Candida auris Antifungal Resistance, Virulence and Susceptibility to a Novel Nitric Oxide-Releasing Microparticle and Its Correlations to Clade Identification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Candida Auris Strains
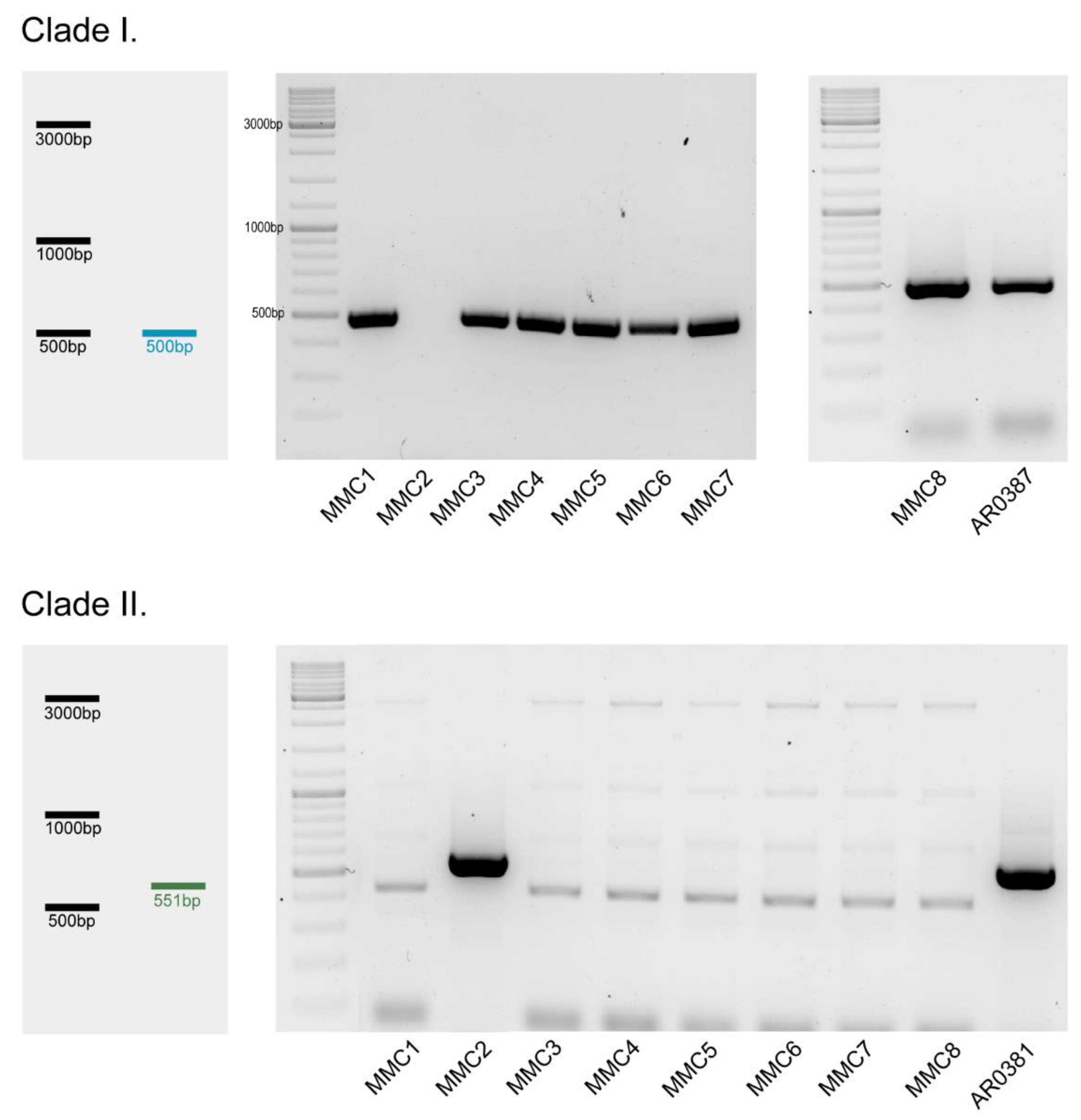
2.2. DNA Extraction and Clade Classification
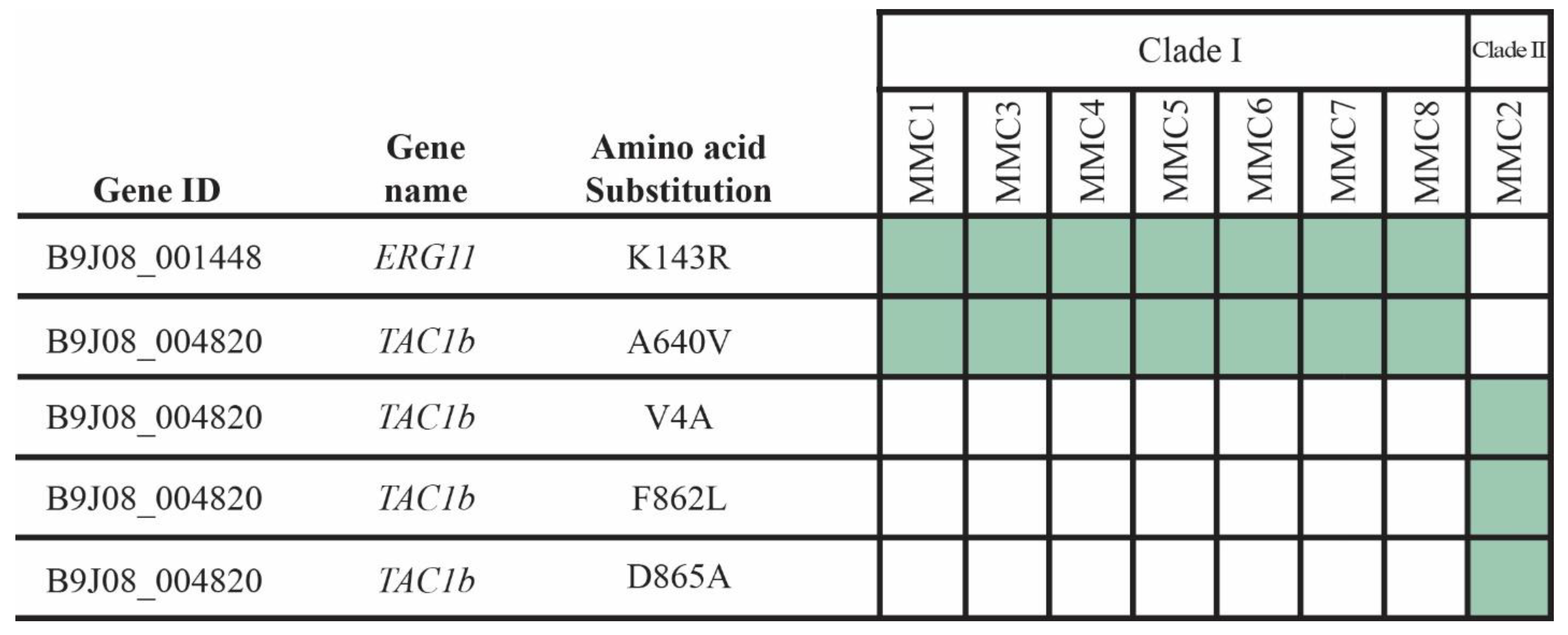
2.3. Sequence Analysis
2.4. Antifungal Microplate Susceptibility Test
2.5. SNO-MP Synthesis and Nitrosation
2.6. SNO-MP Microplate Susceptibility Test
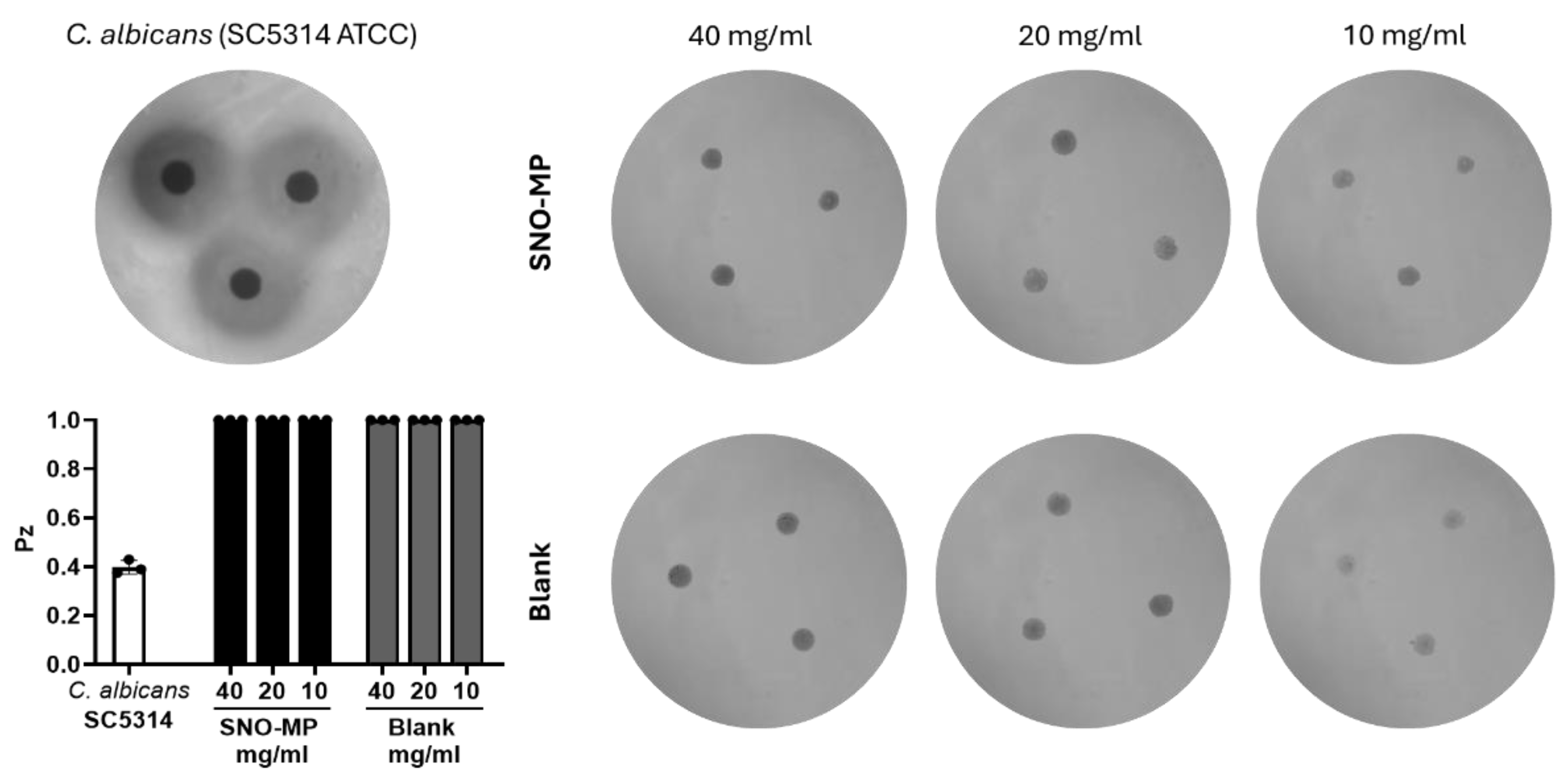
2.7. Hemolysis by SNO-MP
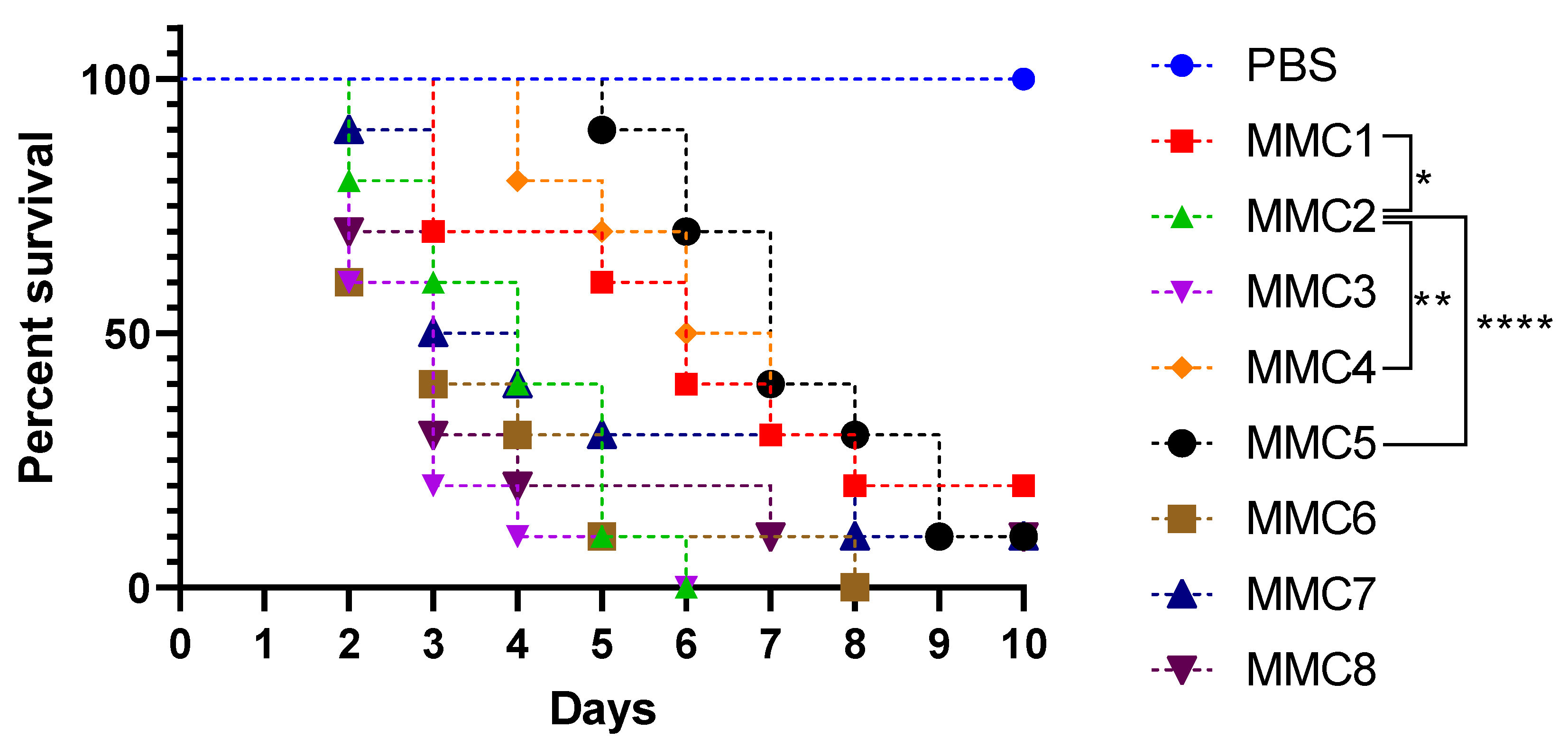
2.8. Galleria Mellonella Infection
3. Results
3.1. Clade Classification
3.2. Antifungal Susceptibility
3.3. Detection of SNPs
3.4. SNO-MP Microplate Susceptibility Test and Hemolytic Activity
3.5. Virulence in Galleria mellonella
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2017, 64, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Spruijtenburg, B.; Badali, H.; Abastabar, M.; Mirhendi, H.; Khodavaisy, S.; Sharifisooraki, J.; Taghizadeh Armaki, M.; de Groot, T.; Meis, J.F. Confirmation of fifth Candida auris clade by whole genome sequencing. Emerg. Microbes Infect. 2022, 11, 2405–2411. [Google Scholar] [CrossRef]
- Suphavilai, C.; Ko, K.K.K.; Lim, K.M.; Tan, M.G.; Boonsimma, P.; Chu, J.J.K.; Goh, S.S.; Rajandran, P.; Lee, L.C.; Tan, K.Y.; et al. Discovery of the sixth Candida auris clade in Singapore. medRxiv 2023. [Google Scholar] [CrossRef]
- Chatterjee, S.; Alampalli, S.V.; Nageshan, R.K.; Chettiar, S.T.; Joshi, S.; Tatu, U.S. Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genomics 2015, 16, 686. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Bing, J.; Hu, T.; Ennis, C.L.; Nobile, C.J.; Huang, G. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020, 16, e1008921. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, J.F.; Gade, L.; Chow, N.A.; Loparev, V.N.; Juieng, P.; Berkow, E.L.; Farrer, R.A.; Litvintseva, A.P.; Cuomo, C.A. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat. Commun. 2018, 9, 5346. [Google Scholar] [CrossRef]
- Chow, N.A.; Muñoz, J.F.; Gade, L.; Berkow, E.L.; Li, X.; Welsh, R.M.; Forsberg, K.; Lockhart, S.R.; Adam, R.; Alanio, A.; et al. Tracing the Evolutionary History and Global Expansion of Candida auris Using Population Genomic Analyses. mBio 2020, 11, e03364-19. [Google Scholar] [CrossRef] [PubMed]
- Maphanga, T.G.; Mpembe, R.S.; Naicker, S.D.; Govender, N.P.; Germs-Sa, F. In Vitro Antifungal Activity of Manogepix and Other Antifungal Agents against South African Candida auris Isolates from Bloodstream Infections. Microbiol. Spectr. 2022, 10, e01717-21. [Google Scholar] [CrossRef]
- Szekely, A.; Borman, A.M.; Johnson, E.M. Candida auris Isolates of the Southern Asian and South African Lineages Exhibit Different Phenotypic and Antifungal Susceptibility Profiles In Vitro. J. Clin. Microbiol. 2019, 57, e02055-18. [Google Scholar] [CrossRef] [PubMed]
- Cleare, L.G.; Li, K.L.; Abuzeid, W.M.; Nacharaju, P.; Friedman, J.M.; Nosanchuk, J.D. NO Candida auris: Nitric Oxide in Nanotherapeutics to Combat Emerging Fungal Pathogen Candida auris. J. Fungi 2020, 6, 85. [Google Scholar] [CrossRef] [PubMed]
- Jeffery-Smith, A.; Taori, S.K.; Schelenz, S.; Jeffery, K.; Johnson, E.M.; Borman, A.; Candida auris Incident Management Team; Manuel, R.; Brown, C.S. Candida auris: A Review of the Literature. Clin. Microbiol. Rev. 2018, 31, e00029-17. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Sharma, C.; Duggal, S.; Agarwal, K.; Prakash, A.; Singh, P.K.; Jain, S.; Kathuria, S.; Randhawa, H.S.; Hagen, F.; et al. New Clonal Strain of Candida auris, Delhi, India. Emerg. Infect. Dis. 2013, 19, 1670–1673. [Google Scholar] [CrossRef] [PubMed]
- Tracking Candida Auris|Candida Auris|Fungal Diseases|CDC. Available online: https://www.cdc.gov/fungal/candida-auris/tracking-c-auris.html (accessed on 14 August 2023).
- Calvo, B.; Melo, A.S.A.; Perozo-Mena, A.; Hernandez, M.; Francisco, E.C.; Hagen, F.; Meis, J.F.; Colombo, A.L. First report of Candida auris in America: Clinical and microbiological aspects of 18 episodes of candidemia. J. Infect. 2016, 73, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.G.; Shin, J.H.; Uh, Y.; Kang, M.G.; Kim, S.H.; Park, K.H.; Jang, H.-C. First Three Reported Cases of Nosocomial Fungemia Caused by Candida auris. J. Clin. Microbiol. 2020, 49, 3139–3142. [Google Scholar] [CrossRef]
- Kathuria, S.; Singh, P.K.; Sharma, C.; Prakash, A.; Masih, A.; Kumar, A.; Meis, J.F.; Chowdhary, A. Multidrug-Resistant Candida auris Misidentified as Candida haemulonii: Characterization by Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry and DNA Sequencing and Its Antifungal Susceptibility Profile Variability by Vitek 2, CLSI Broth Microdilution, and Etest Method. J. Clin. Microbiol. 2015, 53, 1823–1830. [Google Scholar] [CrossRef]
- Chowdhary, A.; Voss, A.; Meis, J.F. Multidrug-resistant Candida auris: ‘new kid on the block’ in hospital-associated infections? J. Hosp. Infect. 2016, 94, 209–212. [Google Scholar] [CrossRef]
- Morales-López, S.E.; Parra-Giraldo, C.M.; Ceballos-Garzón, A.; Martínez, H.P.; Rodríguez, G.J.; Álvarez-Moreno, C.A.; Rodríguez, J.Y. Invasive Infections with Multidrug-Resistant Yeast Candida auris, Colombia. Emerg. Infect. Dis. 2017, 23, 162–164. [Google Scholar] [CrossRef]
- Schwartz, I.S.; Hammond, G.W. First reported case of multidrug-resistant Candida auris in Canada. Can. Commun. Dis. Rep. Releve Mal. Transm. Au Can. 2017, 43, 150–153. [Google Scholar] [CrossRef]
- Vallabhaneni, S. Investigation of the First Seven Reported Cases of Candida auris, a Globally Emerging Invasive, Multidrug-Resistant Fungus—United States, May 2013–August 2016. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 1234–1237. [Google Scholar] [CrossRef] [PubMed]
- Sarma, S.; Kumar, N.; Sharma, S.; Govil, D.; Ali, T.; Mehta, Y.; Rattan, A. Candidemia caused by amphotericin B and Fluconazole resistant Candida auris. Indian J. Med. Microbiol. 2013, 31, 90–91. [Google Scholar] [CrossRef] [PubMed]
- Welsh, R.M.; Bentz, M.L.; Shams, A.; Houston, H.; Lyons, A.; Rose, L.J.; Litvintseva, A.P. Survival, Persistence, and Isolation of the Emerging Multidrug-Resistant Pathogenic Yeast Candida auris on a Plastic Health Care Surface. J. Clin. Microbiol. 2017, 55, 2996–3005. [Google Scholar] [CrossRef] [PubMed]
- Piedrahita, C.T.; Cadnum, J.L.; Jencson, A.L.; Shaikh, A.A.; Ghannoum, M.A.; Donskey, C.J. Environmental Surfaces in Healthcare Facilities are a Potential Source for Transmission of Candida auris and Other Candida Species. Infect. Control Hosp. Epidemiol. 2017, 38, 1107–1109. [Google Scholar] [CrossRef] [PubMed]
- Tsay, S. Notes from the Field: Ongoing Transmission of Candida auris in Health Care Facilities—United States, June 2016–May 2017. MMWR Morb. Mortal. Wkly. Rep. 2017, 66, 514–515. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, S.; Chhillar, A.K.; Sharma, N.; Choudhary, P.; Punia, A.; Balhara, M.; Kaushik, K.; Parmar, V.S. Candida auris and Nosocomial Infection. Curr. Drug Targets 2020, 21, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Sanyaolu, A.; Okorie, C.; Marinkovic, A.; Abbasi, A.F.; Prakash, S.; Mangat, J.; Hosein, Z.; Haider, N.; Chan, J. Candida auris: An Overview of the Emerging Drug-Resistant Fungal Infection. Infect. Chemother. 2022, 54, 236–246. [Google Scholar] [CrossRef]
- Fu, L.; Le, T.; Liu, Z.; Wang, L.; Guo, H.; Yang, J.; Chen, Q.; Hu, J. Different efficacies of common disinfection methods against candida auris and other candida species. J. Infect. Public Health 2020, 13, 730–736. [Google Scholar] [CrossRef]
- Kischkel, B.; Rossi, S.A.; Santos, S.R.; Nosanchuk, J.D.; Travassos, L.R.; Taborda, C.P. Therapies and Vaccines Based on Nanoparticles for the Treatment of Systemic Fungal Infections. Front. Cell. Infect. Microbiol. 2020, 10, 463. [Google Scholar] [CrossRef]
- Nett, J.E.; Andes, D.R. Antifungal Agents: Spectrum of Activity, Pharmacology, and Clinical Indications. Infect. Dis. Clin. N. Am. 2016, 30, 51–83. [Google Scholar] [CrossRef]
- Voltan, A.R.; Quindós, G.; Alarcón, K.P.M.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S.; Chorilli, M. Fungal diseases: Could nanostructured drug delivery systems be a novel paradigm for therapy? Int. J. Nanomedicine 2016, 11, 3715–3730. [Google Scholar] [CrossRef]
- Ahmadi, M.S.; Lee, H.H.; Sanchez, D.A.; Friedman, A.J.; Tar, M.T.; Davies, K.P.; Nosanchuk, J.D.; Martinez, L.R. Sustained Nitric Oxide-Releasing Nanoparticles Induce Cell Death in Candida albicans Yeast and Hyphal Cells, Preventing Biofilm Formation In Vitro and in a Rodent Central Venous Catheter Model. Antimicrob. Agents Chemother. 2016, 60, 2185–2194. [Google Scholar] [CrossRef]
- Macherla, C.; Sanchez, D.; Ahmadi, M.; Vellozzi, E.; Friedman, A.; Nosanchuk, J.; Martinez, L. Nitric Oxide Releasing Nanoparticles for Treatment of Candida Albicans Burn Infections. Front. Microbiol. 2012, 3, 193. [Google Scholar] [CrossRef]
- Costa-Orlandi, C.B.; Martinez, L.R.; Bila, N.M.; Friedman, J.M.; Friedman, A.J.; Mendes-Giannini, M.J.S.; Nosanchuk, J.D. Nitric Oxide-Releasing Nanoparticles Are Similar to Efinaconazole in Their Capacity to Eradicate Trichophyton rubrum Biofilms. Front. Cell. Infect. Microbiol. 2021, 11, 684150. [Google Scholar] [CrossRef] [PubMed]
- Costa-Orlandi, C.B.; Mordorski, B.; Baltazar, L.M.; Mendes-Giannini, M.J.S.; Friedman, J.M.; Nosanchuk, J.D.; Friedman, A.J. Nitric Oxide Releasing Nanoparticles as a Strategy to Improve Current Onychomycosis Treatments. J. Drugs Dermatol. JDD 2018, 17, 717–720. [Google Scholar] [PubMed]
- Biss, M.; Hanna, M.D.; Xiao, W. Isolation of Yeast Nucleic Acids. In Yeast Protocols; Xiao, W., Ed.; Springer: New York, NY, USA, 2014; pp. 15–21. ISBN 978-1-4939-0799-1. [Google Scholar]
- Panel Details|Antimicrobial Resistance Isolate Bank|Antibiotic/Antimicrobial Resistance | CDC. Available online: https://wwwn.cdc.gov/ARIsolateBank/Panel/PanelDetail?ID=2 (accessed on 18 September 2023).
- Narayanan, A.; Selvakumar, P.; Siddharthan, R.; Sanyal, K. ClaID: A Rapid Method of Clade-Level Identification of the Multidrug Resistant Human Fungal Pathogen Candida auris. Microbiol. Spectr. 2022, 10, e0063422. [Google Scholar] [CrossRef]
- M27 Ed4 Broth Dilution Antifungal Susceptibility, Yeasts. (n.d.). Clinical & Laboratory Standards Institute. Available online: https://clsi.org/standards/products/microbiology/documents/m27/ (accessed on 13 July 2023).
- Tar, M.T.; Friedman, J.M.; Draganski, A.; Davies, K.P. Topically delivered nitric oxide acts synergistically with an orally administered PDE5 inhibitor in eliciting an erectile response in a rat model of radical prostatectomy. Int J Impot Res. 2022, 34, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Li, K.L.; Miranda, D.Z.; Cleare, L.G.; Akbar, N.A.; Friedman, J.M.; Draganski, A.; Nosanchuk, J.D.; Abuzeid, W.M. Nitric oxide-generating microparticles: An in vitro evaluation of anti-biofilm efficacy and sinonasal epithelial cell cytotoxicity. Int. Forum Allergy Rhinol. 2022, 13, 954–957. [Google Scholar] [CrossRef] [PubMed]
- WHO Fungal Priority Pathogens List to Guide Research, Development and Public Health Action. Available online: https://www.who.int/publications-detail-redirect/9789240060241 (accessed on 23 May 2023).
- Adams, E.; Quinn, M.; Tsay, S.; Poirot, E.; Chaturvedi, S.; Southwick, K.; Greenko, J.; Fernandez, R.; Kallen, A.; Vallabhaneni, S.; et al. Candida auris in Healthcare Facilities, New York, USA, 2013-2017. Emerg. Infect. Dis. 2018, 24, 1816–1824. [Google Scholar] [CrossRef] [PubMed]
- Welsh, R.M.; Sexton, D.J.; Forsberg, K.; Vallabhaneni, S.; Litvintseva, A. Insights into the Unique Nature of the East Asian Clade of the Emerging Pathogenic Yeast Candida auris. J. Clin. Microbiol. 2019, 57, e00007-19. [Google Scholar] [CrossRef] [PubMed]
- Zamith-Miranda, D.; Heyman, H.M.; Cleare, L.G.; Couvillion, S.P.; Clair, G.C.; Bredeweg, E.L.; Gacser, A.; Nimrichter, L.; Nakayasu, E.S.; Nosanchuk, J.D. Multi-omics Signature of Candida auris, an Emerging and Multidrug-Resistant Pathogen. mSystems 2019, 4, e00257-19. [Google Scholar] [CrossRef] [PubMed]
- Rybak, J.M.; Cuomo, C.A.; Rogers, P.D. The molecular and genetic basis of antifungal resistance in the emerging fungal pathogen Candida auris. Curr Opin Microbiol. 2022, 70, 102208. [Google Scholar] [CrossRef]
- Valdez, A.F.; Zamith-Miranda, D.; Nimrichter, L.; Nosanchuk, J.D. Micro- and nanoparticles as platforms for the treatment of fungal infections: Present and future perspectives. Futur. Microbiol. 2023, 18, 1007–1011. [Google Scholar] [CrossRef] [PubMed]
- Cabrales, P.; Han, G.; Nacharaju, P.; Friedman, A.J.; Friedman, J.M. Reversal of hemoglobin-induced vasoconstriction with sustained release of nitric oxide. American Journal of Physiology. Heart Circ. Physiol. 2011, 300, H49–H56. [Google Scholar] [CrossRef] [PubMed]
- Jani, V.P.; Friedman, J.M.; Cabrales, P. Nitric oxide releasing nanoparticles reduce inflammation in a small animal model of ARDS. Biomed. Pharmacother. 2022, 148, 112705. [Google Scholar] [CrossRef]
- Tar, M.; Cabrales, P.; Navati, M.; Adler, B.; Nacharaju, P.; Friedman, A.J.; Friedman, J.; Davies, K.P. Topically applied NO-releasing nanoparticles can increase intracorporal pressure and elicit spontaneous erections in a rat model of radical prostatectomy. J. Sex. Med. 2014, 11, 2903–2914. [Google Scholar] [CrossRef] [PubMed]
- Rossow, J.; Ostrowsky, B.; Adams, E.; Greenko, J.; McDonald, R.; Vallabhaneni, S.; Forsberg, K.; Perez, S.; Lucas, T.; A Alroy, K.; et al. Factors Associated With Candida auris Colonization and Transmission in Skilled Nursing Facilities With Ventilator Units, New York, 2016–2018. Clin Infect Dis. 2021, 72, e753–e760. [Google Scholar] [CrossRef] [PubMed]
- Honorato, L.; de Araujo, J.F.D.; Ellis, C.C.; Piffer, A.C.; Pereira, Y.; Frases, S.; Araújo, G.R.d.S.; Pontes, B.; Mendes, M.T.; Pereira, M.D.; et al. Extracellular Vesicles Regulate Biofilm Formation and Yeast-to-Hypha Differentiation in Candida albicans. mBio 2022, 13, e0030122. [Google Scholar] [CrossRef] [PubMed]
- Vargas, G.; Rocha, J.D.B.; Oliveira, D.L.; Albuquerque, P.C.; Frases, S.; Santos, S.S.; Nosanchuk, J.D.; Gomes, A.M.O.; Medeiros, L.C.A.S.; Miranda, K.; et al. Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Cell Microbiol. 2015, 17, 389–407. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, J.J.A.; Honorato, L.; de Oliveira, D.F.C.; Gonçalves, R.A.; Guimarães, A.; Miranda, K.; Nimrichter, L. Silver chitosan nanocomposites as a potential treatment for superficial candidiasis. Med. Mycol. 2021, 59, 993–1005. [Google Scholar] [CrossRef]
| Minimal Inhibitory Concentration (μg/mL) | |||
|---|---|---|---|
| Strain | Amphotericin B | Caspofungin | Fluconazole |
| MMC1 | 1 | 0.25 | >256 |
| MMC2 | 0.25 | 0.25 | 4 |
| MMC3 | 1 | 0.25 | >256 |
| MMC4 | 1 | 0.25 | >256 |
| MMC5 | 1 | 0.25 | >256 |
| MMC6 | 1 | 0.25 | >256 |
| MMC7 | 1 | 0.25 | >256 |
| MMC8 | 1 | 0.25 | >256 |
| Minimal Inhibitory Concentration (mg/mL) | |||
|---|---|---|---|
| Strain | SNO-MP | Strain | SNO-MP |
| MMC1 | 20 | MMC5 | 20 |
| MMC2 | 5 | MMC6 | 20 |
| MMC3 | 20 | MMC7 | 20 |
| MMC4 | 20 | MMC8 | 20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valdez, A.F.; Bohner, F.; Goldman, J.P.; Jaquiery, A.B.; Amaral, E.V.C.d.; Correa-Junior, D.; Draganski, A.; Nimrichter, L.; Nosanchuk, J.D.; Zamith-Miranda, D. Candida auris Antifungal Resistance, Virulence and Susceptibility to a Novel Nitric Oxide-Releasing Microparticle and Its Correlations to Clade Identification. Microbiol. Res. 2025, 16, 15. https://doi.org/10.3390/microbiolres16010015
Valdez AF, Bohner F, Goldman JP, Jaquiery AB, Amaral EVCd, Correa-Junior D, Draganski A, Nimrichter L, Nosanchuk JD, Zamith-Miranda D. Candida auris Antifungal Resistance, Virulence and Susceptibility to a Novel Nitric Oxide-Releasing Microparticle and Its Correlations to Clade Identification. Microbiology Research. 2025; 16(1):15. https://doi.org/10.3390/microbiolres16010015
Chicago/Turabian StyleValdez, Alessandro F., Flora Bohner, Joshua P. Goldman, Ali B. Jaquiery, Eduardo V. C. do Amaral, Dario Correa-Junior, Andrew Draganski, Leonardo Nimrichter, Joshua D. Nosanchuk, and Daniel Zamith-Miranda. 2025. "Candida auris Antifungal Resistance, Virulence and Susceptibility to a Novel Nitric Oxide-Releasing Microparticle and Its Correlations to Clade Identification" Microbiology Research 16, no. 1: 15. https://doi.org/10.3390/microbiolres16010015
APA StyleValdez, A. F., Bohner, F., Goldman, J. P., Jaquiery, A. B., Amaral, E. V. C. d., Correa-Junior, D., Draganski, A., Nimrichter, L., Nosanchuk, J. D., & Zamith-Miranda, D. (2025). Candida auris Antifungal Resistance, Virulence and Susceptibility to a Novel Nitric Oxide-Releasing Microparticle and Its Correlations to Clade Identification. Microbiology Research, 16(1), 15. https://doi.org/10.3390/microbiolres16010015








