Comparison of Gut Microbiota in Overwintering Bees: Apis cerana vs. Apis mellifera
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Acquisition
2.2. DNA Extraction
2.3. Bioinformatics Analysis
3. Results
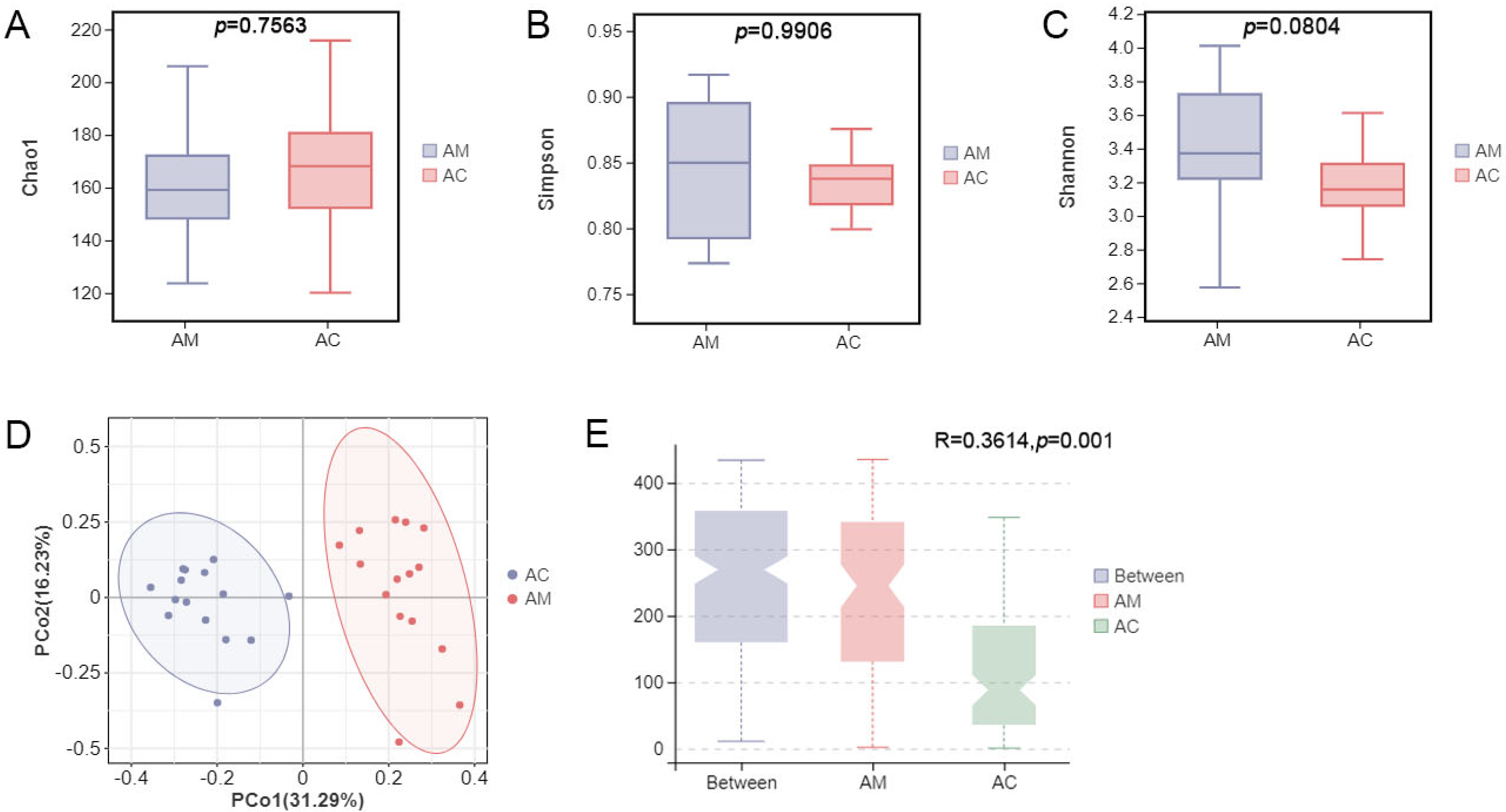
3.1. Structural Diversity of the Gut Microbiota
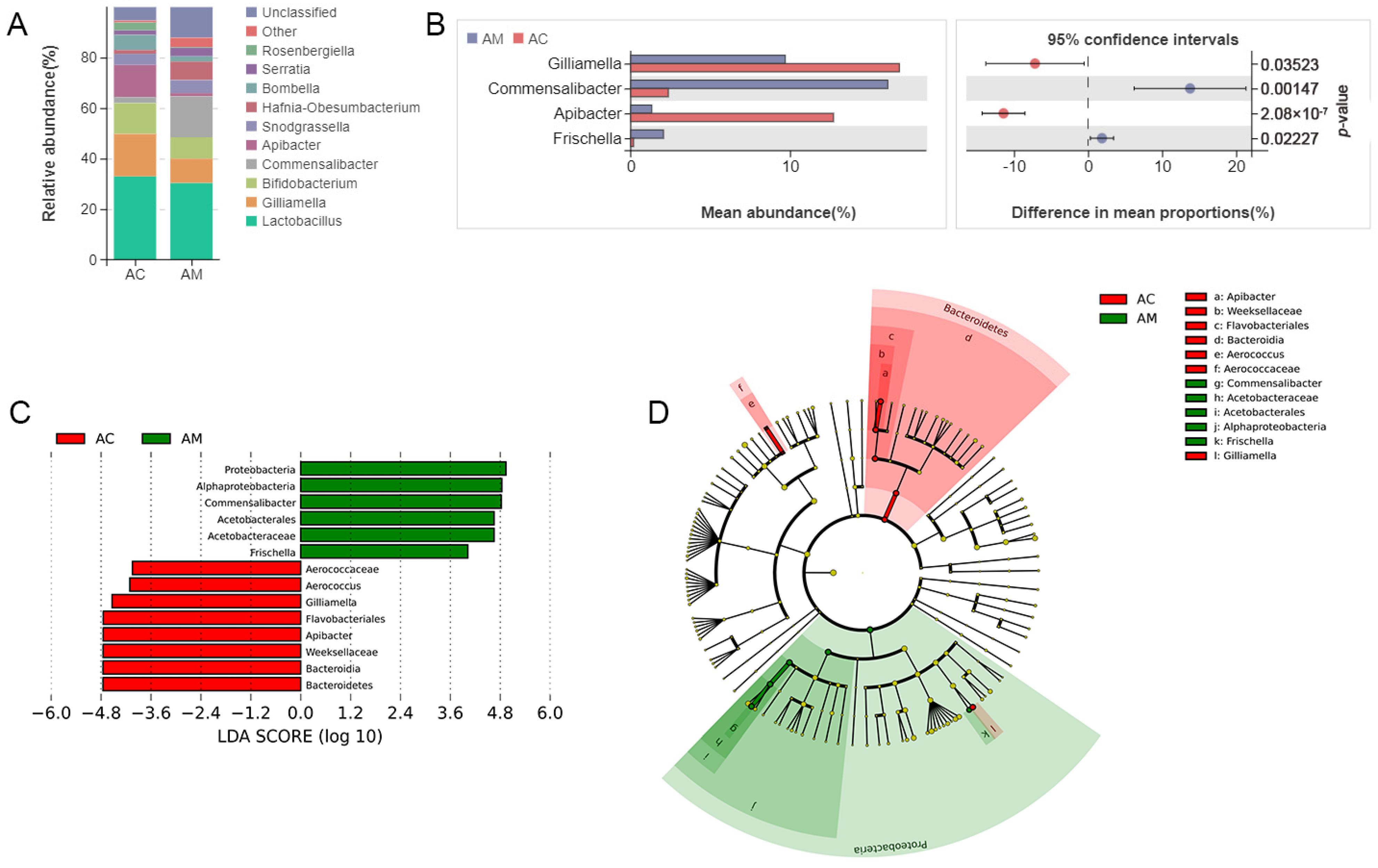
3.2. Composition of the Gut Microbiota
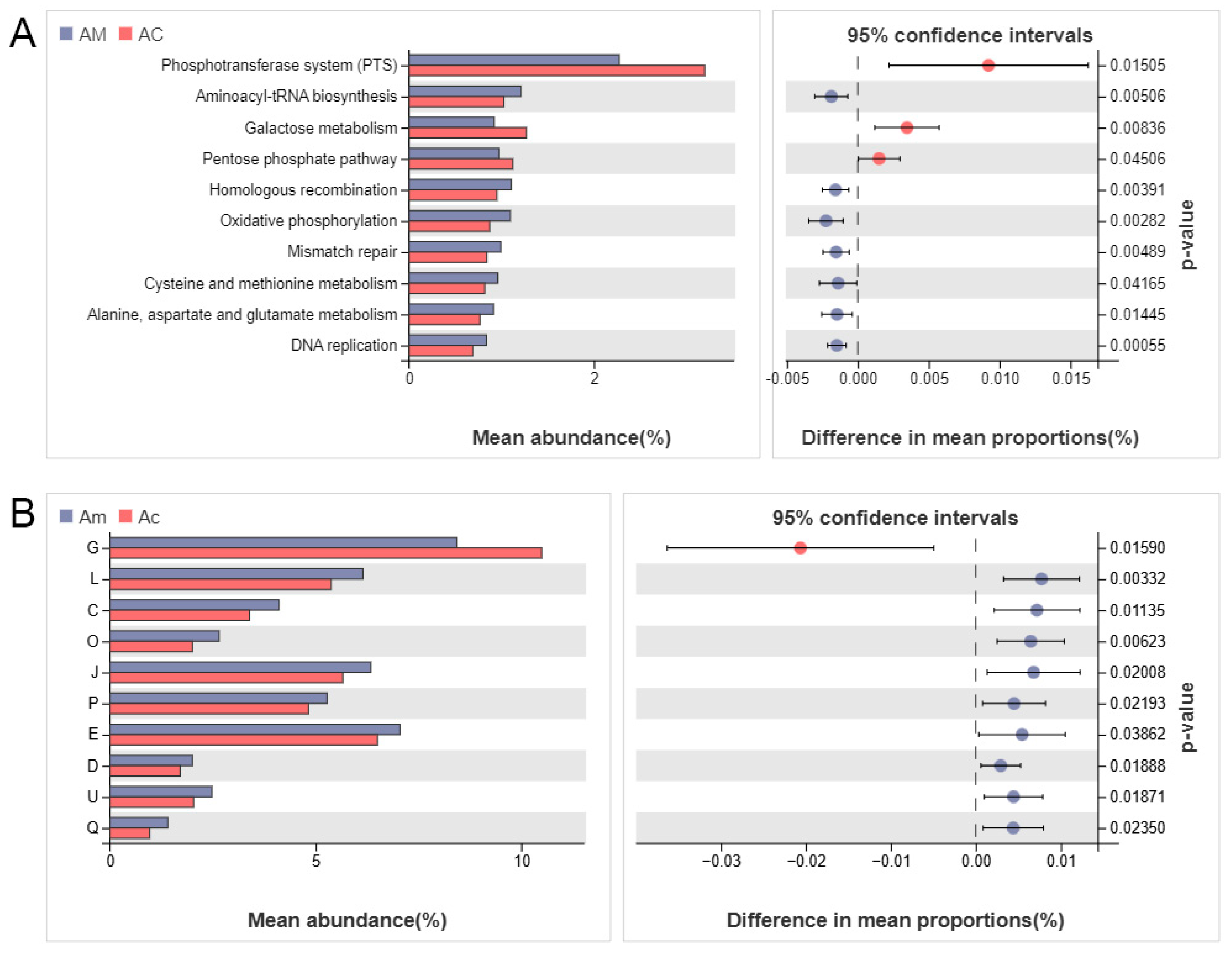
3.3. Metagenomic Analysis Revealed Differential Functional Features
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Potts, S.G.; Imperatriz-Fonseca, V.; Ngo, H.T.; Aizen, M.A.; Biesmeijer, J.C.; Breeze, T.D.; Dicks, L.V.; Garibaldi, L.A.; Hill, R.; Settele, J.; et al. Safeguarding Pollinators and Their Values to Human Well-Being. Nature 2016, 540, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.M.; Vaissière, B.E.; Cane, J.H.; Steffan-Dewenter, I.; Cunningham, S.A.; Kremen, C.; Tscharntke, T. Importance of Pollinators in Changing Landscapes for World Crops. Proc. R. Soc. B Biol. Sci. 2007, 274, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Tawfik, A.I.; Ahmed, Z.H.; Abdel-Rahman, M.F.; Moustafa, A.M. Influence of Winter Feeding on Colony Development and the Antioxidant System of the Honey Bee, Apis mellifera. J. Apic. Res. 2020, 59, 752–763. [Google Scholar] [CrossRef]
- Knoll, S.; Pinna, W.; Varcasia, A.; Scala, A.; Cappai, M.G. The Honey Bee (Apis mellifera L., 1758) and the Seasonal Adaptation of Productions. Highlights on Summer to Winter Transition and Back to Summer Metabolic Activity. A Review. Livest. Sci. 2020, 235, 104011. [Google Scholar] [CrossRef]
- Döke, M.A.; Frazier, M.; Grozinger, C.M. Overwintering Honey Bees: Biology and Management. Curr. Opin. Insect Sci. 2015, 10, 185–193. [Google Scholar] [CrossRef]
- Li, C.; Tang, M.; Li, X.; Zhou, X. Community Dynamics in Structure and Function of Honey Bee Gut Bacteria in Response to Winter Dietary Shift. MBio 2022, 13, e0113122. [Google Scholar] [CrossRef]
- Liu, Z.; Yao, P.; Guo, X.; Xu, B. Two Small Heat Shock Protein Genes in Apis cerana Cerana: Characterization, Regulation, and Developmental Expression. Gene 2014, 545, 205–214. [Google Scholar] [CrossRef]
- Xu, K.; Niu, Q.; Zhao, H.; Du, Y.; Jiang, Y. Transcriptomic Analysis to Uncover Genes Affecting Cold Resistance in the Chinese Honey Bee (Apis cerana Cerana). PLoS ONE 2017, 12, e0179922. [Google Scholar] [CrossRef]
- Omholt, S.W.; Amdam, G.V. Epigenetic Regulation of Aging in Honeybee Workers. Sci. Aging Knowl. Environ. 2004, 2004, pe28. [Google Scholar] [CrossRef]
- Lee, S.; Kalcic, F.; Duarte, I.F.; Titera, D.; Kamler, M.; Mrna, P.; Hyrsl, P.; Danihlik, J.; Dobes, P.; Kunc, M.; et al. 1H NMR Profiling of Honey Bee Bodies Revealed Metabolic Differences between Summer and Winter Bees. Insects 2022, 13, 193. [Google Scholar] [CrossRef]
- Kunc, M.; Dobeš, P.; Hurychová, J.; Vojtek, L.; Poiani, S.B.; Danihlík, J.; Havlík, J.; Titěra, D.; Hyršl, P. The Year of the Honey Bee (Apis mellifera L.) with Respect to Its Physiology and Immunity: A Search for Biochemical Markers of Longevity. Insects 2019, 10, 244. [Google Scholar] [CrossRef] [PubMed]
- Kunert, K.; Crailsheim, K. Seasonal Changes in Carbohydrate, Lipid and Protein Content in Emerging Worker Honeybees and Their Mortality. J. Apic. Res. 1988, 27, 13–21. [Google Scholar] [CrossRef]
- Maes, P.W.; Rodrigues, P.A.P.; Oliver, R.; Mott, B.M.; Anderson, K.E. Diet-Related Gut Bacterial Dysbiosis Correlates with Impaired Development, Increased Mortality and Nosema Disease in the Honeybee (Apis mellifera). Mol. Ecol. 2016, 25, 5439–5450. [Google Scholar] [CrossRef]
- Zheng, H.; Steele, M.I.; Leonard, S.P.; Motta, E.V.S.; Moran, N.A. Honey Bees as Models for Gut Microbiota Research. Lab Anim. 2018, 47, 317–325. [Google Scholar] [CrossRef]
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.W.; Soh, E.J.Y.; Ascher, J.S.; Jaffé, R.; Moran, N.A. Dynamic Microbiome Evolution in Social Bees. Sci. Adv. 2017, 3, e1600513. [Google Scholar] [CrossRef]
- Anderson, K.E.; Ricigliano, V.A.; Mott, B.M.; Copeland, D.C.; Floyd, A.S.; Maes, P. The Queen’s Gut Refines with Age: Longevity Phenotypes in a Social Insect Model. Microbiome 2018, 6, 108. [Google Scholar] [CrossRef]
- Ellegaard, K.M.; Engel, P. Genomic Diversity Landscape of the Honey Bee Gut Microbiota. Nat. Commun. 2019, 10, 446. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.E.; Ricigliano, V.A. Honey Bee Gut Dysbiosis: A Novel Context of Disease Ecology. Curr. Opin. Insect Sci. 2017, 22, 125–132. [Google Scholar] [CrossRef]
- Kešnerová, L.; Emery, O.; Troilo, M.; Liberti, J.; Erkosar, B.; Engel, P. Gut Microbiota Structure Differs between Honeybees in Winter and Summer. ISME J. 2020, 14, 801–814. [Google Scholar] [CrossRef]
- Liu, P.; Zhu, Y.; Ye, L.; Shi, T.; Li, L.; Cao, H.; Yu, L. Overwintering Honeybees Maintained Dynamic and Stable Intestinal Bacteria. Sci. Rep. 2021, 11, 22233. [Google Scholar] [CrossRef]
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global Pollinator Declines: Trends, Impacts and Drivers. Trends Ecol. Evol. 2010, 25, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.D.; Evans, J.D.; Pettis, J. Colony Losses, Managed Colony Population Decline, and Colony Collapse Disorder in the United States. J. Apic. Res. 2010, 49, 134–136. [Google Scholar] [CrossRef]
- Goodrich, B.K.; Goodhue, R.E. Are All Colonies Created Equal? The Role of Honey Bee Colony Strength in Almond Pollination Contracts. Ecol. Econ. 2020, 177, 106744. [Google Scholar] [CrossRef]
- Dillon, R.J.; Dillon, V.M. The Gut Bacteria of Insects: Nonpathogenic Interactions. Annu. Rev. Entomol. 2004, 49, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Freitak, D.; Vogel, H.; Ping, L.; Shao, Y.; Cordero, E.A.; Andersen, G.; Westermann, M.; Heckel, D.G.; Boland, W. Complexity and Variability of Gut Commensal Microbiota in Polyphagous Lepidopteran Larvae. PLoS ONE 2012, 7, e36978. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Moran, N.A. Gut Microbial Communities of Social Bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef]
- Martinson, V.G.; Moy, J.; Moran, N.A. Establishment of Characteristic Gut Bacteria during Development of the Honeybee Worker. Appl. Environ. Microbiol. 2012, 78, 2830–2840. [Google Scholar] [CrossRef]
- Kapheim, K.M.; Rao, V.D.; Yeoman, C.J.; Wilson, B.A.; White, B.A.; Goldenfeld, N.; Robinson, G.E. Caste-Specific Differences in Hindgut Microbial Communities of Honey Bees (Apis mellifera). PLoS ONE 2015, 10, e0123911. [Google Scholar] [CrossRef]
- Jeyaprakash, A.; Hoy, M.A.; Allsopp, M.H. Bacterial Diversity in Worker Adults of Apis mellifera Capensis and Apis mellifera Scutellata (Insecta: Hymenoptera) Assessed Using 16S RRNA Sequences. J. Invertebr. Pathol. 2003, 84, 96–103. [Google Scholar] [CrossRef]
- Sabree, Z.L.; Hansen, A.K.; Moran, N.A. Independent Studies Using Deep Sequencing Resolve the Same Set of Core Bacterial Species Dominating Gut Communities of Honey Bees. PLoS ONE 2012, 7, e41250. [Google Scholar] [CrossRef]
- Cox-Foster, D.L.; Conlan, S.; Holmes, E.C.; Palacios, G.; Evans, J.D.; Moran, N.A.; Quan, P.L.; Briese, T.; Hornig, M.; Geiser, D.M.; et al. A Metagenomic Survey of Microbes in Honey Bee Colony Collapse Disorder. Science 2007, 318, 283–287. [Google Scholar] [CrossRef]
- Ludvigsen, J.; Rangberg, A.; Avershina, E.; Sekelja, M.; Kreibich, C.; Amdam, G.; Rudi, K. Shifts in the Midgut/Pyloric Microbiota Composition within a Honey Bee Apiary throughout a Season. Microbes Environ. 2015, 30, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, X.; Su, Q.; Tang, M.; Zheng, H.; Zhou, X. Genomic Features Underlying the Evolutionary Transitions of Apibacter to Honey Bee Gut Symbionts. Insect Sci. 2022, 29, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Moran, N.A. Apibacter adventoris Gen. Nov., sp. Nov., a Member of the Phylum Bacteroidetes Isolated from Honey Bees. Int. J. Syst. Evol. Microbiol. 2016, 66, 1323–1329. [Google Scholar] [CrossRef]
- Mockler, B.K.; Kwong, W.K.; Moran, N.A.; Koch, H. Microbiome Structure Influences Infection by the Parasite Crithidia Bombi in Bumble Bees. Appl. Environ. Microbiol. 2018, 84, e02335-17. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Steele, M.I.; Moran, N.A. Genome Sequences of Apibacter spp., Gut Symbionts of Asian Honey Bees. Genome Biol. Evol. 2018, 10, 1174–1179. [Google Scholar] [CrossRef]
- Praet, J.; Aerts, M.; de Brandt, E.; Meeus, I.; Smagghe, G.; Vandamme, P. Apibacter mensalis sp. Nov.: A Rare Member of the Bumblebee Gut Microbiota. Int. J. Syst. Evol. Microbiol. 2016, 66, 1645–1651. [Google Scholar] [CrossRef]
- Powell, J.E.; Leonard, S.P.; Kwong, W.K.; Engel, P.; Moran, N.A. Genome-Wide Screen Identifies Host Colonization Determinants in a Bacterial Gut Symbiont. Proc. Natl. Acad. Sci. USA 2016, 113, 13887–13892. [Google Scholar] [CrossRef]
- Ludvigsen, J.; Porcellato, D.; Amdam, G.V.; Rudi, K. Addressing the Diversity of the Honeybee Gut Symbiont Gilliamella: Description of Gilliamella apis sp. Nov., Isolated from the Gut of Honeybees (Apis mellifera). Int. J. Syst. Evol. Microbiol. 2018, 68, 1762–1770. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Cultivation and Characterization of the Gut Symbionts of Honey Bees and Bumble Bees: Description of Snodgrassella alvi Gen. Nov., sp. Nov., a Member of the Family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola Gen. Nov., sp. Nov., a Memb. Int. J. Syst. Evol. Microbiol. 2013, 63, 2008–2018. [Google Scholar] [CrossRef]
- Kwong, W.K.; Engel, P.; Koch, H.; Moran, N.A. Genomics and Host Specialization of Honey Bee and Bumble Bee Gut Symbionts. Proc. Natl. Acad. Sci. USA 2014, 111, 11509–11514. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Kwong, W.K.; Moran, N.A. Frischella perrara Gen. Nov., sp. Nov., a Gammaproteobacterium Isolated from the Gut of the Honeybee, Apis mellifera. Int. J. Syst. Evol. Microbiol. 2013, 63, 3646–3651. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Martinson, V.G.; Moran, N.A. Functional Diversity within the Simple Gut Microbiota of the Honey Bee. Proc. Natl. Acad. Sci. USA 2012, 109, 11002–11007. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Nishida, A.; Kwong, W.K.; Koch, H.; Engel, P.; Steele, M.I.; Moran, N.A. Metabolism of Toxic Sugars by Strains of the Bee Gut Symbiont Gilliamella apicola. MBio 2016, 7, e01326-16. [Google Scholar] [CrossRef] [PubMed]
- Robillard, G.T.; Broos, J. Structure/Function Studies on the Bacterial Carbohydrate Transporters, Enzymes II, of the Phosphoenolpyruvate-Dependent Phosphotransferase System. Biochim. Biophys. Acta Rev. Biomembr. 1999, 1422, 73–104. [Google Scholar] [CrossRef]
- Nguyen, V.H. Genomic Investigations of Diverse Corbiculate Bee Gut-Associated Gilliamella Reveal Conserved Pathways for Energy Metabolism, with Diverse and Variable Energy Sources. Access Microbiol. 2024, 6, 000793.v3. [Google Scholar] [CrossRef]
- Zheng, H.; Perreau, J.; Elijah Powell, J.; Han, B.; Zhang, Z.; Kwong, W.K.; Tringe, S.G.; Moran, N.A. Division of Labor in Honey Bee Gut Microbiota for Plant Polysaccharide Digestion. Proc. Natl. Acad. Sci. USA 2019, 116, 25909–25916. [Google Scholar] [CrossRef]
- Levin, E.; Lopez-Martinez, G.; Fane, B.; Davidowitz, G. Hawkmoths Use Nectar Sugar to Reduce Oxidative Damage from Flight. Science 2017, 355, 733–735. [Google Scholar] [CrossRef]
- Blatt, J.; Roces, F. Haemolymph Sugar Levels in Foraging Honeybees (Apis mellifera Carnica): Dependence on Metabolic Rate and in Vivo Measurement of Maximal Rates of Trehalose Synthesis. J. Exp. Biol. 2001, 204, 2709–2716. [Google Scholar] [CrossRef]
- Lee, F.J.; Rusch, D.B.; Stewart, F.J.; Mattila, H.R.; Newton, I.L.G. Saccharide Breakdown and Fermentation by the Honey Bee Gut Microbiome. Environ. Microbiol. 2015, 17, 796–815. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Gao, L.; Liu, J.; Ji, C.; Dang, X.; Zhou, Z.; Luo, W. Comparison of Gut Microbiota in Overwintering Bees: Apis cerana vs. Apis mellifera. Microbiol. Res. 2024, 15, 2425-2434. https://doi.org/10.3390/microbiolres15040163
Chen H, Gao L, Liu J, Ji C, Dang X, Zhou Z, Luo W. Comparison of Gut Microbiota in Overwintering Bees: Apis cerana vs. Apis mellifera. Microbiology Research. 2024; 15(4):2425-2434. https://doi.org/10.3390/microbiolres15040163
Chicago/Turabian StyleChen, Heng, Lijiao Gao, Jialin Liu, Conghui Ji, Xiaoqun Dang, Zeyang Zhou, and Wenhua Luo. 2024. "Comparison of Gut Microbiota in Overwintering Bees: Apis cerana vs. Apis mellifera" Microbiology Research 15, no. 4: 2425-2434. https://doi.org/10.3390/microbiolres15040163
APA StyleChen, H., Gao, L., Liu, J., Ji, C., Dang, X., Zhou, Z., & Luo, W. (2024). Comparison of Gut Microbiota in Overwintering Bees: Apis cerana vs. Apis mellifera. Microbiology Research, 15(4), 2425-2434. https://doi.org/10.3390/microbiolres15040163





