New Insights into Molecular Characterization, Antimicrobial Resistance and Virulence Factors of Methicillin-Sensitive Coagulase-Positive Staphylococcus spp. from Dogs with Pyoderma and Otitis Externa
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Bacteriological Analyses
2.2. Antimicrobial Susceptibility Patterns
2.3. DNA Extraction and PCR Conditions
2.4. Detection of Genes Encoding Antimicrobial Resistance
2.5. Detection of Virulence Genes
2.6. Genotyping of Capsular Polysaccharide Types
2.7. Biofilm Production Assay of S. aureus Isolates
2.8. MultiLocus Sequence Typing of S. aureus Isolates
3. Results
3.1. Prevalence of Coagulase-Positive staphylococci Species
3.2. Antibiotic Susceptibility Profiles
3.3. Detection of Antimicrobial Resistance Genes
3.4. Virulence Gene Profiles
3.5. Genotyping of Capsular Genes
3.6. Biofilm Production
3.7. MLST Typing of S. aureus Isolates Detected in This Study
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fontana, C.; Favaro, M. Coagulase-Positive and Coagulase-Negative Staphylococci in Human Disease. In Pet-To-Man Travelling Staphylococc, A World in Progress; Elsevier: Amsterdam, The Netherlands, 2018; pp. 25–42. [Google Scholar] [CrossRef]
- Nagase, N.; Sasaki, A.; Yamashita, K.; Shimizu, A.; Wakita, Y.; Kitai, S.; Kawano, J. Isolation and Species Distribution of Staphylococci from Animal and Human Skin. J. Vet. Med. Sci. 2002, 64, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, H.; Melles, D.; Vos, M.; Leeuwen, W.; van Belkum, A.; Verbrugh, H.; Nouwen, J. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2006, 5, 751–762. [Google Scholar] [CrossRef] [PubMed]
- González-Martín, M.; Corbera, J.A.; Suárez-Bonnet, A.; Tejedor-Junco, M.T. Virulence factors in coagulase-positive staphylococci of veterinary interest other than Staphylococcus aureus. Vet. Q. 2020, 40, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Perreten, V.; Kania, S.A.; Bemis, D. Staphylococcus ursi sp. nov., a new member of the ‘Staphylococcus intermedius group’ isolated from healthy black bears. Int. J. Syst. Evol. Microbiol. 2020, 70, 4637–4645. [Google Scholar] [CrossRef] [PubMed]
- Madhaiyan, M.; Wirth, J.S.; Saravanan, V.S. Phylogenomic analyses of the Staphylococcaceae family suggest the reclassification of five species within the genus Staphylococcus as heterotypic synonyms, the promotion of five subspecies to novel species, the taxonomic reassignment of five Staphylococcus species to Mammaliicoccus gen. nov., and the formal assignment of Nosocomiicoccus to the family Staphylococcaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 5926–5936. [Google Scholar] [PubMed]
- Glajzner, P.; Szewczyk, E.M.; Szemraj, M. Pathogenic potential and antimicrobial resistance of Staphylococcus pseudintermedius isolated from human and animals. Folia Microbiol. 2023, 68, 231–243. [Google Scholar] [CrossRef]
- Foster, T.J. The Staphylococcus aureus “superbug”. J. Clin. Investig. 2004, 114, 1693–1696. [Google Scholar] [CrossRef]
- Devriese, L.A.; Vancanneyt, M.; Baele, M.; Vaneechoutte, M.; De Graef, E.; Snauwaert, C.; Cleenwerck, I.; Dawyndt, P.; Swings, J.; Decostere, A.; et al. Staphylococcus pseudintermedius sp. nov., a coagulase-positive species from animals. Int. J. Syst. Evol. Microbiol. 2005, 55, 1569–1573. [Google Scholar] [CrossRef] [PubMed]
- Lowy, F.D. Staphylococcus aureus Infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.A.; Helbig, K.J. The Complex Diseases of Staphylococcus pseudintermedius in Canines: Where to Next? Vet. Sci. 2021, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Fudaba, Y.; Nishifuji, K.; Andresen, L.O.; Yamaguchi, T.; Komatsuzawa, H.; Amagai, M.; Sugai, M. Staphylococcus hyicus exfoliative toxins selectively digest porcine desmoglein 1. Microb. Pathog. 2005, 39, 171–176. [Google Scholar] [CrossRef]
- L’Ecuyer, C.; Jericho, K. Exudative epidermitis in pigs: Etiological studies and pathology. Can. J. Comp. Med. Vet. Sci. 1966, 30, 94. [Google Scholar] [PubMed]
- Roberson, J.; Fox, L.; Hancock, D.; Gay, J.; Besser, T. Prevalence of coagulase-positive staphylococci, other than Staphylococcus aureus, in bovine mastitis. Am. J. Vet. Res. 1996, 57, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.W.; King, R.; Kloos, W. Isolation of Staphylococcus hyicus subsp hyicus from a pig with septic polyarthritis. Am. J. Vet. Res. 1980, 41, 274–276. [Google Scholar] [PubMed]
- Araújo, M.R.; Preis, I.S.; França, S.A.; Paniago, J.G.; Costa, M.C.; Oliveira, J.S.; Ecco, R. Mastitis accompanied by lymphadenitis in a dog caused by Staphylococcus hyicus. Braz. J. Vet. Pathol. 2011, 4, 52–57. [Google Scholar]
- Casanova, C.; Iselin, L.; von Steiger, N.; Droz, S.; Sendi, P. Staphylococcus hyicus bacteremia in a farmer. J. Clin. Microbiol. 2011, 49, 4377–4378. [Google Scholar] [CrossRef] [PubMed]
- Foissac, M.; Lekaditi, M.; Loutfi, B.; Ehrhart, A.; Dauchy, F.-A. Spondylodiscitis and bacteremia due to Staphylococcus hyicus in an immunocompetent man. Germs 2016, 6, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Bertelloni, F.; Cagnoli, G.; Ebani, V.V. Virulence and Antimicrobial Resistance in Canine Staphylococcus spp. Isolates. Microorganisms 2021, 9, 515. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Gao, W.; Wang, X.; Li, Q.; Ma, H.; Li, Z.; Xia, Z. Isolation and identification of multidrug-resistant pathogenic Staphylococcus delphini from racing pigeons. J. Jilin Agric. Univ. 2017, 39, 204–221. [Google Scholar]
- Sudagidan, M.; Aydin, A. Virulence properties of Staphylococcus delphini strains isolated from domestic pigeons. Med. Weter 2012, 68, 231–236. [Google Scholar]
- Stull, J.; Slavić, D.; Rousseau, J.; Weese, J. Staphylococcus delphini and methicillin-resistant Staphylococcus pseudintermedius in horses at a veterinary teaching hospital. J. Equine Vet. Sci. 2012, 32, 5. [Google Scholar] [CrossRef]
- Ronaghinia, A.A.; Nikolaisen, N.K.; Hansen, S.G.; Poulsen, H.H.; Frandsen, H.L.; Struve, T.; Toutain, P.L.; Damborg, P. Validating an empiric sulfadiazine–trimethoprim dosage regimen for treatment of Escherichia coli and Staphylococcus delphini infections in mink (Neovison vison). J. Vet. Pharmacol. Ther. 2021, 44, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Guardabassi, L.; Schmidt, K.R.; Petersen, T.S.; Espinosa-Gongora, C.; Moodley, A.; Agersø, Y.; Olsen, J.E. Mustelidae are natural hosts of Staphylococcus delphini group A. Vet. Microbiol. 2012, 159, 351–353. [Google Scholar] [CrossRef] [PubMed]
- Varaldo, P.E.; Kilpper-Bälz, R.; Biavasco, F.; Satta, G.; Schleifer, K.H. Staphylococcus delphini sp. nov., a coagulase-positive species isolated from dolphins. Int. J. Syst. Evol. Microbiol. 1988, 38, 436–439. [Google Scholar] [CrossRef]
- Magleby, R.; Bemis, D.A.; Kim, D.; Carroll, K.C.; Castanheira, M.; Kania, S.A.; Jenkins, S.G.; Westblade, L.F. First reported human isolation of Staphylococcus delphini. Diagn. Microbiol. Infect. Dis. 2019, 94, 274–276. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococcal Biofilms. Microbiol. Spectr. 2018, 6, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Ocloo, R.; Nyasinga, J.; Munshi, Z.; Hamdy, A.; Marciniak, T.; Soundararajan, M.; Newton-Foot, M.; Ziebuhr, W.; Shittu, A.; Revathi, G.; et al. Epidemiology and antimicrobial resistance of staphylococci other than Staphylococcus aureus from domestic animals and livestock in Africa: A systematic review. Front. Vet. Sci. 2022, 9, 1059054. [Google Scholar] [CrossRef]
- Schlievert, P.M.; Strandberg, K.L.; Lin, Y.C.; Peterson, M.L.; Leung, D.Y.M. Secreted virulence factor comparison between methicillin-resistant and methicillin-sensitive Staphylococcus aureus, and its relevance to atopic dermatitis. J. Allergy Clin. Immunol. 2010, 125, 39–49. [Google Scholar] [CrossRef]
- Melzer, M.; Eykyn, S.J.; Gransden, W.R.; Chinn, S. Is methicillin-resistant Staphylococcus aureus more virulent than methicillin-susceptible S. aureus? A comparative cohort study of British patients with nosocomial infection and bacteremia. Clin. Infect. Dis. 2003, 37, 1453–1460. [Google Scholar] [CrossRef]
- Weese, J.S.; Faires, M.C.; Frank, L.A.; Reynolds, L.M.; Battisti, A. Factors associated with methicillin-resistant versus methicillin-susceptible Staphylococcus pseudintermedius infection in dogs. J. Am. Vet. Med. Assoc. 2012, 240, 1450–1455. [Google Scholar] [CrossRef]
- Stepanovic, S.; Vukovic, D.; Dakic, I.; Savic, B.; Svabic-Vlahovic, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Nocera, F.P.; Ambrosio, M.; Fiorito, F.; Cortese, L.; De Martino, L. On Gram-Positive- and Gram-Negative-Bacteria-Associated Canine and Feline Skin Infections: A 4-Year Retrospective Study of the University Veterinary Microbiology Diagnostic Laboratory of Naples, Italy. Animals 2021, 11, 1603. [Google Scholar] [CrossRef] [PubMed]
- Gharsa, H.; Ben Slama, K.; Gómez-Sanz, E.; Lozano, C.; Zarazaga, M.; Messadi, L.; Boudabous, A.; Torres, C. Molecular characterization of Staphylococcus aureus from nasal samples of healthy farm animals and pets in Tunisia. Vector Borne Zoonotic Dis. 2015, 15, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Gharsa, H.; Ben Slama, K.; Gómez-Sanz, E.; Lozano, C.; Klibi, N.; Jouini, A.; Messadi, L.; Boudabous, A.; Torres, C. Antimicrobial resistance, virulence genes, and genetic lineages of Staphylococcus pseudintermedius in healthy dogs in tunisia. Microb. Ecol. 2013, 66, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Scherer, C.B.; Botoni, L.S.; Coura, F.M.; Silva, R.O.; Santos, R.D.; Heinemann, M.B.; Costa-Val, A.P. Frequency and antimicrobial susceptibility of Staphylococcus pseudintermedius in dogs with otitis externa. Ciência Rural 2018, 48, e20170738. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, J.; Logue, C.M.; Liu, K.; Cao, X.; Zhang, W.; Shen, J.; Wu, C. Methicillin-resistant Staphylococcus pseudintermedius isolated from canine pyoderma in North China. J. Appl. Microbiol. 2012, 112, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, E.; Viçosa, G.N.; Corsini, C.M.M.; Moreira, M.A.S.; Nero, L.A.; Conceição, L.G. Description of Methicillin-resistant Staphylococcus pseudintermedius from canine pyoderma in Minas Gerais state, Brazil. Arq. Bras. Med. Veterinária Zootec. 2016, 68, 299–306. [Google Scholar] [CrossRef]
- Botoni, L.S.; Scherer, C.B.; Silva, R.O.; Coura, F.M.; Heinemann, M.B.; Paes-Leme, F.O.; Costa-Val, A.P. Prevalence and in vitro susceptibility of methicillin-resistant Staphylococcus pseudintermedius (MRSP) from skin and nostrils of dogs with superficial pyoderma. Pesqui. Veterinária Bras. 2016, 36, 1178–1180. [Google Scholar] [CrossRef]
- Melter, O.; Svec, P.; Tkadlec, J.; Doskar, J.; Kinská, H.; Pantucek, R. Characterisation of methicillin-susceptible Staphylococcus pseudintermedius isolates from canine infections and determination of virulence factors using multiplex PCR. Veterinární Med. 2017, 62, 81–89. [Google Scholar] [CrossRef]
- Viegas, F.M.; Santana, J.A.; Silva, B.A.; Xavier, R.G.C.; Bonisson, C.T.; Câmara, J.L.S.; Rennó, M.C.; Cunha, J.L.R.; Figueiredo, H.C.P.; Lobato, F.C.F.; et al. Occurrence and characterization of methicillin-resistant Staphylococcus spp. in diseased dogs in Brazil. PLoS ONE 2022, 17, e0269422. [Google Scholar] [CrossRef]
- Boost, M.V.; O’Donoghue, M.M.; James, A. Prevalence of Staphylococcus aureus carriage among dogs and their owners. Epidemiol. Infect. 2008, 136, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Rafatpanah, S.; Rad, M.; Movassaghi, A.R.; Khoshnegah, J. Clinical, bacteriological and histopathological aspects of first-time pyoderma in a population of Iranian domestic dogs: A retrospective study. Iran. J. Vet. Res. 2020, 21, 130–135. [Google Scholar]
- Murugesan, A.C.; Ramachandran, M.; Varughese, H.S.; Kumaragurubaran, K. Staphylococcus coagulans possesses many virulence factors of Staph. aureus and Staph. pseudintermedius. J. Appl. Microbiol. 2023, 134, 1. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, I.M.; Guimarães, L.; da Silva, I.T.; Fonseca, C.; Assumpção, Y.; Lima dos Santos, A.L.; Antunes, M.; Pesset, C.; Ferreira, E.; Penna, B. High prevalence of Panton–Valentine Leucocidin among Staphylococcus coagulans isolated from dogs in Rio de Janeiro. J. Appl. Microbiol. 2023, 134, 12. [Google Scholar] [CrossRef] [PubMed]
- Palomino-Farfán, J.A.; Vega, L.G.A.; Espinoza, S.Y.C.; Magallanes, S.G.; Moreno, J.J.S. Methicillin-resistant Staphylococcus schleiferi subspecies coagulans associated with otitis externa and pyoderma in dogs. Open Vet. J. 2021, 11, 364–369. [Google Scholar]
- Costa, S.S.; Oliveira, V.; Serrano, M.; Pomba, C.; Couto, I. Phenotypic and Molecular Traits of Staphylococcus coagulans Associated with Canine Skin Infections in Portugal. Antibiotics 2021, 10, 518. [Google Scholar] [CrossRef] [PubMed]
- János, D.; Viorel, H.; Ionica, I.; Corina, P.; Tiana, F.; Roxana, D. Carriage of Multidrug Resistance Staphylococci in Shelter Dogs in Timisoara, Romania. Antibiotics 2021, 10, 801. [Google Scholar] [CrossRef] [PubMed]
- Vanni, M.; Tognetti, R.; Pretti, C.; Crema, F.; Soldani, G.; Meucci, V.; Intorre, L. Antimicrobial susceptibility of Staphylococcus intermedius and Staphylococcus schleiferi isolated from dogs. Res. Vet. Sci. 2009, 87, 192–195. [Google Scholar] [CrossRef]
- Souza, S.S.R.; Smith, J.T.; Bruce, S.A. Multi-host infection and phylogenetically diverse lineages shape the recombination and gene pool dynamics of Staphylococcus aureus. BMC Microbiol. 2023, 23, 235. [Google Scholar] [CrossRef]
- Rubin, J.E.; Chirino-Trejo, M. Prevalence, sites of colonization, and antimicrobial resistance among Staphylococcus pseudintermedius isolated from healthy dogs in Saskatoon, Canada. J. Vet. Diagn. Investig. 2011, 23, 351–354. [Google Scholar] [CrossRef]
- Tabatabaei, S.; Najafifar, A.; Askari Badouei, M.; Zahraei Salehi, T.; Ashrafi Tamai, I.; Khaksar, E.; Abbassi, M.S.; Ghazisaeedi, F. Genetic characterisation of methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in pets and veterinary personnel in Iran: New insights into emerging methicillin-resistant S. pseudintermedius (MRSP). J. Glob. Antimicrob. Resist. 2019, 16, 6–10. [Google Scholar] [CrossRef]
- O’Neill, A.J.; McLaws, F.; Kahlmeter, G.; Henriksen, A.S.; Chopra, I. Genetic basis of resistance to fusidic acid in staphylococci. Antimicrob. Agents Chemother. 2007, 51, 1737–1740. [Google Scholar] [CrossRef] [PubMed]
- Norström, T.; Lannergård, J.; Hughes, D. Genetic and phenotypic identification of fusidic acid-resistant mutants with the small-colony-variant phenotype in Staphylococcus aureus. Antimicrob. Agents Chemother. 2007, 51, 4438–4446. [Google Scholar] [CrossRef] [PubMed]
- Frosini, S.M.; Bond, R.; Rantala, M.; Grönthal, T.; Rankin, S.; O’Shea, K.; Timofte, D.; Schmidt, V.; Lindsay, J.; Loeffler, A. Genetic resistance determinants to fusidic acid and chlorhexidine in variably susceptible staphylococci from dogs. BMC Microbiol. 2019, 19, 81. [Google Scholar] [CrossRef]
- Lim, Y.J.; Hyun, J.E.; Hwang, C.Y. Identification of fusidic acid resistance in clinical isolates of Staphylococcus pseudintermedius from dogs in Korea. Vet. Dermatol. 2020, 31, 267.e62. [Google Scholar] [CrossRef] [PubMed]
- Frosini, S.M.; Bond, R.; Loeffler, A.; Larner, J. Opportunities for topical antimicrobial therapy: Permeation of canine skin by fusidic acid. BMC Vet. Res. 2017, 13, 345. [Google Scholar] [CrossRef] [PubMed]
- Seyedi Marghaki, F.; Kalantar-Neyestanaki, D.; Safaari, F.; Fasihi, Y.; Moradi, M. Frequency of aminoglycoside-resistance genes in methicillin resistant Staphylococcus aureus isolated from clinical specimens. J. Maz. Univ. Med. Sci. 2017, 27, 112–117. [Google Scholar]
- Choi, S.M.; Kim, S.H.; Kim, H.J.; Lee, D.G.; Choi, J.H.; Yoo, J.H.; Kang, J.H.; Shin, W.S.; Kang, M.W. Multiplex PCR for the detection of genes encoding aminoglycoside modifying enzymes and methicillin resistance among Staphylococcus species. J. Korean Med. Sci. 2003, 18, 631–636. [Google Scholar] [CrossRef]
- Gold, R.; Cohen, N.; Lawhon, S. Amikacin resistance in Staphylococcus pseudintermedius isolated from dogs. J. Clin. Microbiol. 2014, 52, 3641–3646. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.J.; Rather, P.N.; Hare, R.S.; Miller, G.H. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 1993, 57, 138–163. [Google Scholar] [CrossRef]
- Lord, J.; Millis, N.; Jones, R.D. Patterns of antimicrobial, multidrug and methicillin resistance among Staphylococcus spp. isolated from canine specimens submitted to a diagnostic laboratory in Tennessee, USA: A descriptive study. BMC Vet. Res. 2022, 18, 91. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M. Biofilm formation: A clinically relevant microbiological process. Clin. Infect. Dis. 2001, 33, 1387–1392. [Google Scholar] [CrossRef]
- Azmi, K.; Qrei, W.; Abdeen, Z. Screening of genes encoding adhesion factors and biofilm production in methicillin resistant strains of Staphylococcus aureus isolated from Palestinian patients. BMC Genom. 2019, 20, 578. [Google Scholar] [CrossRef]
- Verdier, I.; Durand, G.; Bes, M.; Taylor, K.L.; Lina, G.; Vandenesch, F.; Fattom, A.I.; Etienne, J. Identification of the capsular polysaccharides in Staphylococcus aureus clinical isolates by PCR and agglutination tests. J. Clin. Microbiol. 2007, 45, 725–729. [Google Scholar] [CrossRef]
- Bardiau, M.; Caplin, J.; Detilleux, J.; Graber, H.; Moroni, P.; Taminiau, B.; Mainil, J.G. Existence of two groups of Staphylococcus aureus strains isolated from bovine mastitis based on biofilm formation, intracellular survival, capsular profile and agr-typing. Vet. Microbiol. 2016, 185, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, D.; Shi, L.; Cai, R.; Li, C.; Yan, H. Association between agr type, virulence factors, biofilm formation and antibiotic resistance of Staphylococcus aureus isolates from pork production. Front. Microbiol. 2018, 9, 1876. [Google Scholar] [CrossRef] [PubMed]
- Ando, E.; Monden, K.; Mitsuhata, R.; Kariyama, R.; Kumon, H. Biofilm formation among methicillin-resistant Staphylococcus aureus isolates from patients with urinary tract infection. Acta Medica Okayama 2004, 58, 207–214. [Google Scholar] [PubMed]
- Anderson, M.J.; Schaaf, E.; Breshears, L.M.; Wallis, H.W.; Johnson, J.R.; Tkaczyk, C.; Sellman, B.R.; Sun, J.; Peterson, M.L. Alpha-Toxin Contributes to Biofilm Formation among Staphylococcus aureus Wound Isolates. Toxins 2018, 10, 157. [Google Scholar] [CrossRef] [PubMed]
- Ferry, T.; Perpoint, T.; Vandenesch, F.; Etienne, J. Virulence determinants in Staphylococcus aureus and their involvement in clinical syndromes. Curr. Infect. Dis. Rep. 2005, 7, 420. [Google Scholar] [CrossRef]
- Wiseman, G.M.; Caird, J.D.; Fackrell, H.B. Trypsin-mediated activation of the alpha-haemolysin of Staphylococcus aureus. J. Med. Microbiol. 1975, 8, 29–38. [Google Scholar] [CrossRef]
- Moraveji, Z.; Tabatabaei, M.; Shirzad Aski, H.; Khoshbakht, R. Characterization of hemolysins of Staphylococcus strains isolated from human and bovine, southern Iran. Iran. J. Vet. Res. 2014, 15, 326–330. [Google Scholar] [PubMed]
- Alonzo, F.; Kozhaya, L.; Rawlings, S.A.; Reyes-Robles, T.; DuMont, A.L.; Myszka, D.G.; Landau, N.R.; Unutmaz, D.; Torres, V.J. CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature 2013, 493, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Morinaga, N.; Kaihou, Y.; Noda, M. Purification, cloning and characterization of variant LukE-LukD with strong leukocidal activity of staphylococcal bi-component leukotoxin family. Microbiol. Immunol. 2003, 47, 81–90. [Google Scholar] [CrossRef]
- Diep, B.A.; Gill, S.R.; Chang, R.F.; Phan, T.H.; Chen, J.H.; Davidson, M.G.; Lin, F.; Lin, J.; Carleton, H.A.; Mongodin, E.F.; et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 2006, 367, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Gravet, A.; Rondeau, M.; Harf-Monteil, C.; Grunenberger, F.; Monteil, H.; Scheftel, J.M.; Prévost, G. Predominant Staphylococcus aureus isolated from antibiotic-associated diarrhea is clinically relevant and produces enterotoxin A and the bicomponent toxin LukE-lukD. J. Clin. Microbiol. 1999, 37, 4012–4019. [Google Scholar] [CrossRef] [PubMed]
- Gravet, A.; Couppié, P.; Meunier, O.; Clyti, E.; Moreau, B.; Pradinaud, R.; Monteil, H.; Prévost, G. Staphylococcus aureus isolated in cases of impetigo produces both epidermolysin A or B and LukE-LukD in 78% of 131 retrospective and prospective cases. J. Clin. Microbiol. 2001, 39, 4349–4356. [Google Scholar] [CrossRef]
- Tseng, C.W.; Biancotti, J.C.; Berg, B.L.; Gate, D.; Kolar, S.L.; Muller, S.; Rodriguez, M.D.; Rezai-Zadeh, K.; Fan, X.; Beenhouwer, D.O.; et al. Increased susceptibility of humanized NSG mice to Panton-Valentine leukocidin and Staphylococcus aureus skin infection. PLoS Pathog. 2015, 11, e1005292. [Google Scholar] [CrossRef]
- Azarian, T.; Cella, E.; Baines, S.L.; Shumaker, M.J.; Samel, C.; Jubair, M.; Pegues, D.A.; David, M.Z. Genomic Epidemiology and Global Population Structure of Exfoliative Toxin A-Producing Staphylococcus aureus Strains Associated with Staphylococcal Scalded Skin Syndrome. Front. Microbiol. 2021, 12, 663831. [Google Scholar] [CrossRef]
- Gómez-Sanz, E.; Torres, C.; Lozano, C.; Zarazaga, M. High diversity of Staphylococcus aureus and Staphylococcus pseudintermedius lineages and toxigenic traits in healthy pet-owning household members. Underestimating normal household contact? Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Nakaminami, H.; Okamura, Y.; Tanaka, S.; Wajima, T.; Murayama, N.; Noguchi, N. Prevalence of antimicrobial-resistant staphylococci in nares and affected sites of pet dogs with superficial pyoderma. J. Vet. Med. Sci. 2021, 83, 214–219. [Google Scholar] [CrossRef]
- Wedley, A.L.; Dawson, S.; Maddox, T.W.; Coyne, K.P.; Pinchbeck, G.L.; Clegg, P.; Jamrozy, D.; Fielder, M.D.; Donovan, D.; Nuttall, T.; et al. Carriage of Staphylococcus species in the veterinary visiting dog population in mainland UK: Molecular characterisation of resistance and virulence. Vet. Microbiol. 2014, 170, 81–88. [Google Scholar] [CrossRef]
- Yoon, J.W.; Lee, G.J.; Lee, S.Y.; Park, C.; Yoo, J.H.; Park, H.M. Prevalence of genes for enterotoxins, toxic shock syndrome toxin 1 and exfoliative toxin among clinical isolates of Staphylococcus pseudintermedius from canine origin. Vet. Dermatol. 2010, 21, 484–489. [Google Scholar] [CrossRef]
- Bardiau, M.; Detilleux, J.; Farnir, F.; Mainil, J.G.; Ote, I. Associations between properties linked with persistence in a collection of Staphylococcus aureus isolates from bovine mastitis. Vet. Microbiol. 2014, 169, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Garbacz, K.; Żarnowska, S.; Piechowicz, L.; Haras, K. Pathogenicity potential of Staphylococcus pseudintermedius strains isolated from canine carriers and from dogs with infection signs. Virulence 2013, 4, 255–259. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ruzauskas, M.; Couto, M.; Pavilonis, A.; Klimiene, I.; Siugzdiniene, R.; Virgailis, M.; Vaskeviciute, L.; Anskiene, L.; Pomba, C. Characterization of Staphylococcus pseudintermedius isolated from diseased dogs in Lithuania. Pol. J. Vet. Sci. 2016, 19, 7–14. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pitchenin, L.C.; Brandão, L.N.S.; Rosa, J.M.A.; Kagueyama, F.C.; da Silva Alves, A.; Rocha, Í.S.M.; Nakazato, L.; Dutra, V. Occurrence of toxin genes in Staphylococcus pseudintermedius from diseased dogs and other domestic and wild species. J. Infect. Dev. Ctries. 2017, 11, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Hritcu, O.M.; Schmidt, V.M.; Salem, S.E.; Maciuca, I.E.; Moraru, R.F.; Lipovan, I.; Mareş, M.; Solcan, G.; Timofte, D. Geographical Variations in Virulence Factors and Antimicrobial Resistance Amongst Staphylococci Isolated from Dogs from the United Kingdom and Romania. Front. Vet. Sci. 2020, 7, 414. [Google Scholar] [CrossRef] [PubMed]
- Somayaji, R.; Priyantha, M.; Rubin, J.; Church, D. Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin: Report of 24 cases. Diagn. Microbiol. Infect. Dis. 2016, 85, 471–476. [Google Scholar] [CrossRef]
- Robb, A.R.; Wright, E.D.; Foster, A.M.E.; Walker, R.; Malone, C. Skin infection caused by a novel strain of Staphylococcus pseudintermedius in a Siberian husky dog owner. JMM Case Rep. 2017, 4, 3. [Google Scholar] [CrossRef]
- Bannoehr, J.; Guardabassi, L. Staphylococcus pseudintermedius in the dog: Taxonomy, diagnostics, ecology, epidemiology and pathogenicity. Vet. Dermatol. 2012, 23, 253-e52. [Google Scholar] [CrossRef]
- Edwards, V.M.; Deringer, J.R.; Callantine, S.D.; Deobald, C.F.; Berger, P.H.; Kapur, V.; Stauffacher, C.V.; Bohach, G.A. Characterization of the canine type C enterotoxin produced by Staphylococcus intermedius pyoderma isolates. Infect. Immun. 1997, 65, 2346–2352. [Google Scholar] [CrossRef] [PubMed]
- Bunikowski, R.; Mielke, M.E.A.; Skarabis, H.; Worm, M.; Anagnostopoulos, I.; Kolde, G.; Wahn, U.; Renz, H. Evidence for a disease-promoting effect of Staphylococcus aureus–derived exotoxins in atopic dermatitis. J. Allergy Clin. Immunol. 2000, 105, 814–819. [Google Scholar] [CrossRef] [PubMed]
- Beceiro, A.; Tomás, M.; Bou, G. Antimicrobial resistance and virulence: A successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef]
- Monecke, S.; Coombs, G.; Shore, A.C.; Coleman, D.C.; Akpaka, P.; Borg, M.; Chow, H.; Ip, M.; Jatzwauk, L.; Jonas, D. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE 2011, 6, e17936. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Raji, A.; Garaween, G.; Soge, O.; Rey-Ladino, J.; Al-Kattan, W.; Shibl, A.; Senok, A. Antimicrobial resistance and virulence markers in methicillin sensitive Staphylococcus aureus isolates associated with nasal colonization. Microb. Pathog. 2016, 93, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Japoni-Nejad, A.; Rezazadeh, M.; Kazemian, H.; Fardmousavi, N.; van Belkum, A.; Ghaznavi-Rad, E. Molecular characterization of the first community-acquired methicillin-resistant Staphylococcus aureus strains from Central Iran. Int. J. Infect. Dis. 2013, 17, e949–e954. [Google Scholar] [CrossRef] [PubMed]
- Shady, H.M.A.; Bakr, A.E.A.; Hashad, M.E.; Alzohairy, M.A. Staphylococcus aureus nasal carriage among outpatients attending primary health care centers: A comparative study of two cities in Saudi Arabia and Egypt. Braz. J. Infect. Dis. 2015, 19, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Senok, A.; Ehricht, R.; Monecke, S.; Al-Saedan, R.; Somily, A. Molecular characterization of methicillin-resistant Staphylococcus aureus in nosocomial infections in a tertiary-care facility: Emergence of new clonal complexes in Saudi Arabia. New Microbes New Infect. 2016, 14, 13–18. [Google Scholar] [CrossRef]
- Senok, A.C.; Somily, A.M.; Slickers, P.; Raji, M.A.; Garaween, G.; Shibl, A.; Monecke, S.; Ehricht, R. Investigating a rare methicillin-resistant Staphylococcus aureus strain: First description of genome sequencing and molecular characterization of CC15-MRSA. Infect. Drug Resist. 2017, 10, 307. [Google Scholar] [CrossRef]
- Raji, M.A.; Garaween, G.; Ehricht, R.; Monecke, S.; Shibl, A.M.; Senok, A. Genetic characterization of Staphylococcus aureus isolated from retail meat in Riyadh, Saudi Arabia. Front. Microbiol. 2016, 7, 911. [Google Scholar] [CrossRef]
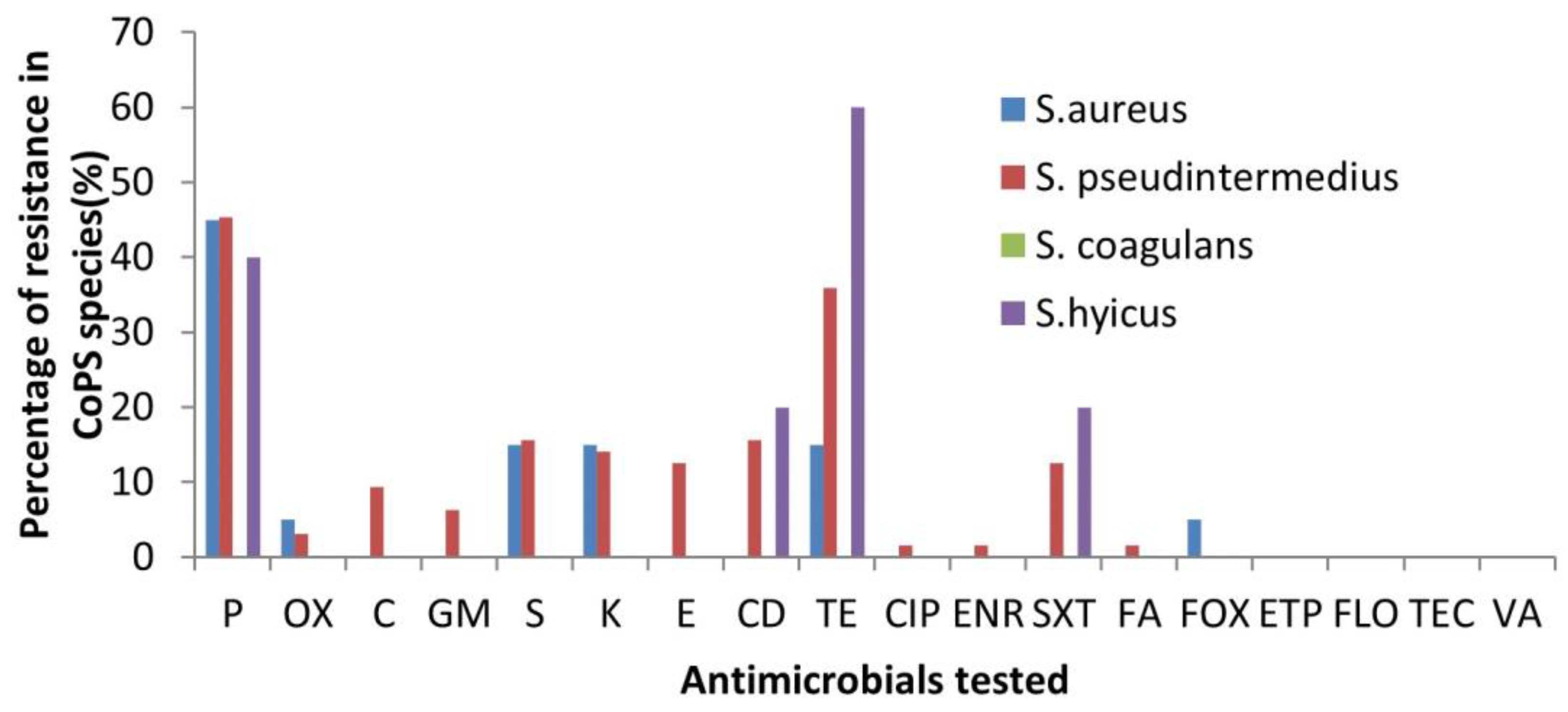
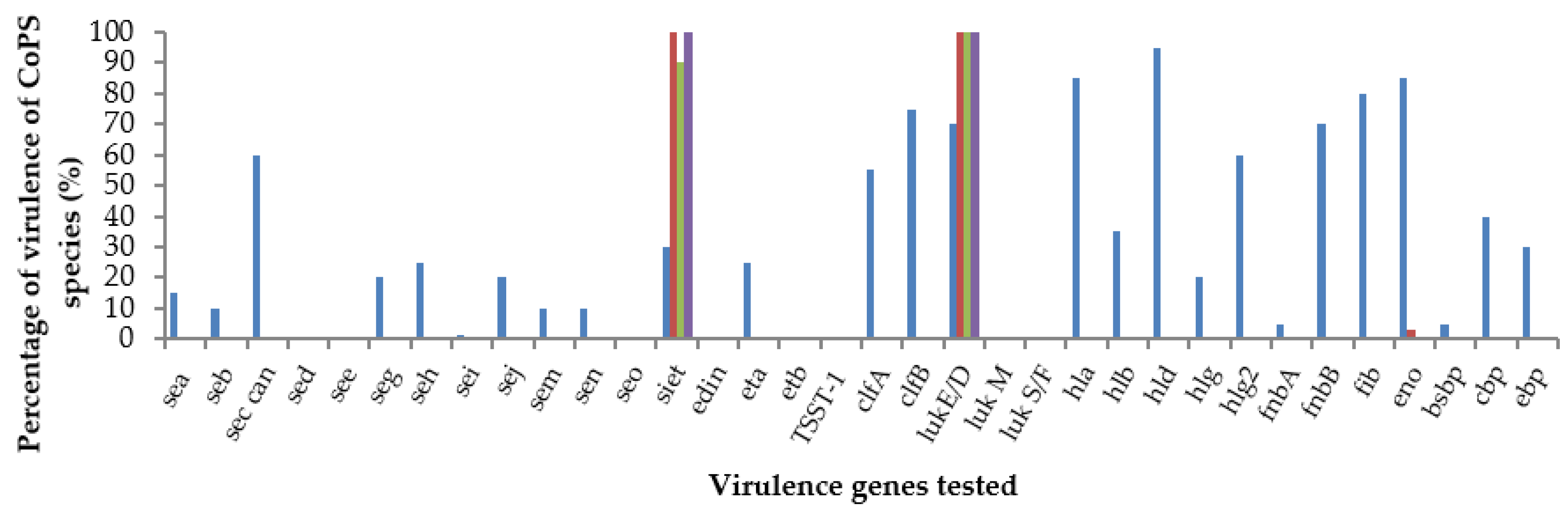
| Site | S. aureus (n) | S. pseudintermedius (n) | S. coagulans (n) | S. hyicus (n) | Total |
|---|---|---|---|---|---|
| N (n = 20) | 4 | 13 | 1 | 2 | 20 |
| E (n = 20) | 1 | 15 | 4 | 0 | 20 |
| O (n = 20) | 7 | 10 | 3 | 0 | 20 |
| S (n = 14) | 4 | 8 | 1 | 1 | 14 |
| R (n = 25) | 4 | 18 | 1 | 2 | 25 |
| Total (n = 99) | 20 (20.2%) | 64 (64.64%) | 10 (10.1%) | 5 (5%) |
| Species (Number of Isolates) | Antimicrobial Resistance | Virulence Genes (Number of Isolates) | |
|---|---|---|---|
| Phenotype (Number of Isolates) | Genotype (Number of Isolates) | ||
| S. pseudintermedius (64) | P (10) | blaZ (9) | siet, (4) |
| T (7) | TetM (6) | lukE/D (2) | |
| Cl (1) | blaZ, TetM (6) | siet, lukE/D (58) | |
| Sul (1) | blaZ, aph(3)-IIIa (1) | siet, lukE/D, eno (2) | |
| P, T (4) | blaZ, TetM, ant (6)-Ia (3) | ||
| P, S (1) | blaZ, Cat (PC221), TetM, ermB, aph(3)-IIIa (1) | ||
| Cl, Sul (1) | blaZ, Cat (PC221), TetM, ermB, aph(3)-IIIa, ant (6)-Ia (1) | ||
| E, Cl (1) | Cat (PC221), ermB, aph(3)-IIIa, ant (6)-Ia (1) | ||
| S, Cl (1) | |||
| T, Sul (1) | |||
| P, S, T (1) | |||
| P, K, S, T (1) | |||
| P, G, K, T (3) | |||
| P, Ch, S, E, Cl, T (1) | |||
| P, E, Cl, Cip, En, Sul, Fus (1) | |||
| P, Ch, G, S, K, E, Cl (1) | |||
| P, Ch, S, K, E, Cl, T (2) | |||
| P, O, Ch, S, K, E, Cl, T (1) | |||
| P, Ch, S, K, E, Cl, T, Sul (1) | |||
| Susceptible (24) | |||
| S. hyicus (5) | P (1) | blaZ (1) | siet, lukE/D (5) |
| P,T (1) | tetM (1) | ||
| P, Cl, T (1) | blaZ, tetM (1) | ||
| T, Sul (1) | |||
| Susceptible (1) | |||
| CODE | Animal | Sites | Disease | Treatment | ST/CC | Phenotypic Resistance | Antibiotic Resistance Genes | Virulence-Associated Genes | Capsular Type | Biofilm Production |
|---|---|---|---|---|---|---|---|---|---|---|
| S1 | 1 | nose | Otitis | yes | 36548 | P | blaZ | seccan, seh, clfA, clfB, lukDE, hla, hld, mphlg2, fnA, fnbB, fib, cbp, eno, ebp | 5 | high |
| S2 | 2 | skin | Pyoderma | no | 36549 | P, O | blaZ | seb seccan, seh, clfA, lukDE, hla, hld, mphg2, fnbB, fib, eno | 8 | moderate |
| S3 | 2 | mouth | 36549 | seb, seh, clfA, clfB, lukDE, hla, hld, mphlg2, fnbB, fib, eno, ebp | 8 | no | ||||
| S4 | 3 | mouth | Pyoderma | yes | 36549 | P | blaZ | sea, seg, sei, sej, sem, sen, siet, clfA, clfB, hla, hld, mphlg, mphlg2, fib, cbp, eno, ebp | 8 | no |
| S5 | 4 | mouth | Pyoderma or otitis | 36550 | P | blaZ | sea, seccansiet, clfA, clfB, lukDE, hla, hld, mphlg2, fnbB, fib, bsp, eno | 8 | moderate | |
| S6 | 5 | nose | Otitis | no | 36550 | P | blaZ | clfA, clfB, lukDE, hlb, hld, mphlg2, fnbB, fib, eno, ebp | 8 | |
| S7 | 6 | mouth | Pyoderma | yes | 36550 | seccansej, siet, clfA, clfB, lukDE, hla, hlb, hld, mphlg2, fnbB, fib, eno, ebp | 8 | no | ||
| S8 | 6 | skin | 36551 | sej, clfA, clfB, lukDE, hla, hlb, hld, mphlg2, fib, eno, ebp | 5 | no | ||||
| S9 | 6 | 36551 | FOX | seccan, siet, clfB, lukDE, hla, hlb, hld, mphlg2, eta, fnbB, fib, eno, ebp | 5 | high | ||||
| S10 | 6 | nose | 36551 | siet, clfA, clfB, lukDE, hla, hlb, hld, mphlg2, eta, fib, eno | 5 | moderate | ||||
| S11 | 7 | skin | Pyoderma | no | 36551 | siet, clfA, clfB, lukDE, hla, hld, fib, cbp, eno | 8 | no | ||
| S12 | 7 | mouth | 36551 | seccan, clfA, clfB, hld, mphlg2, fnbB, fib, eno | 8 | moderate | ||||
| S13 | 8 | anus | Pyoderma | no | 5896/CC15 | P, O, S, K, T | aph(3) IIIa | seg, seh, lukDE, hla, hld, eta, fnbB, fib, eno | 5 | high |
| S14 | 8 | mouth | 5896/CC15 | P, S, K, T | blaZ, aph(3) IIIa | seccan, sem, lukE/D, hla, hld, hlg2, eta, fnbB, fib, eno, cbp | 5 and 8 | moderate | ||
| S15 | 9 | nose | ? | ? | 5896/CC15 | P | blaZ | seccan, seg, seh, sen, clfB, hla, hld | 5 | moderate |
| S16 | 10 | skin | Otitis | yes | 5896/CC15 | seccan, clfB, hla, hld, fnbB, cbp | N.T | moderate | ||
| S17 | 10 | ear | 5896/CC15 | seccan, clfB, lukE/D, hlb, hld, hlg, fnbB, cbp, eno | 5 | moderate | ||||
| S18 | 10 | nose | 5896/CC15 | P, S, K | seccan, hla, hld, mphg, hlg2, fnbB, fib | 8 | moderate | |||
| S19 | 10 | anus | 5896/CC15 | sea, seccan, seg, sej, clfB, lukE/D, hlb, hld, hlg fnbB, eno | N.T | moderate | ||||
| S20 | 11 | mouth | ? | ? | 5896/CC15 | P | blaZ | lukE/D, hla, fib, eno, ebp | 8 | weak |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben Chehida, F.; Tombari, W.; Gharsa, H.; Rabia, Y.; Ferhi, S.; Jrad, M.; Messadi, L. New Insights into Molecular Characterization, Antimicrobial Resistance and Virulence Factors of Methicillin-Sensitive Coagulase-Positive Staphylococcus spp. from Dogs with Pyoderma and Otitis Externa. Microbiol. Res. 2024, 15, 1208-1224. https://doi.org/10.3390/microbiolres15030081
Ben Chehida F, Tombari W, Gharsa H, Rabia Y, Ferhi S, Jrad M, Messadi L. New Insights into Molecular Characterization, Antimicrobial Resistance and Virulence Factors of Methicillin-Sensitive Coagulase-Positive Staphylococcus spp. from Dogs with Pyoderma and Otitis Externa. Microbiology Research. 2024; 15(3):1208-1224. https://doi.org/10.3390/microbiolres15030081
Chicago/Turabian StyleBen Chehida, Faten, Wafa Tombari, Haythem Gharsa, Youssef Rabia, Sana Ferhi, Maha Jrad, and Lilia Messadi. 2024. "New Insights into Molecular Characterization, Antimicrobial Resistance and Virulence Factors of Methicillin-Sensitive Coagulase-Positive Staphylococcus spp. from Dogs with Pyoderma and Otitis Externa" Microbiology Research 15, no. 3: 1208-1224. https://doi.org/10.3390/microbiolres15030081
APA StyleBen Chehida, F., Tombari, W., Gharsa, H., Rabia, Y., Ferhi, S., Jrad, M., & Messadi, L. (2024). New Insights into Molecular Characterization, Antimicrobial Resistance and Virulence Factors of Methicillin-Sensitive Coagulase-Positive Staphylococcus spp. from Dogs with Pyoderma and Otitis Externa. Microbiology Research, 15(3), 1208-1224. https://doi.org/10.3390/microbiolres15030081






