Abstract
Antimicrobial resistance is one of the most concerning public health issues in Vietnam due to antibiotic-resistant Gram-negative bacteria, including carbapenem-resistant organisms (CROs), which might appear in remote areas in Vietnam. At this center, we performed a cross-sectional study and collected fecal samples from humans (20 healthcare workers (HCWs) and 67 residents) and 175 environmental samples, with rectal and environment swabs, as data for this study, from September 2022 to December 2022. We conducted microbiological testing, including the MELAB Chromogenic CARBA agar plates, blood agar plates, and the BD PhoenixTM Automated Microbiology System to screen, isolate, and identify bacterial species and phenotypic tests for the antimicrobial susceptibility of carbapenem-resistant bacteria and the whole-genome sequencing for 12 CROs chosen to confirm the CRO transmission between HCWs and residents. The study findings showed that the prevalence and risk factors associated with CRO colonization were detected in HCWs, residents, and the environment in the Center of Care and Protection of Orphan Children. CRO transmission happened between HCWs and residents detected with WGS analysis. Whole-genome sequencing (WGS) plays a significant role in CRO control and prevention and reduces CRO transmission/colonization in this center and other healthcare settings.
1. Introduction
Antimicrobial resistance is one of the most concerned public health problems [1] due to antibiotic-resistant bacteria, including Gram-negative bacteria (GNB), which increase morbidity and mortality worldwide [2,3,4].
The rates of antimicrobial-resistance Gram-negative bacteria (GNB), including carbapenem-resistant Enterobacteriaceae (CRE) and carbapenem-resistant organisms (CROs), such as carbapenem-resistant Pseudomonas aeruginosa and carbapenem-resistant Acinetobacter baumannii [5], have increased during the last ten years, causing the difficulty of choosing antibiotics that are available for treatment, which the WHO considers a priority for developing new drugs [6].
Carbapenem-resistant organisms containing the Carbapenemases hydrolyze many β-lactam antibiotics and confer significant antimicrobial resistance, such as the class A KPC enzyme in K. pneumoniae widely in the USA and some endemic parts of Europe, then the class B metallo-β-lactamases commonly in Pseudomonas aeruginosa and the Enterobacteriaceae family worldwide, and, finally, class D OXA-type enzymes in A. baumanii and the Enterobacteriaceae family worldwide, particularly in Europe and North Africa [7].
The mechanisms of carbapenem-resistant Gram-negative bacteria include the loss of porins, the presence of β-lactamases in the periplasmic space, an increase in expressing the transmembrane efflux pump, the presence of antibiotic-modifying enzymes, target site mutation and a decrease in the antibiotics binding to their site of action, ribosomal mutations or modifications, and a mutation in the lipopolysaccharide. These mechanisms cause a reduction in the drug movement through the cell membrane, remove a drug from the bacterium, or make the antibiotics difficult to adhere to their target, etc. [7].
Some studies showed one isolate producing two or more carbapenemases has increased [8,9]. As a result, diagnosing and treating the infections due to this isolate becomes more difficult. The carbapenem-resistant genes appearing on mobile genetic elements allow for dissemination between Gram-negative species [10].
CRE, which belong to Gram-negative bacteria (GNB), often reside in the gastrointestinal tracts of critically ill patients and spread to patients through the healthcare activities of HCWs [11,12]. Next, many studies screened the CRE carriage of carbapenem-resistant K. pneumoniae and Acinetobacter baumannii in patients in ICUs, with a culture of rectal swabs [13,14,15], showing that risk factors of carbapenem-resistant Gram-negative organisms, including CRE and CROs, are related to previous CRE/CRO colonization [16], past antibiotic use (for example, Cephalosporins) [17], past carbapenem use [18,19], past mechanical ventilation use [20], past intensive care unit stay, dialysis, past catheter inserts, past long-term stay in hospital, comorbidities and intubation use [21,22].
In Vietnam, from 2007 to 2013, ten studies were performed in ICUs in hospitals that participated in these studies. The prevalence of carbapenem-resistant A. baumannii increased from 69% (N = 170) to 89% (N = 167), as described in a Table: epidemiology and the prevalence of carbapenem-resistant Gram-negative bacteria in Vietnam [23]. The samples in these studies were collected from patients with infections in the intensive care unit (ICU) of hospitals. So, these infections acquired in the hospitals were due to A. bamannii. One study of the community-acquired pulmonia (CAP) performed in three hospitals and three medical centers in Vinh Long province, South Vietnam, from April 2018 to May 2019, where the prevalence of the CAP caused by A. baumannii was 28.1 (N = 32) [24]. A point prevalence survey at Vietnamese pediatric ICUs showed that the main risk factors for hospital-acquired infection included an age of less than seven months, intubation use, and infection detected at admission [25].
Whole-genome sequencing (WGS) is a clinically effective tool for determining the relatedness of isolates in an outbreak investigation. It combines epidemiology data, for example, hospital admission dates, past surgical operation history, and procedures performed on patients. As a result, it detects transmission events by activities related to prevention and infection control [26]. WGS becomes a valuable tool for comprehending sources of infection and transmission routes of pathogens associated with healthcare activities. It can show which transmission sources are humans (HCWs or patients) or the hospital environment [27].
However, before we performed the study of CRO colonization/transmission at the Center of Care and Protection of Orphan Children, there was no research on CROs and risk factors related to CRO colonization in HCWs or residents (orphans) in the rural region at this center where residents (orphans) have spent nearly the entirety of their lives because they were abandoned children since they were babies. Orphans have lived in this center since they were babies. They have had many chances to contact other orphans and HCWs. Therefore, risk factors and contamination of CROs due to contact with healthcare activities for a long time can happen. For the reasons above, the objectives of this study were
- (1)
- to determine the prevalence of CRO colonization in HCWs, residents, and the environment;
- (2)
- to determine the CRO contamination/transmission between HCWs and residents; and
- (3)
- to show the risk factors of CRO colonization in HCWs and residents in this center.
2. Materials and Methods
2.1. Definitions
CRE: Carbapenem-resistant Enterobacteriaceae (i.e., Escherichia coli, Klebsiella pneumoniae) are resistant to carbapenems, including all carbapenem resistance mechanisms (i.e., production of an AmpC/ESBL plus porin mutations or carbapenemase production).
CPE: Carbapenemase-producing Enterobacteriaceae have a genetic element that codes for carbapenemase production (e.g., NDM, KPC).
CRO: Carbapenem-resistant organisms include all Gram-negative organisms and all resistance mechanisms.
CPO: Carbapenemase-producing organisms include all Gram-negative bacteria that produce a genetically coded carbapenemase [28].
2.2. Design
A cross-section study was implemented in South Vietnam from September 2022 to December 2022 at the Center of Care and Protection of Orphan Children, whose area is about 2000 m2, including a building consisting of three floors in which the first and general floors are sections used as bedrooms and a patient-care section. On the general floor, there is a section used for preparing food in the kitchen section. There is a small chicken and vegetable farming section to supply food for feeding patients, almost children, residing in this center because they are orphans.
The majority of children in this center have a disability related to cerebral palsy; many of the children have experienced complications of immobility and infection, are admitted to local hospitals, and are exposed to broad-spectrum antibiotics. This center has been operating since 2000, and caring for orphan children of the range of ages from 4 to 26 years old. In this study, we proposed the word of residents to replace the word of the orphans, who have lived at this center for two reasons: (1) the range of their age is from 4 to 26 years old, and (2) they have lived at this center since they were abandoned babies.
All 20 healthcare workers (HCWs) and 67 residents, (including children) participated in this study. We observed and determined 303 environmental samples in the Center of Care and Protection of Orphan Children. But we randomly chose some samples in the environment, based on the Yamane formula [29], which is Where n is the sample size required for the study, depending on N. N is the total number of environmental samples observed and determined in our study site, the Center of Care and Protection of Orphan Children, and e = 0.05, allowable error (%). In our study, N was 307. Hence, the sample size of the random environment required for our study was 175 (n).
The number of randomly chosen environmental samples is present in Table S1 (Supplementary Tables).
This study received approval from the Board of Directors on Ethics in Biomedical Research at the Thien-Phuoc-Nhan-Ai Center of Care and Protection of Disabled Children, located in Cu Chi District, a rural area in HCM City, Vietnam, under the approval number 026/2565. The written consent forms were obtained from HCWs and the guardians of children.
Most of the residents at the center have a physical and (or) mental disability. The residents’ guardians participated in interviews to collect information. The HCWs at the Center are Catholic Sisters who provide nursing and personal care (wound care, bathing, feeding, etc.) in 8–10 h shifts and six days a week. All residents and staff of the center participated in this study (20 HCWs and 67 residents (most of them were children).
Exclusion criteria included non-consent, known CRO/CPO colonization or infection during 6 months before this study, or active infection at the conducting study time. However, none of the residents or HCWs fulfilled any exclusion criteria. Hence, all participated in our study.
The participant and laboratory codes were allocated for residents, HCWs, and isolates to track the data related to participants and isolates.
2.3. Sample Collection
Instruments and methods used for fecal collection are as follows:
Fecal collection: Tools used to collect specimens include sampling swabs and the Amies Transport Medium.
The sampling swab is a method to take a sample. After taking a sample, we transported it in the Amies Transport Medium.
The procedure of fecal collection includes the following steps:
- Open the swab package and remove a sampling swab tube from this package.
- Then, hold the tube cap, and remove a cotton swab stick inside. Only touch the cap.
- Do not touch a cotton swab stick. Insert a sterile rectal cotton sampling swab approximately 1 cm into the anal canal.
- Then, rotate slowly for 10 s. Take the amount of feces collected on a cotton swab that is enough to perform the appropriate microbiological tests.
- Next, put the rectal swab specimens into the Amies-Transport Medium.
- Finally, these fecal specimens must be stored in a container with dry ice and transported to the Lab Department of Hung Vuong Hospital as soon as possible within 6 h after collecting rectal swab specimens.
Environmental sample collection:
The tool used to collect the environmental samples is the environmental sampling swab.
Environmental samples are the samples collected from environmental surfaces and furnishings in the center, including the following steps:
- The sterile rectal cotton sampling swab was a tool used to collect the environmental sample.
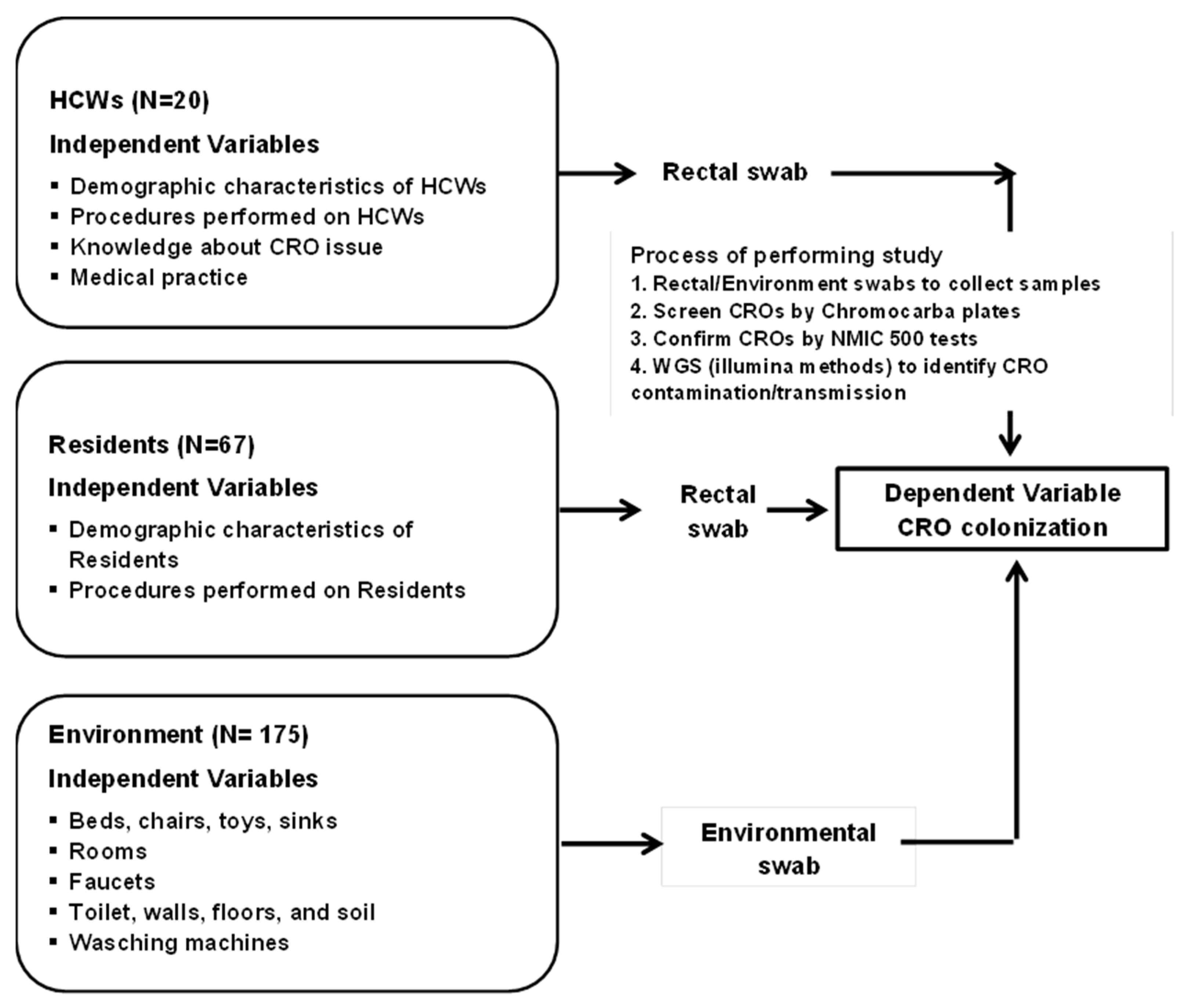
- An approximate area (10 × 10 cm) of environmental surfaces or furnishings was a standard for collecting environmental samples. After that, they were put into the Amies Transport Medium, stored in a container with dry ice, and transported to the Lab Department of Hung Vuong Hospital as soon as possible, within 6 h after sample collection (Figure 1).
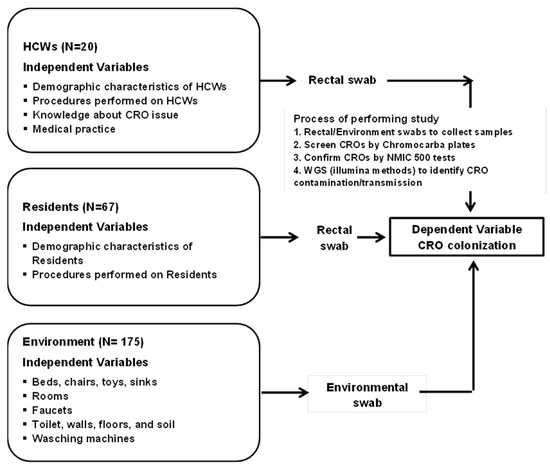 Figure 1. Conceptual framework and process of screening, confirming, and identifying CROs.
Figure 1. Conceptual framework and process of screening, confirming, and identifying CROs.
Demographic characteristics of HCWs/residents are listed, such as age (years), previous antibiotic treatment, chronic disease, previous hospital stay, past surgical intervention, duration of treatment, and professional work (based on number of years and hours per day).
Procedures performed on HCWs/residents: Peripheral IV catheter insertion and urinary catheter insertion.
Knowledge/Attitude and behavior of HCWs in the prevention and control of CRO issues: A questionnaire list evaluates the knowledge/attitude of HCWs before and after training in the prevention, infection control, and medical practice in the prevention and control of CRO colonization.
2.4. Microbiology Methods
The sample swabs were plated onto MELAB Chromogenic CARBA agar plates and incubated at 37 °C. Purity cultures of suspect colonies were performed on blood agar and species identification followed by Card NID, antimicrobial susceptibility testing, and the phenotypic carbapenemase detection by using the NMIC-500 CPO Detect panel of the BD Phoenix TM Automated Microbiology System is a product of the manufacturer of BD Diagnostic Systems, Sparks, MD, USA.
The reference strains in the NMIC 500 test to confirm susceptibilities are E. coli ATCC® 25922, K. pneumoniae ATCC BAA-1705™, and P. aeruginosa ATCC™ 27853.
2.5. Whole-Genome Sequencing (WGS)
We discovered thirty-six CRO isolates in HCWs, residents, and the environment. However, we chose twelve CROs detected from HCWs and residents who had close contact with HCWs (HCWs feed food or bathe residents, etc.). CROs belonged to the same species with a high ratio of identical antibiotic-resistant phenotypes between CRO isolates and the number of CROs chosen to perform WGS available to our financial resources. Hence, we selected twelve CRO isolates from HCWs and residents, as described in Table 1 and Table 2, to determine whether the potential CRO transmission/contamination occurred between HCWs and residents in these chosen CROs. We extracted and measured DNA concentration in Vietnam. Next, we sent the samples to Charles River Laboratories in Australia for sequencing. The information on 12 CROs at Charles River Laboratories in Australia was presented in Part (a) of Table 1.

Table 1.
Process of performing whole-genome sequencing for 12 randomly chosen CROs in 36 CROs detected in our study.

Table 2.
Determination of contamination/transmission with SNPs and OrthoANI values from S67 to S73_isolate.
2.6. At Charles River Laboratories in Australia: Process of Performing WGS
The genomic DNA was quantified with Qubit High-sensitivity assays using a Qubit 4.0 Fluorometer (Thermo Fisher Scientific, Waltham, 168 3rd Ave, MA, USA). The samples were purified using AMPure XP (Beckman Coulter, 250 South Kraemer Boulevard Brea, CA, USA) with a 1 × volume ratio to elute samples with a low EDTA Tris-HCl buffer. Samples were normalized to 50–200 ng and processed using the Illumina DNA Library Prep kit (Illumina Inc., San Diego, CA 92122, USA) according to the manufacturer’s protocols using 5 PCR cycles for indexing. The performance of the quantification and size estimation of the libraries relied on both the Qubit 4.0 Fluorometer (Thermo Fisher Scientific, USA) and the 4200 Tape Station System (Agilent, 5301 Stevens Creek Blvd. Santa Clara, CA 95051, USA). The samples were normalized to 4 nM and then pooled into a new microfuge tube. The pooled library was diluted to 1 nM and sequenced on the MiniSeq Sequencer (2 × 150 bp paired-end reads) (Illumina Inc, San Diego, CA 92122, USA) using a MiniSeq High Output 300 Cycle flow cell.
Antibiograms of 12 CROs are shown in Supplementary Tables.
2.7. Statistical Analysis
After the data were collected and cleaned, we transferred these data to the SPSS software version 23.0 for analysis. Descriptive statistics were employed to describe the independent variables, including the characteristics related to demography in HCWs and residents, antibiotic use, treatment, medical procedures, the length of hospital stay, the number of hospital admission times related to CRO colonization, and chronic diseases, detected in HCWs and residents. Descriptive statistics were also employed to determine the prevalence of CROs in HCWs, residents, the environment, and risk factors associated with CRO colonization. The adjusted odds ratio with a 95% confidence interval was considered significant with statistical significance at p < 0.05.
3. Results
3.1. Results of Whole-Genome Sequencing of 12 CROs Analyzed at Charles River Laboratories in Australia
We sent 12 DNA samples for whole bacterial genome sequencing at Charles River Laboratories in Australia, where our team assembled and annotated the genomes to give provisional (sensu lato) species-level taxonomic classification, as shown in Part (b) in Table 2, and performed the assembly of genomes by using Unicycler v0.4.8 with default settings and a minimum contig size of 300 nt. We performed the annotation of genomes by using the RASTtk tool kit.
Continuously, we conducted genome sequencing for the 6276067, 6276068, 6276070, 6276071, and 6276073_sample because
- (1)
- There might be some infections due to transmission/contamination events within an orphanage.
- (2)
- Antibiotic profiling of these isolates demonstrated high resistance.
Hence, determining the source of this resistance is a crucial issue in our study.
In detail, we used 25 antibiotics to determine the susceptibility or resistance of CROs in our research, as described in Table S2 (Supplementary Tables).
We showed the ratios of identical antibiotic-resistant phenotypes of the isolates with the same species below.
The ratios of identical antibiotic-resistant phenotypes were as follows:
- ▪
- 80% (20/25) between E. coli of C4003 (6276066) and H006 (6276065);
- ▪
- 92% (23/25) between A. baumannii of C3022 (6276070) and H009 (6276073);
- ▪
- 92% (23/25) between A. baumannii of C3011 (6276071) and H009 (6276073);
- ▪
- 100% (25/25) between A. baumannii of C3022 (6276070) and C3011 (6276071).
3.2. Transmission/Contamination Tracking by SNP (Single-Nucleotide Polymorphism) Count
3.2.1. Transmission/Contamination Between the Isolates, Including 6276070, 6276071, and 6276073_Isolate
Firstly, we compared the genomics and performed the MALDI-based typing of five samples (6276067, 6276068, 6276070, 6276071, and 6276073_isolate). The results after performing the WGS for five isolates are described in Part (c) of Table 2.
Secondly, we tracked transmission of the same species, including the 6276067, 6276069, 6276070, 6276071, 6276072, and 6276073_sample, by counting SNPs between samples using the Snippy 4.6.0 and an Acinetobacter nosocomialis M2 reference genome (GCF_005281455.1_ASM528145v1). The number of genomes of Acinetobacter spp., including A. nosocomialis, is approximately 3,940,614.
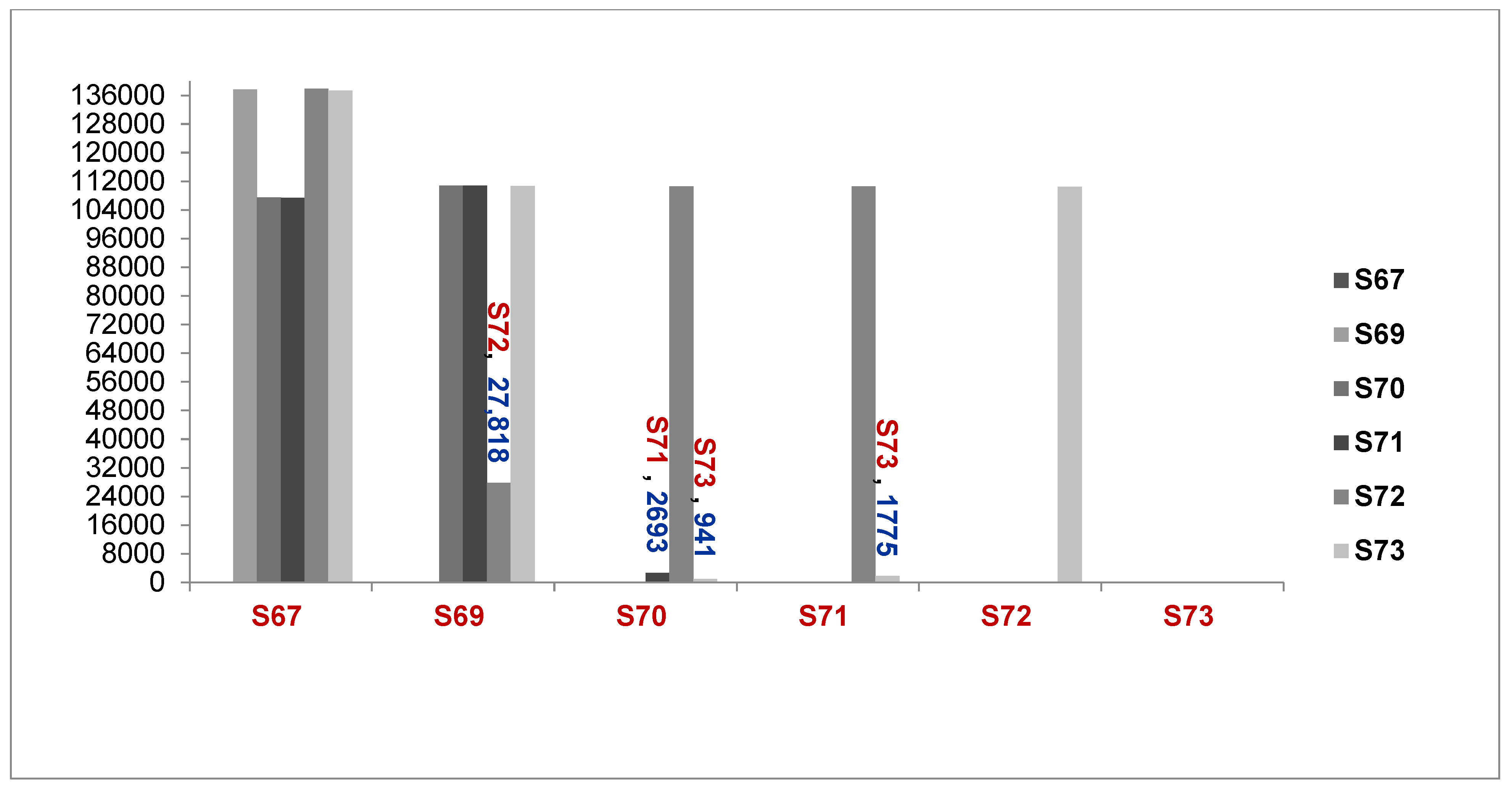
The WGS results showed that the number of SNPs was different between Acinetobacter isolates, including from the 6276067_isolate to the 6276073_isolate, as described in Figure 2 below.
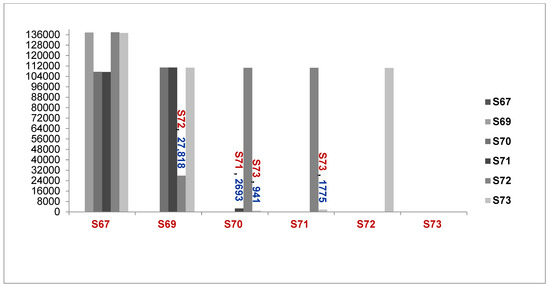
Figure 2.
Comparison of the number of genomes different between samples (S67 to S73, A. baumannii and A. seifertii) (6276067 = S67, 6276069 = S69, 6276070 = S70, 6276071 = S71, 6276072 = S72, 6276073 = S73, A. seifertii, A. baumannii).
Explaination Comparison of Number of Genomes between samples:
1. S67 compared with S69, S70, S71, S72 and S73; 2. S69 compared with S70, S71, S72 and S73; 3. S70 compared with S71, S72 and S73; 4. S71 compared with to S72 and S73; 5 S72 compared with S73.
Following Lee’s study, when the ratio of the ANI (average nucleotide identity) of two or more strains with the same species is over >95%, two or more species belong to an identical clone [30]. The results in Part (a) of Table 2 showed that the genome-difference ratio of two isolates of the same species was less than 5%. Hence, they belonged to an identical strain.
These results in Part (a) of Table 2 confirmed that the 6276067_isolate is A. seifertii, the 6276069_isolate is A. baumannii, the 6276072_isolate is A. baumannii, and 6276073, 6276071, and 6276070_isolate are identical clones (6276073_isolate = H009, 6276071_isolate = C3011, and 6276070_isolate = C3022). The result presented in Part (a) of Table 2 showed that 6276070, 6276071, and 6276073 were closely related between a healthcare worker (H009) and residents (children) (C3022 and C3011) by a contamination event.
To improve the taxonomic accuracy of typing three Acinetobacter spp. isolates, including the 6276071, 6276070, and 6276073_isolate, we calculated the ratio of average nucleotide identity (ANI) of these three isolates by using OrthoANI.
The ratios of the ANI between 6276070, 6276071, 6276073_isolate and A. nosocomialis were over 95% [30], as described in Part (b) of Table 2. In contrast, the ratios of the ANI between the 6276070, 6276071, and 6276073_isolate, compared with A. seifertii and A. baumannii, were less than 95%. So, the 6276070, 6276071, and 6276073_isolate were A. nosocomialis sensu stricto as the ANI is over 95% [30] and belonged to an identical clone. Thus, the 6276070 (C3022), 6276071 (C3011), and 6276073 (H009) were associated with a contamination/transmission event.
Part (b) of Table 2 showed that the 6276073_isolate (H009), which had the ratio of genome identity compared with the 6276071_isolate (C3011), was 99.347% and, compared to the 6276070_isolate (C3022), was 98.8262%. It meant that the 6276073_isolate (H009) had the number of SNPs identical to the 6276071_isolate (C3011) more than the 6276070_isolate (C3022) in the content of their genomes.
3.2.2. Transmission/Contamination Between the Isolates, Including the 6276065, 6276066 Isolates
To determine the transmission/contamination of two E. coli isolates belonging to the 6276065 (H006) isolate and 6276066 (C4003) isolate, we selected more random outgroups of Escherichia coli (FORC_044 and strain_2313) and used K12 isolate as the reference genome for SNP calling. All strains, including the 6276064, 6276065, 6276066, 6276074_isolate, FORC_044, and 2313_strain, have from 40,000 to 80,000 different SNPs pairwise between any two genomes. Hence, they are unrelated isolates.
Next, two isolates (6276065 (H006) and 6276066 (C4003)) had only 16 different SNPs, and the number of genomes of E. coli was about 4,608,319 genomes. The SNP difference between 6276065 and 6276066 was 0.0003% (16/4,608,319). As a result, the ratio of the genomic identity between 6276065 and 6276066 was 99.9997% (100–0.00034%). This result is evidence of a cross-contamination event between H006 and C4003.
The results of WGS showed that there was clear evidence of transmissions/contaminations
- (1)
- between the 6276070 (C3022), 6276071 (C3011), and 6276073 (H009) isolate;
- (2)
- between the 6276065 (H006) and 6276066 (C4003) isolate.
3.3. Prevalence of CROs in HCWs, Residents, and Environment
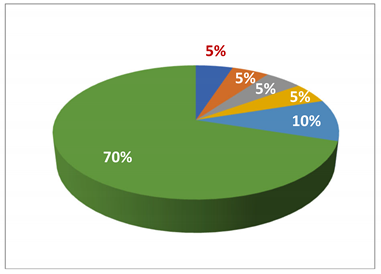
In HCWs, four CREs (one E. cloacae (5%, 1/20), three E. coli isolates (15%, 3/20)), and two CROs (two A. baumannii isolates (10%, 2/20)) were identified in the HCWs, as shown in Table 3.

Table 3.
Prevalence of carbapenem-resistant organisms in Center of Care and Protection of Orphan Children.
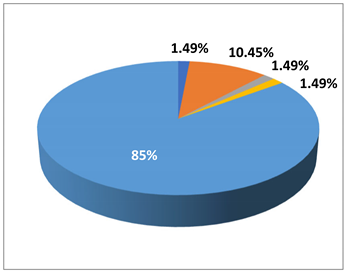
In residents, 11 CRO isolates, in which a resident (child) was colonized, with two CREs (one E. coli (class D) and one K. pneumoniae), were identified in residents as 1.49% (1/67) in E. coli (class D), and K. pneumoniae, 10.45% (7/67) in A. baumannii), 1.49% (1/67) in B. cepacia complex, and 1.49% (1/67) in P. aeruginosa, respectively. The prevalence of CROs in residents was 14.93% (10/67).
In addition, 19 CRO isolates (10.9%, 19/175), including 4.57%, (8/175) in A. baumannii, 2.29%, (4/175) in A. faecalis, 0.57%, (1/175) in P. aeruginosa, 2.29%, (4/175) in P. putida and 1.14%, (2/175) in S. maltophilia, were identified in environmental samples.
3.4. Characteristics (Demographics) of Independent Variables Associated with HCWs and Residents
3.4.1. Demographic Characteristics of HCWs and Residents
The HCWs and residents who participated in this study were 87 (N1 = 20 HCWs and N2 = 67 residents). All HCWs were female 100%, while the ratio of female residents was 26.9%.
For HCWs’ age, an independent variable, with its arrangement, was from 20 to 66 years old (Max = 66, Min = 20). The maximum age of the residents was 26 years old. But almost all HCWs were over or equal to 27 years old, and only one case was 20 years old.
The professions of HCWs included child carer (n = 12, 60%), food provider (2, 10%), registered nurse (2, 10%), pharmacist (1, 5%), and other professions.
The proportion of HCWs with a duration of years for professional work under five years was 50%, and 50% of HCWs have worked for more than five years, while more than 67% of residents have resided for over five years in the Center of Care and Protection of Orphan Children. This result implied that HCWs and residents have lived, worked, and had many opportunities in close contact for a long time.
A total of 100% of HCWs have worked seven days per week, and 90% worked over 8 h per day. This remarkable finding showed that HCWs had much time to contact residents when performing healthcare practice for residents in this healthcare setting. Thus, CRO transmission happened, such as an evitable event, while HCWs cared for and treated patients at this center, as shown in Part (a) in another Table 4.

Table 4.
Characteristics related to demography, antibiotic use, treatment, and medical procedures performed on HCWs and residents.
3.4.2. Demographic Characteristics of HCWs and Residents Related to Antibiotic Use
In many studies, the trend of antibiotic overuse to treat bacterial infections induced bacteria-antibiotic resistance and disseminated the strains of resistant bacteria [2,31]. Previous studies showed the more antibiotic consumption was, the more risk of infection caused by CROs was. [32,33].
When comparing the antibiotic use between HCWs and orphans, we recognized that HCWs consumed antibiotics more significantly than residents (χ2 = 31.0743, p < 0.0001, OR = 23.028571 (95% CI: 6.314, 83.996)), as shown in the Cell 7, Part (b) of Table 4. Next, the number of antibiotic-use times for the last time before the study participation in HCWs from 1 to 10 was 65% (13/20). In contrast, residents only used antibiotics, from one to two times with 7.5% (5/67), as described in the Cell 8, Part (b) in Table 4.
Moreover, the duration of last antibiotic use by HCWs was from 14 to 180 days, before study participation, with 65% (13/20) of HCWs using antibiotics. Most were from 14 to 120 days, with 60% of HCWs (12/20), while the duration of last antibiotic use by residents was from 14 to 90 days, before study participation, with 7.5% (5/67) of patients using the antibiotics, as shown in the Cell 10, Part (b) in Table 4. It implied that the trend of antibiotic use, the duration of antibiotic use, and times for using antibiotics around one year before participating in this research in HCWs were more remarkable than in residents.
Specifically, in our study result, the first antibiotics were amoxicillin plus clavulanate acid or amoxicillin for HCWs. Contrarily, amoxicillin/clavulanate acid and azithromycin were for residents. There was an outstanding difference in the number of antibiotic-use days and the proportion of HCWs and residents using antibiotics to treat infectious diseases (IDs) for the first time. For example, the total number of days of antibiotic use for the first time in HCWs was 75 days compared with 25 days in residents. The number of HCWs using antibiotics from 2 to 30 days was 13 (55%), while that of residents using antibiotics was 5 (7.46%), as shown in Cell 11, Part (b) in Table 4.
Next, HCWs did not use antibiotics for the second time, but, in residents, three cases used antibiotics for five days for each one with 4.48% (3/67), as described in Cell 12, Part (b) of Table 4. When we compared two proportions of not using antibiotics between HCWs and residents, there was no difference in not using antibiotics the second time between HCWs, with 100% (20/20), and orphan children, with 95.52% (64/67), as shown in Cell 14, Section 14, Part (b) in Table 4.
3.4.3. Demographic Characteristics of HCWs and Residents Related to Length of Hospital Stay and Number of Hospital Admission Times
The factors that contributed to the appearance of multi-drug resistant organisms, including carbapenemase-resistant organisms, had to list the number of admission times and the length of hospital stay, as reported in the previous studies [34,35].
Hence, the characteristics should be analyzed as follows: the number of times and days, reasons for hospital admission, and the duration of the last hospital stay. In the Center of Care and Protection of Orphan Children, we recognized that the number of past hospital-admission times outside of the Center in HCWs was one time, with 15% (3/20), as shown in Cell 15, Part (c) of Table 4, but, that in residents was from one to many times, with 16.4% (11/67), as described in Cell 15, Part (c) of Table 4. The reasons for hospital admission in HCWs outside of the Center of Care and Protection of Orphan Children were knee ligament tear, laryngeal fibroids, and pneumonia, with one case for each reason, as shown in Cell 16, Part (c) of Table 4. However, one case of a sacral decubitus ulcer was the reason for the residents. The duration of the last hospitalization outside the Orphanage Care and Protection Center before participating in this study was from 60 to 365 days, occupied by 15% of cases (3/20) in HCWs, while that in residents was 10 days and occupied 1.49% of cases (1/67), as shown in Cell 17, Part (c) of Table 4. The number of days for last admission at hospitals outside the Center of Care and Protection of Orphan Children in HCWs was from 4 to 5 days, occupied by 15% (3/20), while that in residents for ten days was 1.5% (1/67), as shown in Cell 18, Part (c) of Table 4
The characteristics associated with chronic diseases were critically different between HCWs and residents, with χ2 (1, N = 87) = 8.1614, p = 0.004279, OR = 0.2144 (0.0708 to 0.6493). This result suggested the number of cases of chronic diseases in residents more than in HCWs. It also suggested chronic diseases in HCWs, including chronic tonsillitis, laryngeal fibroids, and laryngitis, were distinctive to most chronic diseases in residents: mental and physical disabilities, such as cerebral palsy, epilepsy, Down syndrome, speech impairment, and hyperactivity, as described in Part (d) of Table 4.
Some studies showed that children with cerebral palsy have often experienced a severe neurological impairment associated with a motor impairment. So, these children have not performed daily activities such as eating some food, drinking water, and maintaining personal hygiene [36]. This children group was particularly susceptible to lower respiratory infection [36,37] caused by multi-drug MDROs, including CPO (P. aeruginosa) [38].
3.4.4. Demographic Characteristics of HCWs and Children Related to Procedures Performed on HCWs and Residents
The other factors associated with CRO colonization or infection were the procedures performed in the past, such as past operations, the number of times for past operations, the duration of past operations [39], and invasive urinary or intravenous catheter use [40,41].
The proportions of past operations before participating in this study in HCWs and residents were 5% (N = 20) and 1.50% (N = 67), respectively, as described in Cell 22, Part (e) of Table 4. Similarly, the number of times for past operations last year before participating in this study in HCWs and residents were 5% (N = 20) and 1.50% (N = 67), respectively, as shown in Cell 23, Part (e) of Table 4.
The proportions of peripheral-catheter use before participating in this study of HCWs and residents were 10% (N = 20) and 1.50% (N = 67), as presented in Cell 25, Part (e) in Table 4. The remarkable thing was the urinary catheter procedure but without indication for HCWs and residents in our study. The number of days of using the peripheral intra-catheter in HCWs was from 1 to 5 days, with two cases (10%), but, in residents for 10 days, with one case (1.50%), as shown in Cell 26, Part (e) in Table 4. The urinary catheter use was not performed on HCWs and children, as reported in Cell 27, Part (e) of Table 4.
The number of times for past operations, the number of days, and times of peripheral intra-catheter and urinary catheter use occupied proportions no more than 10% for all characteristics of medical procedures performed on HCWs and residents, as shown in Part (e) of Table 4.
3.5. Risk Factors Associated with CROs in HCWs and Residents
From the results of the WGS of 12 CROs, we showed crucial data, including the following:
- CRO-transmission events between HCWs and residents: one case of E. coli transmission was between an HCW (H006) and one resident, a child (C4003), and another case of A. nosocomialis transmission between an HCW (H009), and two residents (C3022 and (C3011), who were children.
- In twelve CRO isolates detected in 11 participants, we detected two CREs, including one E. coli and one K pneumonia isolate colonized in a child (C4003).
- A total of 11 participants included five HCWs: H006, H007, H008, and H009 working in the first floor and H019 working in both the general and first floor. Five of six residents have resided on the first floor and one (C3002) on the general floor, as described in Table 2 and Table 4. HCWs participating in performing the WGS were the child carers shown in Table S2 (Supplementary Tables).
- The age of all residents was less than 27 years old.
- Residents have lived together in the Center of Care and Protection of Orphan Children for a long time. So, residents have spent a long duration in close contact between themselves and between them and HCWs.
To determine risk factors associated with CRO colonization, we analyzed relevant data as follows:
- The study group included 87 participants (20 HCWs and 67 residents) in a unity group. This data suggested risk factors can occur when HCWs and residents have worked and resided for a long time in the Center of Care and Protection of Orphan Children. Additionally, there have been a lot of occasions for interaction between HCWs and residents during long-term care or treatment. All factors can contribute to developing the risk factors of CRO contamination.
- The CRO colonization was due to the association of two or many characteristics activating together, for instance, an age higher than 27 years old and working or residing in the Center of Care and Protection of Orphan Children for more than four years, etc.
- We divided 87 participants into two sub-groups: one belongs to the HCW sub-group, and another belongs to the resident subgroup, based on a 27-year-old value (Table 5).
 Table 5. Risk factors related to CRO colonization detected in HCWs and children (N = 87, HCWs = 20, residents = 67).
Table 5. Risk factors related to CRO colonization detected in HCWs and children (N = 87, HCWs = 20, residents = 67).
3.5.1. Risk Factors Associated with CROs in a Group, Including HCWs and Residents
Based on the crucial data analyzed above, we detected in the group of 87 that CRO colonization was not significantly different between HCWs (20) and residents (67), between HCWs older than 27 years old compared with the rest of the 87 participants, between HCWs older than 27 years old and using antibiotics compared with the rest of 87 participants, as shown in Cell 1, Cell 2, and Cell 3 in Table 6. However, the CRO colonization in HCWs older than 27 years old and working on the first floor was significantly different compared to residents, with χ2 (1, N = 87) = 7.522, p = 0.006, OR = 6.000 (1.488 to 24.192), as shown in Cell 4 in Table 6.

Table 6.
Risk factors related to CRO colonization detected in children (N = 67).
Next, HCWs older than 27 years old have worked for more than four years in professional work or worked for more than one year at the first floor or worked for more than four years and more than eight hours for healthcare activity per day at the Center of Orphan Children have risk factors colonized with CROs significantly with χ2 (1, N = 87) = 5.025, p = 0.025, OR = 4.156 (1.117 to 15.462), χ2 (1, N = 87) = 7.522, p = 0.006, OR = 6 (1.488 to 24.192), and χ2 (1, N = 87) = 5.025, p = 0.025, OR = 4.156 (1.117 to 15.462), respectively, as shown in Cell 5, 7, and 8 in Table 5.
In particular, as described in Cell 3, Part (a) of Table 4, and Cell 6 of Table 5, child carers (n = 12) had the risk of being colonized with CROs significantly, compared with the rest of the participants in our study, with χ2 (1, N = 87) = 5.025, p = 0.025, OR = 4.156 (1.117 to 15.462).
Moreover, in our study, we detected that child carers who have worked for more than two years at the Center consumed antibiotics more than two times and more than four days each time to treat an infectious disease the last time before participating in our study had the risk of being colonized with CROs with 6.071 compared with the remaining group, with χ2 (1, N = 87) = 9.084, p = 0.003, OR = 6.071 (3.762 to 9.800), as described in Cell 11 in Table 5.
In particular, we discovered that the group of 79 participants, including 67 residents and 12 child carers who worked more than four years at the Center, consumed the antibiotics more than two times and more than four days each time last time to treat the infectious disease before participating in our study and had the risk of being colonized with CROs significantly with χ2 (1, N = 87) = 8.755, p = 0.003, OR = 5.923 (3.608 to 9.723), compared with the remaining group, as shown in Cell 12 in Table 5.
The results shown in Cells 11 and 12 of Table 5 determined that child carers had CRO contamination more than the rest of the participants.
Procedures such as past operation, peripheral intra-catheter, and urinary catheter use before participating in our study were not risk factors associated with CRO colonization, as shown in Cells 9 and 10 of Table 5.
From the analyses and pieces of evidence proven above, the WGS was an effective tool in investigating, estimating, and determining the risk factors associated with CRO colonization and preventing and controlling the contamination and transmission of CROs at the Center of Care and Protection of Orphan Children where we performed this study. Thus, WGS is a valuable tool in controlling and preventing multidrug-resistant organisms, including CROs, in all healthcare settings.
3.5.2. Risk Factors Associated with CROs in Two Different Groups, Including HCWs and Residents
In this case, HCWs and residents belonged to two separate groups, including one for 20 HCWs and another for 67 residents.
If there were only HCWs, we could not analyze the risk factors associated with CRO colonization because the number of HCWs participating in our study was 20 HCWs, which is less than the standard sample size necessary for analyzing the risk factors, which was 30.
If there were only residents to participate in our study, we detected one risk factor associated with CROs in the residents younger than nine years old and residing for less than or equal to seven and a half years in the Center of Care and Protection of Orphan Children, which was significantly different from the rest of group (group = 67), with χ2 (1, N = 67) = 6.385, p = 0.012, OR = 7.714 (1.296 to 45.905), as shown in Cell 13 of Table 6.
Moreover, for antibiotic use and antibiotic consumption lasting more than five days for the first time of infectious disease treatment, the residents younger than nine years old and residing in the Center of Care and Protection of Orphan Children less than seven and a half years had the risk of CRO colonization more than 6.7 and 6.2 times compared to the rest of residents, respectively, as shown in Cell 15 and 16 of Table 6 (8).
In particular, impressively, the residents younger than nine years old and residing at the Center of Care and Protection of Orphan Children for less than seven and a half years, who experienced 2 times of antibiotic use to treat infectious diseases, with more than five days for the first time, had the risk of CRO colonization more than 6.7 times, compared to the rest of residents, as described in Cell 14 of Table 6
Similarly, as the risk factors of CRO colonization detected in the group of 87 participants (20 HCWs and 67 residents), the risk factors of CRO colonization detected in the group of 67 participants, based on the WGS results, were a combination of many factors, such as age, the duration of residence in the Center of Orphan Children, the location of professional work/residence, and the number of times and days of antibiotic use in the first time for infectious disease treatment. So, the analyses above showed that the risk factors were an association of two or many demography and medical procedure characteristics in the group of 87 or 67 participants.
4. Discussion
We detected the evidence of CRO contamination or transmission between E.coli or A.nosocomialis in HCWs and residents with the WGS tool, as shown in Part 3.2: Transmission/Contamination tracking by SNP (single-nucleotide polymorphism) count. It was the first evidence of CRO contamination/transmission detection between HCWs and residents in a healthcare setting in a remote local center in the Cu Chi district in Vietnam.
The previous studies in Vietnam did not show evidence of CRO colonization or transmission between HCWs and patients in the healthcare setting in Vietnam [42]. Even the studies in other countries did not certainly prove that HCWs were a source of CRO transmission because there was no clear evidence to this sensitive issue [43,44,45].
Our WGS results show CRO contamination/transmission in the Center for Care and Protection of Orphan Children. The origin of the problem could happen in caring for residents or might be due to cleaning the environment and even food processing. It was a potential issue in the community near the Center of Care and Protection of Orphan Children.
Hence, to prevent the CRO spread in the Center of Care and Protection of Orphan Children, a hand hygiene program should be consolidated and prepared with alcohol-based hand-rub bottles at the sites appropriate for HCWs, residents, and even visitors contacting, for example, at the entrance of the resident rooms, reception room, kitchen, and the area for residents playing. Alcohol hand-rub bottles at these sites are favorable for using these products and contribute to removing CRO colonization on the hands of HCWs and residents.
The environment cleaning guide is a substantial priority in reducing the environmental CROs and preventing CRO spread in HCWs and residents who have lived in the Center of Care and Protection of Orphan Children.
Additionally, we were concerned about residents living around the Center of Care and Protection of Orphan Children, where CROs were found in HCWs, residents, and the environment and transmitted between HCWs and residents. Based on our study results about CRO colonization at this center, we plan to perform a project to determine the prevalence and evaluate the spread level of CRO colonization in this local community and show the solutions available to control and prevent it. It is the top priority of our group and the community health managers in this community.
The WGS results and data related to 12 CROs, as described in Table 1, performed with WGS, including characteristics, for example, such as age more than 27 years old and working more than four years, or working more than one year, at floor 1, or working more than four years, and working more than 8 h per day as described in Table 5, were the valuable data to analyze risk factors associated with CRO colonization.
Firstly, we confirmed the WGS results were a significant guide to discovering the risk factors related to CRO colonization in a study group, including 20 HCWs, 67 children, and the age of 27 years old, such as a particular cut point to divide this study group into a group belonging to HCWs with ages older or equal to 27 years old and the other group belonging to residents.
Second, when we separately analyzed the resident group to find the risk factors, we detected one factor related to CRO colonization in residents under nine years old and residing less than or equal to seven and a half years in the Center of Care and Protection of Orphan Children. Another risk factor of CRO colonization was detected in residents who were less than 9 years old, resided in the center for less than 7.5 years, consumed antibiotics more than two times, and spent more than 5 days during the first time of antibiotic use. These results suggested that antibiotic consumption and antibiotic consumption for more than five days were potentially related to CRO colonization in residents under nine years old and residing at the Center for less than or equal to seven and a half years.
With the WGS tool in detecting risk factors, in the group of 87 participants (HCWs = 20, residents = 67) and another group (N = 67 residents), we recognized that the risk factors analyzed in the group of 87 showed a significant relationship between residents and HCWs, who had the risk of CRO contamination/colonization more than residents, as shown in Table 5: Risk factors related to CRO colonization detected in HCWs and children (N = 87, HCWs = 20, residents = 67). The risk factors of CRO contamination/colonization detected in the group of 87 participants are reasonable and available to the current medical practice at the Center of Care and Protection of Orphan Children. We suggest that a project of prevention and control of CRO contamination/colonization should be performed in current conditions of this center. The WGS is a beneficial and effective tool for detecting CRO risk factors in our study.
Some studies detected the carriage rate of extended-spectrum β-lactamase–producing Gram-negative bacteria (ESBLs) in HCWs in the ICUs [46], and the prolonged care facilities [47,48] significantly changed from 3.5% to 21.4%.
A study at the US National Institutes of Health Clinical Center from November 2013 to February 2015 showed that healthcare personnel (HCP) or microbiology laboratory staff had a history of regularly close patient contact; they would have a prevalence of ESBL colonization of 4% (15/379), higher than staff without frequent contact history with patients or bacterial specimens, with 2.9% (11/376). However, the difference between the prevalence of ESBL in HCWs and the control group was not statistically significant, with P = 0.55. There were no HCPs colonized with CROs. This result suggested that ESBL can reside on HCWs through close contact between HCWs and patients. It also implied that HCWs are “victims” of ESBL colonization during healthcare for ESBL colonized patients [49].
In Vietnam, a prospective study investigated characteristics of antibiotic resistance of intestinal Gram-negative bacteria (GNB) detected in HCWs at the adult ICU at the Hospital for Tropical Diseases (HTD) for two months at the beginning of October 28, 2019. The study results showed that, among 40 HCWs participating in this study, the prevalence of HCWs carrying ESBL/AmpC β-lactamase-producing Escherichia coli was 65% (26/40), and only one HCW colonized with Acinetobacter baumannii. A total of 25% of HCWs (N = 40) were ESBL-persistent and frequent carriers [42]. Our study results showed that CRO colonization appeared at the remote Centers of Care and Protection of Orphan Children, and multi-drug-resistant organisms (MDROs), such as ESBL-producing organisms, were detected in HCWs in other studies, including the studies in Vietnam. Hence, it suggested that CRO colonization or multi-drug resistant organisms can appear in HCWs in Vietnam. An essential project should be conducted at the Central and Local Hospitals in Vietnam to screen and determine the prevalence, risk factors, and CRO colonization in HCWs at hospitals and prevent CRO colonization/transmission between HCWs and patients, including children.
5. Conclusions
Our study showed that CROs appeared in HCWs, residents, and the environment in the Center of Care and Protection of Orphan Children. Risk factors of CRO colonization detected in HCWs were associated with an age of more than 27 years, professional working time of more than four years or working site at floor 1, and working time over or equal to 8 h per day, etc. Moreover, CRO transmission could have happened in our study because we detected CRE in an HCW and a child (E. coli) or CROs (A. baumannii) in an HCW and two children. So, whole-genome sequencing will be an effective and beneficial tool to determine CRO contamination/transmission and risk factors associated with CRO colonization. A hand hygiene program and a clear and effective plan will be the best choices to prevent and control CRO colonization/transmission in this center.
6. Limitation of Study
This study was performed at one center of care and protection in a local district in South Vietnam with 87 participants. The study results could show a part of the problem of CRO colonization/transmission in healthcare settings for orphan children. However, it was the first detection of CRO colonization and even CRO transmission in a local healthcare setting in Vietnam. Our study contributes to determining the microbiological characteristics related to pathogenic CRO and is a good reference for future studies about CRO colonization in healthcare settings in Vietnam.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/microbiolres16010028/s1, Table S1: Distribution of environmental samples, number of random collected environmental samples, and the number and the ratio of CROs detected; Table S2: Antibiograms of 12 CROs suspected with close relations based on the identical rate of antibiotic-resistant phylotype/BD; and Table S2 (cont): Antibiograms of 12 CROs suspected with close relations based on the identical rate of antibiotic-resistant phylotype/BD.
Author Contributions
Conceptual framework, methodology, design, data acquisition, analyses, and interpretation of study findings, V.K.N.; participating in performing the antimicrobial susceptibility testing and phenotypic carbapenemase detection using the NMIC-500 CPO Detect panel of the BD Phoenix TM Automated Microbiology System, H.P.P.N.; review, editing, supervision, and funding acquisition, P.T.N., P.N., E.A. and K.I.; preparing and performing WGS for 12 DNA of 12 CRO isolates, L.C. All authors have read and agreed to the published version of the manuscript.
Funding
The authors gratefully acknowledge the financial support “Invitation Research” provided by the Faculty of Public Health, Thammasat University, the Thammasat University Research Unit in Modern Microbiology and Public Health Genomics. Specially, Prof Eugene Athan supported us with funding to perform WGS for our study.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Board of Directors on Ethics in Biomedical Research at THIEN PHUOC NHAN AI CENTER OF CARE AND PROTECTION OF DISABLED CHILDREN (protocol code 026/2565 and date of approval: HCM City, 19 April 2021.
Informed Consent Statement
Written informed consent was obtained from 20 healthcare workers and 67 residents involved in this study to publish this paper.
Data Availability Statement
Data and materials are available on request.
Acknowledgments
The authors sincerely thank HCWs and children for their participation in our study. We gratefully acknowledge the Director and Vice Director of Hung Vuong Hospital, Hoang Thi Diem Tuyet, and Phan Thi Hang, respectively, for their support. We thank Pham Nguyen Huu Phuc, Tran Dang Thang, Cao Thang Long, Nguyen Thi Tu Tam, Nguyen Thi Hien, and Ta Qui Tan for their support and help in conducting the microbiology tests at the Microbiology Unit of Hung Vuong Hospital. We also thank Tran Van Hung, Lam Kim Dung, and Hai Yen for supporting our project. The authors gratefully acknowledge the financial support “Invitation Research” provided by the Faculty of Public Health, Thammasat University, the Thammasat University Research Unit in Modern Microbiology and Public Health Genomics. Specially, Eugene Athan supported us with funding to perform WGS for our study.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations and Acronyms
| 1. Abbreviations of antibiotics | ||
| AK: Amikacin; | CRO: Ceftriaxone; | NOR: Norfloxacin; |
| AM: Ampicillin; | CXM: Cefuroxime; | TZP: Piperacillin/tazobactam; |
| AMC: Amoxicillin-clavulanic acid; | CIP: Ciprofloxacin, | TGC: Tigecylin; |
| AZM: Aztreonam; | CST: Colistin; | SXT: Trimethoprim-sulphamethoxazole; |
| CZ: Cefazolin; | FO: Fosfomycin; | I: Intermediate; |
| FEP: Cefepime; | GEN: Gentamicin; | R: Resistance; |
| FOX: Cefoxitin; | LVX: Levofloxacin; | X: MICs of antibiotics are in the range of sensitive or intermediate; |
| CAZ: Ceftazidime; | MIN: Minocycline; | N: is an antibiotic not recommended for the treatment of infections; |
| CZA: Ceftazidime/avibactam; | NFN: Nitrofurantoin. | |
| 2. Abbreviations related to study | ||
| ANI: Average nucleotide identity | B. cepacia: Burkholderia cepacia | |
| CPE: Carbapenemase-producing Enterobacteriaceae; | A. baumannii: Acinetobacter baumannii: | |
| CPO: Carbapenemase-producing organisms; | A. faecalis: Alcaligenes faecalis; | |
| CRE: Carbapenem-resistant Enterobacteriaceae; | E. cloacae: Enterobacter cloacae; | |
| CRO: Carbapenem-resistant organisms; | E. coli: Escherichia coli; | |
| ESBL: Extended-spectrum β-lactamase; | K. pneumoniae: Klebsiella pneumoniae | |
| HCW: Healthcare Workers; | P. aeruginosa: Pseudomonas aeruginosa ; | |
| HCP: Healthcare personnel; | P. putida: Pseudomonas putida | |
| NICU: Neonatal Intensive Care Unit; | S. maltophilia: Stenotrophomonas maltophilia; | |
| PICU: Pediatric Intensive Care Unit | ||
| SICU: Surgical Intensive Care Unit | ||
| SNP: Single nucleotide polymorphism; | ||
| WGS: Whole-Genome Sequencing | ||
References
- WHO. Antimicrobial Resistance Global Report on Surveillance: 2014 Summary. 2014. Available online: https://www.who.int/publications/i/item/WHO-HSE-PED-AIP-2014.2 (accessed on 23 November 2024).
- CDC. ANTIBIOTIC RESISTANCE THREATS in the United States, 2013. 2013. Available online: https://www.cdc.gov/antimicrobial-resistance/media/pdfs/ar-threats-2013-508.pdf (accessed on 23 November 2024).
- Public Health Agency of Canada. Antimicrobial resistance and use in Canada: A federal framework for action. Can. Commun. Dis. Rep. 2014, 40, 2–5. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Antimicrobial Resistance Surveillance in Europe 2015; Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net); ECDC: Stockholm, Sweden, 2017; Available online: https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2015 (accessed on 23 November 2024).
- Nordmann, P.; Poirel, L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019, 69, S521–S528. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; Hooper, D.C. Hospital-acquired infections due to gram-negative bacteria. New Engl. J. Med. 2010, 362, 1804–1813. [Google Scholar] [CrossRef] [PubMed]
- Giakkoupi, P.; Papagiannitsis, C.C.; Miriagou, V.; Pappa, O.; Polemis, M.; Tryfinopoulou, K.; Tzouvelekis, L.S.; Vatopoulos, A.C. An update of the evolving epidemic of blaKPC-2-carrying Klebsiella pneumoniae in Greece (2009-10). J. Antimicrob. Chemother. 2011, 66, 1510–1513. [Google Scholar] [CrossRef]
- Porres-Osante, N.; Azcona-Gutiérrez, J.M.; Rojo-Bezares, B.; Undabeitia, E.; Torres, C.; Sáenz, Y. Emergence of a multiresistant KPC-3 and VIM-1 carbapenemase-producing Escherichia coli strain in Spain. J. Antimicrob. Chemother. 2014, 69, 1792–1795. [Google Scholar] [CrossRef]
- Reyes, J.A.; Melano, R.; Cárdenas, P.A.; Trueba, G. Mobile genetic elements associated with carbapenemase genes in South American Enterobacterales. Braz. J. Infect. Dis. 2020, 24, 231–238. [Google Scholar] [CrossRef]
- Harris, A.D.; Kotetishvili, M.; Shurland, S.; Johnson, J.A.; Morris, J.G.; Nemoy, L.L.; Johnson, J.K. How important is patient-to-patient transmission in extended-spectrum beta-lactamase Escherichia coli acquisition. Am. J. Infect. Control 2007, 35, 97–101. [Google Scholar] [CrossRef]
- Harris, A.D.; Perencevich, E.N.; Johnson, J.K.; Paterson, D.L.; Morris, J.G.; Strauss, S.M.; Johnson, J.A. Patient-to-patient transmission is important in extended-spectrum beta-lactamase-producing Klebsiella pneumoniae acquisition. Clin. Infect. Dis. 2007, 45, 1347–1350. [Google Scholar] [CrossRef]
- Adler, A.; Navon-Venezia, S.; Moran-Gilad, J.; Marcos, E.; Schwartz, D.; Carmeli, Y. Laboratory and clinical evaluation of screening agar plates for detection of carbapenem-resistant Enterobacteriaceae from surveillance rectal swabs. J. Clin. Microbiol. 2011, 49, 2239–2242. [Google Scholar] [CrossRef]
- Kochar, S.; Sheard, T.; Sharma, R.; Hui, A.; Tolentino, E.; Allen, G.; Landman, D.; Bratu, S.; Augenbraun, M.; Quale, J. Success of an infection control program to reduce the spread of carbapenem-resistant Klebsiella pneumoniae. Infect. Control Hosp. Epidemiol. 2009, 30, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Enfield, K.B.; Huq, N.N.; Gosseling, M.F.; Low, D.J.; Hazen, K.C.; Toney, D.M.; Slitt, G.; Zapata, H.J.; Cox, H.L.; Lewis, J.D.; et al. Control of simultaneous outbreaks of carbapenemase-producing enterobacteriaceae and extensively drug-resistant Acinetobacter baumannii infection in an intensive care unit using interventions promoted in the Centers for Disease Control and Prevention 2012 carbapenemase-resistant Enterobacteriaceae Toolkit. Infect. Control Hosp. Epidemiol. 2014, 35, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhou, M.; Yan, Q.; Jian, Z.; Liu, W.; Li, H. Risk factors for carbapenem-resistant Enterobacterales infection among hospitalized patients with previous colonization. J. Clin. Lab. Anal. 2022, 36, e24715. [Google Scholar] [CrossRef]
- Kaye, K.S.; Cosgrove, S.; Harris, A.; Eliopoulos, G.M.; Carmeli, Y. Risk factors for emergence of resistance to broad-spectrum cephalosporins among Enterobacter spp. Antimicrob. Agents Chemother. 2001, 45, 2628–2630. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.M.; Yuan, Z.; Zhou, H.Y. Risk factors for carbapenem-resistant Klebsiella pneumoniae infection relative to two types of control patients: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 2020, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Harris, A.D.; Smith, D.; Johnson, J.A.; Bradham, D.D.; Roghmann, M.C. Risk factors for imipenem-resistant Pseudomonas aeruginosa among hospitalized patients. Clin. Infect. Dis. 2002, 34, 340–345. [Google Scholar] [CrossRef]
- Dizbay, M.; Tunccan, O.G.; Sezer, B.E.; Hizel, K. Nosocomial imipenem-resistant Acinetobacter baumannii infections: Epidemiology and risk factors. Scand. J. Infect. Dis. 2010, 42, 741–746. [Google Scholar] [CrossRef]
- Yigit, H.; Queenan, A.M.; Anderson, G.J.; Domenech-Sanchez, A.; Biddle, J.W.; Steward, C.D.; Alberti, S.; Bush, K.; Tenover, F.C. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001, 45, 1151–1161. [Google Scholar] [CrossRef]
- Palacios-Baena, Z.R.; Giannella, M.; Manissero, D.; Rodríguez-Baño, J.; Viale, P.; Lopes, S.; Wilson, K.; McCool, R.; Longshaw, C. Risk factors for carbapenem-resistant Gram-negative bacterial infections: A systematic review. Clin. Microbiol. Infect. 2021, 27, 228–235. [Google Scholar] [CrossRef]
- Hsu, L.Y.; Apisarnthanarak, A.; Khan, E.; Suwantarat, N.; Ghafur, A.; Tambyah, P.A. Carbapenem-Resistant Acinetobacter baumannii and Enterobacteriaceae in South and Southeast Asia. Clin. Microbiol. Rev. 2017, 30, 1–22. [Google Scholar] [CrossRef]
- Tran, H.D.; Bach Nguyen, Y.T.; Thanh Tran, T.; Thu Le, T.T.; Thu Nguyen, H.T.; Minh Nguyen, C.; Bach Le, H.T.; Ngoc Phan, T.T.; Thanh Vo, T.T.; Ngoc Bui, H.T.; et al. Community-acquired pneumonia-causing bacteria and antibiotic resistance rate among Vietnamese patients: A cross-sectional study. Medicine 2022, 101, e30458. [Google Scholar] [CrossRef]
- Le, N.K.; Hf, W.; Vu, P.D.; Khu, D.T.K.; Le, H.T.; Hoang, B.T.N.; Vo, V.T.; Lam, Y.M.; Vu, D.T.V.; Nguyen, T.H.; et al. High prevalence of hospital-acquired infections caused by gram-negative carbapenem resistant strains in Vietnamese pediatric ICUs: A multi-centre point prevalence survey. Medicine 2016, 95, e4099. [Google Scholar] [CrossRef] [PubMed]
- Price, J.R.; Didelot, X.; Crook, D.W.; Llewelyn, M.J.; Paul, J. Whole genome sequencing in the prevention and control of Staphylococcus aureus infection. J. Hosp. Infect. 2013, 83, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Eyre, D.W. Infection prevention and control insights from a decade of pathogen whole-genome sequencing. J. Hosp. Infect. 2022, 122, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Gniadek, T.J.; Carroll, K.C.; Simner, P.J. Carbapenem-Resistant Non-Glucose-Fermenting Gram-Negative Bacilli: The Missing Piece to the Puzzle. J. Clin. Microbiol. 2016, 54, 1700–1710. [Google Scholar] [CrossRef]
- Israel, G.D. Determining Sample Size; Institute of Food and Agricultural Sciences (IFAS), University of Florida: Gainesville, FL, USA, 1992; Available online: https://www.gjimt.ac.in/web/wp-content/uploads/2017/10/2_Glenn-D.-Israel_Determining-Sample-Size.pdf (accessed on 8 December 2024).
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Read, A.F.; Woods, R.J. Antibiotic resistance management. Evol. Med. Public Health 2014, 2014, 147. [Google Scholar] [CrossRef]
- Sedláková, M.H.; Urbánek, K.; Vojtová, V.; Suchánková, H.; Imwensi, P.; Kolář, M. Antibiotic consumption and its influence on the resistance in Enterobacteriaceae. BMC Res. Notes 2014, 7, 454. [Google Scholar] [CrossRef]
- Lee, H.S.; Loh, Y.X.; Lee, J.J.; Liu, C.S.; Chu, C. Antimicrobial consumption and resistance in five Gram-negative bacterial species in a hospital from 2003 to 2011. J. Microbiol. Immunol. Infect. 2015, 48, 647–654. [Google Scholar] [CrossRef]
- Moghnieh, R.; Abdallah, D.; Jadayel, M.; Zorkot, W.; El Masri, H.; Dib, M.J.; Omar, T.; Sinno, L.; Lakkis, R.; Jisr, T. Epidemiology, risk factors, and prediction score of carbapenem resistance among inpatients colonized or infected with 3rd generation cephalosporin resistant Enterobacterales. Sci. Rep. 2021, 11, 14757. [Google Scholar] [CrossRef]
- Wilson, G.M.; Suda, K.J.; Fitzpatrick, M.A.; Bartle, B.; Pfeiffer, C.D.; Jones, M.; Rubin, M.A.; Perencevich, E.; Evans, M.; Evans, C.T. Risk Factors Associated with Carbapenemase-Producing Carbapenem-Resistant Enterobacteriaceae Positive Cultures in a Cohort of US Veterans. Clin. Infect. Dis. 2021, 73, 1370–1378. [Google Scholar] [CrossRef]
- Lin, J.L.; Van Haren, K.; Rigdon, J.; Saynina, O.; Song, H.; Buu, M.C.; Thakur, Y.; Srinivas, N.; Asch, S.M.; Sanders, L.M. Pneumonia Prevention Strategies for Children with Neurologic Impairment. Pediatrics 2019, 144, e20190543. [Google Scholar] [CrossRef]
- Allen, J.; Brenner, M.; Hauer, J.; Molloy, E.; McDonald, D. Severe Neurological Impairment: A delphi consensus-based definition. Eur. J. Paediatr. Neurol. 2020, 29, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Çokyaman, T.; Kasap, T.; Çelik, T. Accompanying Infections in Hospitalized Children with Neurological Disease. J. Curr. Pediatr. 2022, 20, 17–26. [Google Scholar] [CrossRef]
- Schechner, V.; Kotlovsky, T.; Kazma, M.; Mishali, H.; Schwartz, D.; Navon-Venezia, S.; Schwaber, M.J.; Carmeli, Y. Asymptomatic rectal carriage of blaKPC producing carbapenem-resistant Enterobacteriaceae: Who is prone to become clinically infected? Clin. Microbiol. Infect. 2013, 19, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Correa, L.; Martino, M.D.; Siqueira, I.; Pasternak, J.; Gales, A.C.; Silva, C.V.; Camargo, T.Z.; Scherer, P.F.; Marra, A.R. A hospital-based matched case-control study to identify clinical outcome and risk factors associated with carbapenem-resistant Klebsiella pneumoniae infection. BMC Infect. Dis. 2013, 13, 80. [Google Scholar] [CrossRef]
- Feldman, N.; Adler, A.; Molshatzki, N.; Navon-Venezia, S.; Khabra, E.; Cohen, D.; Carmeli, Y. Gastrointestinal colonization by KPC-producing Klebsiella pneumoniae following hospital discharge: Duration of carriage and risk factors for persistent carriage. Clin. Microbiol. Infect. 2013, 19, E190–E196. [Google Scholar] [CrossRef]
- Duong, B.T.; Duong, M.C.; Campbell, J.; Nguyen, V.M.H.; Nguyen, H.H.; Bui, T.B.H.; Nguyen, V.V.C.; McLaws, M.L. Antibiotic-Resistant Gram-negative Bacteria Carriage in Healthcare Workers Working in an Intensive Care Unit. Infect. Chemother. 2021, 53, 546–552. [Google Scholar] [CrossRef]
- Majumdar, S.; Kirby, A.; Berry, N.; Williams, C.; Hassan, I.; Eddleston, J.; Burnie, J.P. An outbreak of imipenem-resistant Pseudomonas aeruginosa in an intensive care unit. J. Hosp. Infect. 2004, 58, 160–161. [Google Scholar] [CrossRef]
- Ambrogi, V.; Cavalié, L.; Mantion, B.; Ghiglia, M.J.; Cointault, O.; Dubois, D.; Prère, M.F.; Levitzki, N.; Kamar, N.; Malavaud, S. Transmission of metallo-β-lactamase-producing Pseudomonas aeruginosa in a nephrology-transplant intensive care unit with potential link to the environment. J. Hosp. Infect. 2016, 92, 27–29. [Google Scholar] [CrossRef]
- Landelle, C.; Legrand, P.; Lesprit, P.; Cizeau, F.; Ducellier, D.; Gouot, C.; Bréhaut, P.; Soing-Altrach, S.; Girou, E.; Brun-Buisson, C. Protracted outbreak of multidrug-resistant Acinetobacter baumannii after intercontinental transfer of colonized patients. Infect. Control Hosp. Epidemiol. 2013, 34, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Abdel Rahman, A.T.; Hafez, S.F.; Abdelhakam, S.M.; Ali-Eldin, Z.A.; Esmat, I.M.; Elsayed, M.S.; Aboul-Fotouh, A. Antimicrobial resistant bacteria among health care workers in intensive care units at Ain Shams University Hospitals. J. Egypt. Soc. Parasitol. 2010, 40, 71–83. [Google Scholar] [PubMed]
- March, A.; Aschbacher, R.; Pagani, E.; Sleghel, F.; Soelva, G.; Hopkins, K.L.; Doumith, M.; Innocenti, P.; Burth, J.; Piazzani, F.; et al. Changes in colonization of residents and staff of a long-term care facility and an adjacent acute-care hospital geriatric unit by multidrug-resistant bacteria over a four-year period. Scand. J. Infect. Dis. 2014, 46, 114–122. [Google Scholar] [CrossRef]
- Adler, A.; Baraniak, A.; Izdebski, R.; Fiett, J.; Salvia, A.; Samso, J.V.; Lawrence, C.; Solomon, J.; Paul, M.; Lerman, Y.; et al. A multinational study of colonization with extended spectrum β-lactamase-producing Enterobacteriaceae in healthcare personnel and family members of carrier patients hospitalized in rehabilitation centres. Clin. Microbiol. Infect. 2014, 20, O516–O523. [Google Scholar] [CrossRef] [PubMed]
- Decker, B.K.; Lau, A.F.; Dekker, J.P.; Spalding, C.D.; Sinaii, N.; Conlan, S.; Henderson, D.K.; Segre, J.A.; Frank, K.M.; Palmore, T.N. Healthcare personnel intestinal colonization with multidrug-resistant organisms. Clin. Microbiol. Infect. 2018, 24, e81–e82. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).