. 2020, 47, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Sprenger, G.A. From scratch to value: Engineering Escherichia coli wild type cells to the production of L-phenylalanine and other fine chemicals derived from chorismate. Appl. Microbiol. Biotechnol. 2007, 75, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Flores, S.; Gosset, G.; Flores, N.; de Graaf, A.A.; Bolivar, F. Analysis of carbon metabolism in Escherichia coli strains with an inactive phosphotransferase system by (13)C labeling and NMR spectroscopy. Metab. Eng. 2002, 4, 124–137. [Google Scholar] [CrossRef]
- Guo, L.; Ding, S.; Liu, Y.; Gao, C.; Hu, G.; Song, W.; Liu, J.; Chen, X.; Liu, L. Enhancing tryptophan production by balancing precursors in Escherichia coli. Biotechnol. Bioeng. 2022, 119, 983–993. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, Y.; Ding, D.; Wen, J.; Zhu, B.; Zhang, D. Genetic engineering of Escherichia coli to improve L-phenylalanine production. BMC Biotechnol. 2018, 18, 5. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Cherepanov, P.P.; Wackernagel, W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 1995, 158, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Kuhlman, T.E.; Cox, E.C. Site-specific chromosomal integration of large synthetic constructs. Nucleic Acids Res. 2010, 38, e92. [Google Scholar] [CrossRef] [PubMed]
- Lerner, C.G.; Inouye, M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990, 18, 4631. [Google Scholar] [CrossRef] [PubMed]
- Gu, P.; Yang, F.; Li, F.; Liang, Q.; Qi, Q. Knocking out analysis of tryptophan permeases in Escherichia coli for improving L-tryptophan production. Appl. Microbiol. Biotechnol. 2013, 97, 6677–6683. [Google Scholar] [CrossRef] [PubMed]
- Gu, P.; Yang, F.; Kang, J.; Wang, Q.; Qi, Q. One-step of tryptophan attenuator inactivation and promoter swapping to improve the production of L-tryptophan in Escherichia coli. Microb. Cell Fact. 2012, 11, 30. [Google Scholar] [CrossRef] [PubMed]
- Doroshenko, V.; Airich, L.; Vitushkina, M.; Kolokolova, A.; Livshits, V.; Mashko, S. YddG from Escherichia coli promotes export of aromatic amino acids. FEMS Microbiol. Lett. 2007, 275, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Li, R.; Gao, J.; Fan, X.; Li, Q.; Gu, P. Different performance of Escherichia coli mutants with defects in the phosphoenolpyruvate: Carbohydrate phosphotransferase system under low glucose condition. 3 Biotech 2019, 9, 50. [Google Scholar] [CrossRef]
- Gosset, G. Improvement of Escherichia coli production strains by modification of the phosphoenolpyruvate:sugar phosphotransferase system. Microb. Cell Fact. 2005, 4, 14. [Google Scholar] [CrossRef]
- Cases, I.; Velazquez, F.; de Lorenzo, V. The ancestral role of the phosphoenolpyruvate-carbohydrate phosphotransferase system (PTS) as exposed by comparative genomics. Res. Microbiol. 2007, 158, 666–670. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, X. Construction of pyruvate producing strain with intact pyruvate dehydrogenase and genome-wide transcription analysis. World J. Microbiol. Biotechnol. 2017, 33, 59. [Google Scholar] [CrossRef]
- Hernandez-Montalvo, V.; Martinez, A.; Hernandez-Chavez, G.; Bolivar, F.; Valle, F.; Gosset, G. Expression of galP and glk in a Escherichia coli PTS mutant restores glucose transport and increases glycolytic flux to fermentation products. Biotechnol. Bioeng. 2003, 83, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Kurgan, G.; Onyeabor, M.; Holland, S.C.; Taylor, E.; Schneider, A.; Kurgan, L.; Billings, T.; Wang, X. Directed evolution of Zymomonas mobilis sugar facilitator Glf to overcome glucose inhibition. J. Ind. Microbiol. Biotechnol. 2022, 49, kuab066. [Google Scholar] [CrossRef]
- Shimada, T.; Nakazawa, K.; Tachikawa, T.; Saito, N.; Niwa, T.; Taguchi, H.; Tanaka, K. Acetate overflow metabolism regulates a major metabolic shift after glucose depletion in Escherichia coli. FEBS Lett. 2021, 595, 2047–2056. [Google Scholar] [CrossRef] [PubMed]
- Enjalbert, B.; Cocaign-Bousquet, M.; Portais, J.C.; Letisse, F. Acetate exposure determines the diauxic behavior of Escherichia coli during the glucose-acetate transition. J. Bacteriol. 2015, 197, 3173–3181. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, N.; Saini, D.; Guleria, R.; Mukherjee, K.J. Designing an Escherichia coli strain for phenylalanine overproduction by metabolic engineering. Mol. Biotechnol. 2017, 59, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Baez-Viveros, J.L.; Osuna, J.; Hernandez-Chavez, G.; Soberon, X.; Bolivar, F.; Gosset, G. Metabolic engineering and protein directed evolution increase the yield of L-phenylalanine synthesized from glucose in Escherichia coli. Biotechnol. Bioeng. 2004, 87, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.B.; Guo, X.L.; Zhang, M.L.; Huang, Q.G.; Qi, F.; Huang, J.Z. Enhancement of l-phenylalanine production in Escherichia coli by heterologous expression of Vitreoscilla hemoglobin. Biotechnol. Appl. Biochem. 2018, 65, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.P.; Liu, R.X.; Xiao, M.R.; Zhang, L.; Ding, Z.Y.; Gu, Z.H.; Shi, G.Y. A systems level engineered E. coli capable of efficiently producing L-phenylalanine. Process Biochem. 2014, 49, 751–757. [Google Scholar] [CrossRef]
- Liu, S.P.; Xiao, M.R.; Zhang, L.; Xu, J.; Ding, Z.Y.; Gu, Z.H.; Shi, G.Y. Production of l-phenylalanine from glucose by metabolic engineering of wild type Escherichia coli W3110. Process Biochem. 2013, 48, 413–419. [Google Scholar] [CrossRef]
- Ding, R.; Liu, L.; Chen, X.; Cui, Z.; Ao, Z.; Ren, D.; Zhang, L. Introduction of two mutations into AroG increases phenylalanine production in Escherichia coli. Biotechnol. Lett. 2014, 36, 2103–2108. [Google Scholar] [CrossRef]
- Cheng, L.K.; Wang, J.; Xu, Q.Y.; Zhao, C.G.; Shen, Z.Q.; Xie, X.X.; Chen, N. Strategy for pH control and pH feedback-controlled substrate feeding for high-level production of L-tryptophan by Escherichia coli. World J. Microbiol. Biotechnol. 2013, 29, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Jing, K.; Tang, Y.; Yao, C.; Del Rio-Chanona, E.A.; Ling, X.; Zhang, D. Overproduction of L-tryptophan via simultaneous feed of glucose and anthranilic acid from recombinant Escherichia coli W3110: Kinetic modeling and process scale-up. Biotechnol. Bioeng. 2018, 115, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Gerigk, M.; Bujnicki, R.; Ganpo-Nkwenkwa, E.; Bongaerts, J.; Sprenger, G.; Takors, R. Process control for enhanced L-phenylalanine production using different recombinant Escherichia coli strains. Biotechnol. Bioeng. 2002, 80, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Weiner, M.; Trondle, J.; Albermann, C.; Sprenger, G.A.; Weuster-Botz, D. Improvement of constraint-based flux estimation during L-phenylalanine production with Escherichia coli using targeted knock-out mutants. Biotechnol. Bioeng. 2014, 111, 1406–1416. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Ding, D.; Bai, D.; Lin, Y.; Zhu, Y.; Zhang, C.; Zhang, D. Transcriptomics and metabolomics analysis of L-phenylalanine overproduction in Escherichia coli. Microb. Cell Fact. 2023, 22, 65. [Google Scholar] [CrossRef] [PubMed]
- Mahr, R.; von Boeselager, R.F.; Wiechert, J.; Frunzke, J. Screening of an Escherichia coli promoter library for a phenylalanine biosensor. Appl. Microbiol. Biotechnol. 2016, 100, 6739–6753. [Google Scholar] [CrossRef]
- Yakandawala, N.; Romeo, T.; Friesen, A.D.; Madhyastha, S. Metabolic engineering of Escherichia coli to enhance phenylalanine production. Appl. Microbiol. Biotechnol. 2008, 78, 283–291. [Google Scholar] [CrossRef]
- Ojima, Y.; Komaki, M.; Nishioka, M.; Iwatani, S.; Tsujimoto, N.; Taya, M. Introduction of a stress-responsive gene, yggG, enhances the yield of L-phenylalanine with decreased acetic acid production in a recombinant Escherichia coli. Biotechnol. Lett. 2009, 31, 525–530. [Google Scholar] [CrossRef]
- Baez, A.; Cho, K.M.; Liao, J.C. High-flux isobutanol production using engineered Escherichia coli: A bioreactor study with in situ product removal. Appl. Microbiol. Biotechnol. 2011, 90, 1681–1690. [Google Scholar] [CrossRef]



| Name | Relevant Genotype | Reference |
|---|---|---|
| DH5α | F−, endA1, hsdR17 (rK−, mK+), supE44, thi-l, λ−, recA1, gyrA96, ΔlacU169 (Φ80lacZ ΔM15) | Lab stock |
| MG1655 | F−, λ−, rph-1 | Lab stock |
| MG-1 | MG1655 (ΔpoxB) | This study |
| MG-2 | MG1655 (ΔpoxBΔpta) | This study |
| MG-3 | MG1655/pT1 | |
| MG-4 | MG-1/pT1 | This study |
| MG-5 | MG-2/pT1 | This study |
| MPH-1 | MG1655 (ΔpoxBΔptaΔptsI) | This study |
| MPH-2 | MPH-1/pT1 | This study |
| MPH-3 | MPH-1/pT1/pT2 | This study |
| MPH-4 | MPH-1 with tyrA replacement by glf containing plasmid pT1 | This study |
| Name | Relevant Genotype | Reference |
|---|---|---|
| pKD3 | bla, FRT-cat-FRT | [20] |
| pCP20 | bla and cat, helper plasmid | [21] |
| pTKRed | SpcR, IPTG induced λRed enzymes | [22] |
| pCL1920 | SpcR | [23] |
| pTrc99a | bla | Lab stock |
| pT1 | pCL1920-pheAfbr-tktA-aroGfbr-ppsA-yddG | Synthesized by TSINGKE Biological Technology |
| pT2 | pTrc99a-glf | Synthesized by TSINGKE Biological Technology |
| E. coli Strains | Genes Overexpressed in Plasmids | Promoters and Replicon in Plasmids | Host Engineering | Culture Methods | L-Phenylalanine Production | References | |
|---|---|---|---|---|---|---|---|
| Titer (g/L) | Yields (g/g) | ||||||
| Xllp21 | pheA (Thr326Pro), aroF, galP, glk, and aroD | pBR322 replicon and BBa_J23106 promoter | W3110 mutant with L-tyrosine auxotrophic (ΔptsH and tyrR (T495I)) | 5 L fed-batch fermentation | 72.9 | 0.26 | [19] |
| PAPV | aroF, pheAfbr, and vgb from Vitreoscilla | PLPRpromoters and replicon was not indicated | Derived from Escherichia coli K-12, Hfr (PO1), λ−, el4-, tyrA4, relA1, spoT1, thiE1 | 3 L fed-batch fermentation | 44.21 | 0.071 | [37] |
| W14 (pR15BABKG) | aroG15, pheAfbr, aroK, ydiB, yddG, and tyrB | λcIts857 replicon and PLPR promoters | W3110 mutant with L-tyrosine auxotrophic (Δcrr) | 15 L fed-batch fermentation | 47 | 0.252 | [38] |
| W3110 (pNpheABK15) | pheAfbr and aroG15 | pBR322 replicon and PN25 promoter | Wild W3110 | 15 L fed-batch fermentation | 23.8 | 0.154 | [39] |
| W3110 (pQPTABG8/15) | AroG (A202T and M147I) | pBR322 replicon and Ptacpromoter | Wild W3110 | 3 L fed-batch fermentation | 26.78 | 0.231 | [40] |
| WSH-Z06 (pAP-B03) | pheAfbr and aroFwt | p15A replicon and PLPR promoters | W3110 mutant with L-tyrosine auxotrophic | 3 L fed-batch fermentation | 35.38 | 0.238 | [1] |
| MPH-3 | pheAfbr, tktA, aroGfbr, ppsA, and yddG | pBR322 replicon with Ptrc promoter and pSC101 replicon with Plac promoter | MG1655 (ΔpoxBΔptaΔptsI) | 5 L fed-batch fermentation | 19.24 | 0.279 | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, P.; Zhao, S.; Li, C.; Jiang, S.; Zhou, H.; Li, Q. Construction of Recombinant Escherichia coli with a High L-Phenylalanine Production Yield from Glucose. Microbiol. Res. 2023, 14, 1185-1198. https://doi.org/10.3390/microbiolres14030079
Gu P, Zhao S, Li C, Jiang S, Zhou H, Li Q. Construction of Recombinant Escherichia coli with a High L-Phenylalanine Production Yield from Glucose. Microbiology Research. 2023; 14(3):1185-1198. https://doi.org/10.3390/microbiolres14030079
Chicago/Turabian StyleGu, Pengfei, Shuo Zhao, Chengwei Li, Shuixing Jiang, Hao Zhou, and Qiang Li. 2023. "Construction of Recombinant Escherichia coli with a High L-Phenylalanine Production Yield from Glucose" Microbiology Research 14, no. 3: 1185-1198. https://doi.org/10.3390/microbiolres14030079
APA StyleGu, P., Zhao, S., Li, C., Jiang, S., Zhou, H., & Li, Q. (2023). Construction of Recombinant Escherichia coli with a High L-Phenylalanine Production Yield from Glucose. Microbiology Research, 14(3), 1185-1198. https://doi.org/10.3390/microbiolres14030079





