New Variants in the Chloroplast Genome Sequence of Two Colombian Individuals of the Cedar Timber Species (Cedrela odorata L.), Using Long-Read Oxford Nanopore Technology
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and High Molecular Weight (HMW) DNA Extraction
2.2. MinION Sequencing
2.3. Chloroplast Read Extraction and Assembly
2.4. Post-Assembly Polishing and Characterization
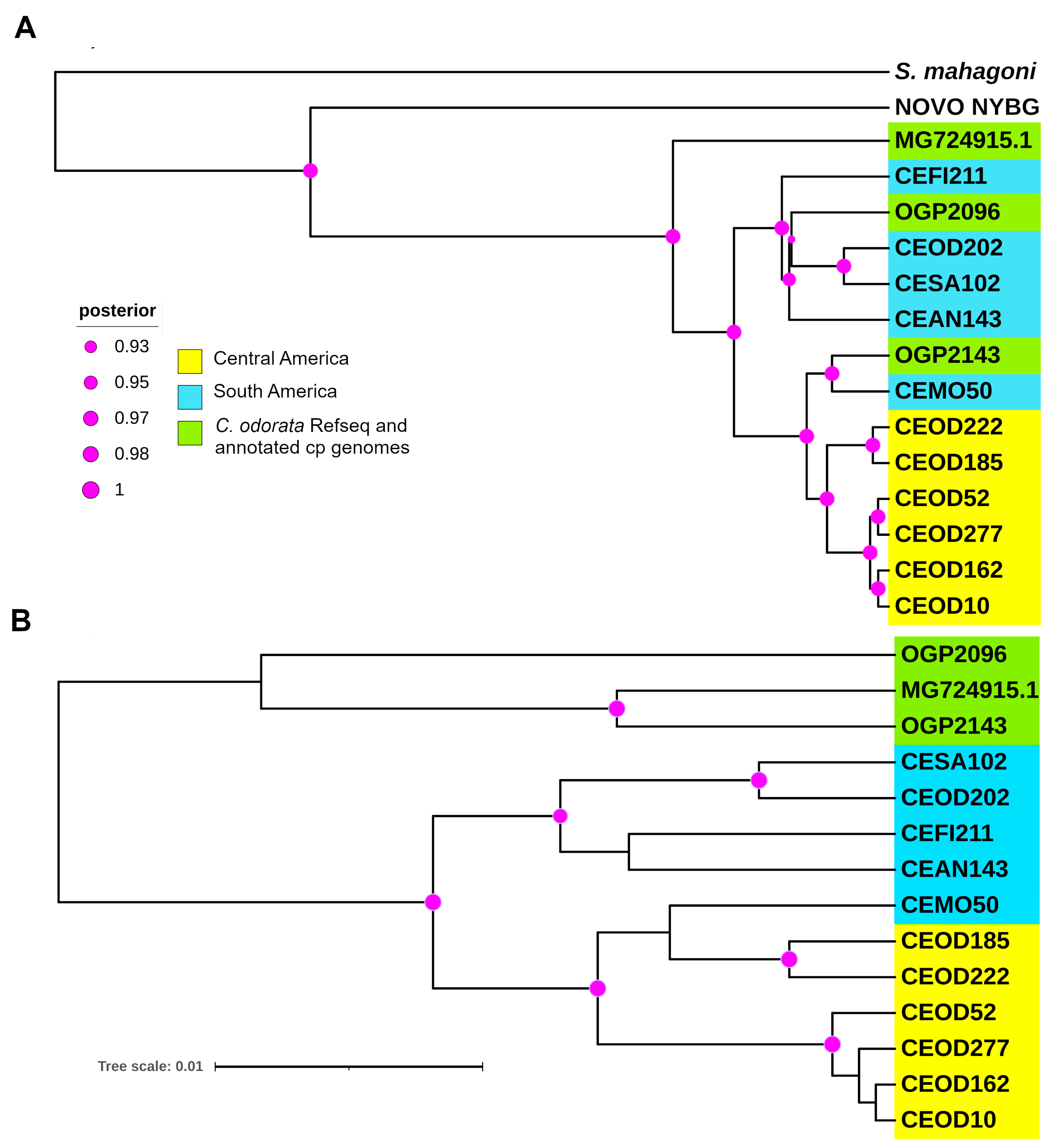
2.5. Variant Discovery and Phylogenetic Analysis
3. Results
3.1. MinION-ONT Whole Genome Sequencing
3.2. Chloroplast Genome Assembly
3.3. Genome Annotation
3.4. Phylogenetics and Structural Variation Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mark, J.; Rivers, M.C. Cedrela odorata, Spanish Cedar. IUCN Red List Threatened Species. 2017, p. e.T32292A68080590. Available online: https://www.iucnredlist.org/species/32292/68080590 (accessed on 1 June 2024).
- Cárdenas, D.; Castaño, N.; Tunjuano, S.; Quintero, L. Plan de Manejo Para La Conservación de Abarco, Caoba, Cedro, Palorosa y Canelo de Los Andaquíes; Instituto Amazónico de Investigaciones Científicas—SINCHI, Ed.; Instituto Amazónico de Investigaciones Científicas—SINCHI: Bogota, Colombia, 2015; ISBN 9789588317878. [Google Scholar]
- Franco, N.; Clavijo, C.; Rojas, J.; Talero, C. Plan de Manejo y Conservación Del Cedro (Cedrela odorata L.) Para La Jurisdicción de La Corporación Autónoma Regional de Cundinamarca CAR; Instituto Amazónico de Investigaciones Científicas—SINCHI: Bogotá, Colombia, 2019. [Google Scholar]
- Molinares, C.; Prada, E.; León, E. Condenando el Bosque: Ilegalidad y Falta de Governanza en la Amazonia Colombiana; Environmental Investigation Agency: London, UK, 2019. [Google Scholar]
- Shaw, J.; Shafer, H.L.; Rayne Leonard, O.; Kovach, M.J.; Schorr, M.; Morris, A.B. Chloroplast DNA Sequence Utility for the Lowest Phylogenetic and Phylogeographic Inferences in Angiosperms: The Tortoise and the Hare IV. Am. J. Bot. 2014, 101, 1987–2004. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Cantino, P.D.; Olmstead, R.G.; Bramley, G.L.C.; Xiang, C.L.; Ma, Z.H.; Tan, Y.H.; Zhang, D.X. A Large-Scale Chloroplast Phylogeny of the Lamiaceae Sheds New Light on Its Subfamilial Classification. Sci. Rep. 2016, 6, 34343. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.J.; Lu, Y.B.; Ye, Q.Q.; Tang, S.Q. Population Genetic Structure and Phylogeography of Camellia flavida (Theaceae) Based on Chloroplast and Nuclear DNA Sequences. Front. Plant Sci. 2017, 8, 718. [Google Scholar] [CrossRef] [PubMed]
- Degen, B.; Fladung, M. Use of DNA-Markers for Tracing Illegal Logging. In Proceedings of the International Workshop “Fingerprinting Methods for the Identification of Timber Origins”, Bonn, Germany, 8–9 October 2007; Volume 321, pp. 6–14. [Google Scholar]
- Cavers, S.; Navarro, C.; Lowe, A.J. Chloroplast DNA Phylogeography Reveals Colonization History of a Neotropical Tree, Cedrela odorata L., in Mesoamerica. Mol. Ecol. 2003, 12, 1451–1460. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.L.; Ci, X.Q.; Liu, Z.F.; Dormontt, E.E.; Conran, J.G.; Lowe, A.J.; Li, J. Assessing Candidate DNA Barcodes for Chinese and Internationally Traded Timber Species. Mol. Ecol. Resour. 2022, 22, 1478–1492. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Villanueva, K.; de Groot, G.A.; Laros, I.; Bovenschen, J.; Bongers, F.; Zuidema, P.A. Genetic Differences among Cedrela odorata Sites in Bolivia Provide Limited Potential for Fine-Scale Timber Tracing. Tree Genet. Genomes 2019, 15, 33. [Google Scholar] [CrossRef]
- Schroeder, H.; Cronn, R.; Yanbaev, Y.; Jennings, T.; Mader, M.; Degen, B.; Kersten, B. Development of Molecular Markers for Determining Continental Origin of Wood from White Oaks (Quercus L. Sect. Quercus). PLoS ONE 2016, 11, e0158221. [Google Scholar] [CrossRef]
- Mader, M.; Pakull, B.; Blanc-Jolivet, C.; Paulini-Drewes, M.; Bouda, Z.H.N.; Degen, B.; Small, I.; Kersten, B. Complete Chloroplast Genome Sequences of Four Meliaceae Species and Comparative Analyses. Int. J. Mol. Sci. 2018, 19, 701. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L.; Fazekas, A.J.; et al. A DNA Barcode for Land Plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef]
- Finch, K.N.; Jones, F.A.; Cronn, R.C. Genomic Resources for the Neotropical Tree Genus Cedrela (Meliaceae) and Its Relatives. BMC Genom. 2019, 20, 58. [Google Scholar] [CrossRef]
- Michael, T.P.; Jupe, F.; Bemm, F.; Motley, S.T.; Sandoval, J.P.; Lanz, C.; Loudet, O.; Weigel, D.; Ecker, J.R. High Contiguity Arabidopsis thaliana Genome Assembly with a Single Nanopore Flow Cell. Nat. Commun. 2018, 9, 541. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Olsen, H.E.; Paten, B.; Akeson, M. The Oxford Nanopore MinION: Delivery of Nanopore Sequencing to the Genomics Community. Genome Biol. 2016, 17, 239. [Google Scholar] [CrossRef]
- Scott, A.D.; Zimin, A.V.; Puiu, D.; Workman, R.; Britton, M.; Zaman, S.; Caballero, M.; Read, A.C.; Bogdanove, A.J.; Burns, E.; et al. A Reference Genome Sequence for Giant Sequoia. G3 Genes|Genomes|Genet. 2020, 10, 3907–3919. [Google Scholar] [CrossRef]
- Giordano, F.; Aigrain, L.; Quail, M.A.; Coupland, P.; Bonfield, J.K.; Davies, R.M.; Tischler, G.; Jackson, D.K.; Keane, T.M.; Li, J.; et al. De novo Yeast Genome Assemblies from MinION, PacBio and MiSeq Platforms. Sci. Rep. 2017, 7, 3935. [Google Scholar] [CrossRef]
- Wang, W.; Schalamun, M.; Morales-Suarez, A.; Kainer, D.; Schwessinger, B.; Lanfear, R. Assembly of Chloroplast Genomes with Long- and Short-Read Data: A Comparison of Approaches Using Eucalyptus pauciflora as a Test Case. BMC Genom. 2018, 19, 977. [Google Scholar] [CrossRef]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-Generation Sequencing Technologies: An Overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore Sequencing Technology, Bioinformatics and Applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef]
- Schalamun, M.; Nagar, R.; Kainer, D.; Beavan, E.; Eccles, D.; Rathjen, J.P.; Lanfear, R.; Schwessinger, B. Harnessing the MinION: An Example of How to Establish Long-Read Sequencing in a Laboratory Using Challenging Plant Tissue from Eucalyptus Pauciflora. Mol. Ecol. Resour. 2019, 19, 77–89. [Google Scholar] [CrossRef]
- Reiling, S.J.; Chen, S.-H.; Ragoussis, I. McGill Nanopore Ligation LibPrep Protocol SQK-LSK109. Protocolos.io 2020. Available online: https://www.protocols.io/view/mcgill-nanopore-ligation-libprep-protocol-sqk-lsk1-bp2l6b1ezgqe/v1 (accessed on 10 May 2021). [CrossRef]
- Jain, M.; Koren, S.; Miga, K.H.; Quick, J.; Rand, A.C.; Sasani, T.A.; Tyson, J.R.; Beggs, A.D.; Dilthey, A.T.; Fiddes, I.T.; et al. Nanopore Sequencing and Assembly of a Human Genome with Ultra-Long Reads. Nat. Biotechnol. 2018, 36, 338–345. [Google Scholar] [CrossRef] [PubMed]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and Processing Long-Read Sequencing Data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef] [PubMed]
- Chaisson, M.J.; Tesler, G. Mapping Single Molecule Sequencing Reads Using Basic Local Alignment with Successive Refinement (BLASR): Application and Theory. BMC Bioinform. 2012, 13, 238. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and Accurate Long-Read Assembly via Adaptive k-Mer Weighting and Repeat Separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Kamath, G.M.; Shomorony, I.; Xia, F.; Courtade, T.A.; Tse, D.N. HINGE: Long-Read Assembly Achieves Optimal Repeat Resolution. Genome Res. 2017, 27, 747–756. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and Open Software for Comparing Large Genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef]
- Loman, N.J.; Quick, J.; Simpson, J.T. A Complete Bacterial Genome Assembled de Novo Using Only Nanopore Sequencing Data. Nat. Methods 2015, 12, 733–735. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and Accurate de Novo Genome Assembly from Long Uncorrected Reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and Accurate Annotation of Organelle Genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Mayor, C.; Brudno, M.; Schwartz, J.R.; Poliakov, A.; Rubin, E.M.; Frazer, K.A.; Pachter, L.S.; Dubchak, I. Vista: Visualizing Global DNA Sequence Alignments of Arbitrary Length. Bioinformatics 2000, 16, 1046–1047. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows—Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; Van Den Beek, M.; Bouvier, D.; Ech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Van der Auwera, G.; O’Connor, B. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra, 1st ed.; O’Reilly Media, Inc., Ed.; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2020; ISBN 978-1491975190. [Google Scholar]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An Advanced Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
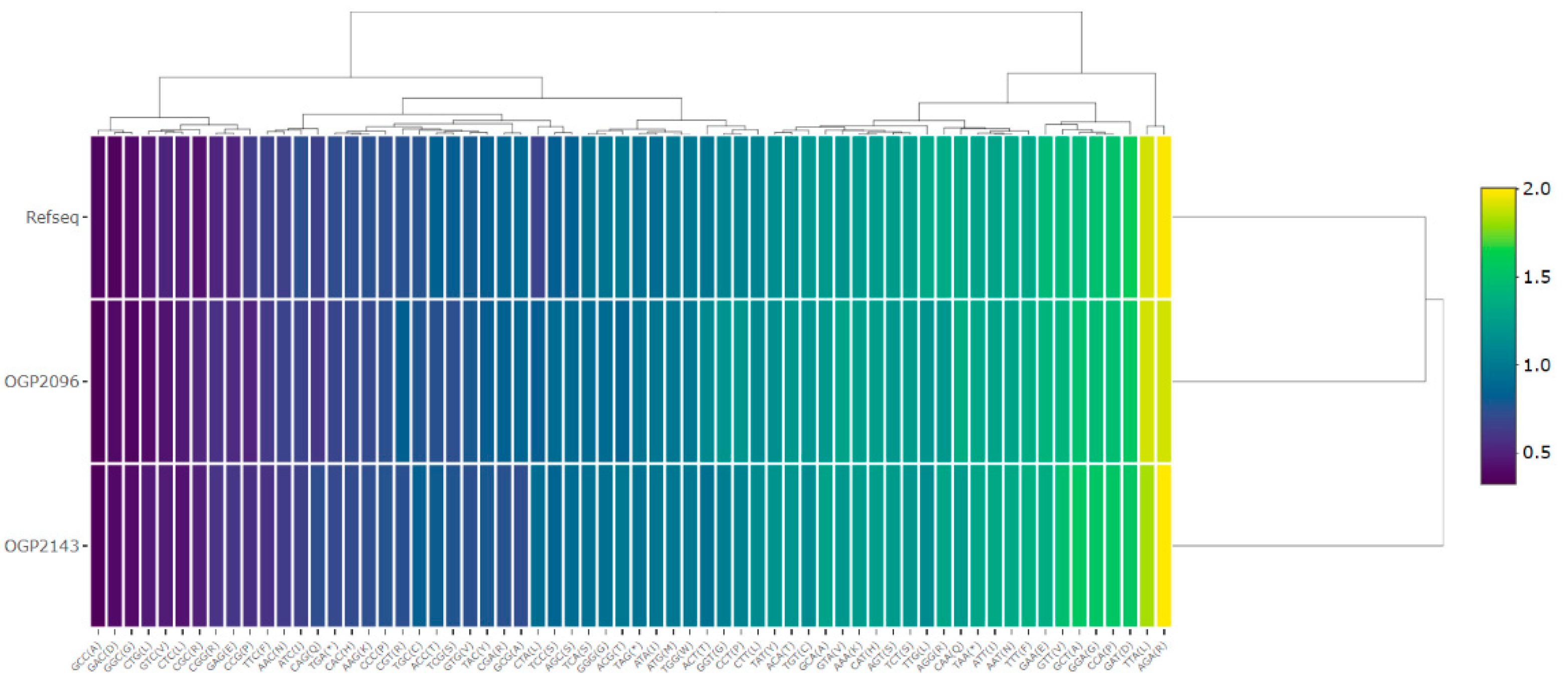
- Sharp, P.M.; Li, W.-H. The Codon Adaptation Index-a Measure of Directional Synonymous Codon Usage Bias, and Its Potential Applications. Nucleic Acids Res. 1986, 14, 4683–4690. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, W.; Zhu, D.; Hong, P.; Zhang, S.; Xiao, S.; Tan, Y.; Chen, X.; Xu, L.; Zong, X.; et al. Chromosome-Scale Genome Assembly of Sweet Cherry (Prunus avium L.) Cv. Tieton Obtained Using Long-Read and Hi-C Sequencing. Hortic. Res. 2020, 7, 122. [Google Scholar] [CrossRef] [PubMed]
- Wöhner, T.W.; Emeriewen, O.F.; Wittenberg, A.H.J.; Schneiders, H.; Vrijenhoek, I.; Halász, J.; Hrotkó, K.; Hoff, K.J.; Gabriel, L.; Lempe, J.; et al. The Draft Chromosome-Level Genome Assembly of Tetraploid Ground Cherry (Prunus fruticosa Pall.) from Long Reads. Genomics 2021, 113, 4173–4183. [Google Scholar] [CrossRef]
- Cavers, S.; Telford, A.; Arenal Cruz, F.; Pérez Castañeda, A.J.; Valencia, R.; Navarro, C.; Buonamici, A.; Lowe, A.J.; Vendramin, G.G. Cryptic Species and Phylogeographical Structure in the Tree Cedrela odorata L. throughout the Neotropics. J. Biogeogr. 2013, 40, 732–746. [Google Scholar] [CrossRef]
- Samji, A.; Eashwarlal, K.; Shanmugavel, S.; Kumar, S.; Warrier, R.R. Chloroplast Genome Skimming of a Potential Agroforestry Species Melia Dubia. Cav and Its Comparative Phylogenetic Analysis with Major Meliaceae Members. 3 Biotech 2023, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Finch, K.N.; Jones, F.A.; Cronn, R.C. Cryptic Species Diversity in a Widespread Neotropical Tree Genus: The Case of Cedrela Odorata. Am. J. Bot. 2022, 109, 1622–1640. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and Challenges in Long-Read Sequencing Data Analysis. Genome Biol. 2020, 21, 30. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Winefield, C.; Bombarely, A.; Prentis, P.; Waterhouse, P. Tools and Strategies for Long-Read Sequencing and De Novo Assembly of Plant Genomes. Trends Plant Sci. 2019, 24, 700–724. [Google Scholar] [CrossRef] [PubMed]
| Reference | Collection Site | Species | Sequence Name |
|---|---|---|---|
| Finch et al. [15] | Southern Bolivia | C. angustifolia | CEAN143 |
| Northern Bolivia | C. fissilis | CEFI211 | |
| Western Colombia | C. montana | CEMO50 | |
| Southern Bolivia | C. saltensis | CESA102 | |
| Nicaragua | C. odorata | COD10 | |
| Costa Rica | COD52 | ||
| Nicaragua | COD162 | ||
| Costa Rica | COD185 | ||
| Venezuela | COD202 | ||
| Panamá | COD222 | ||
| Costa Rica | COD277 | ||
| Mexico | NOVO_NYBG | ||
| Mader et al. [13] | Cuba | C. odorata | MG724915.1 |
| This study | Colombia (Caquetá) | C. odorata | OGP2096 |
| Colombia (Putumayo) | C. odorata | OGP2143 |
| Sample | MinION-ONT | Genome Assembly (kb) | Post-Assembly (kb) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total Sequences (Gb) | No. Reads | Mean Read Length (kb) | Assembler | MuMmer | Racon + Nanopolish | Coverage (%) | ||||||
| Cp Reads | Canu | Hinge | Canu | Hinge | Canu | Hinge | Canu | Hinge | ||||
| OGP2096 | 2.6 | 594,819 | 4.37 | 1437 | 180 | 190 | 138 | 157 | 156 | 159 | 86 | 100 |
| OGP2143 | 1.0 | 110,484 | 9.67 | 884 | NA | 182 | NA | 157 | NA | 159 | NA | 100 |
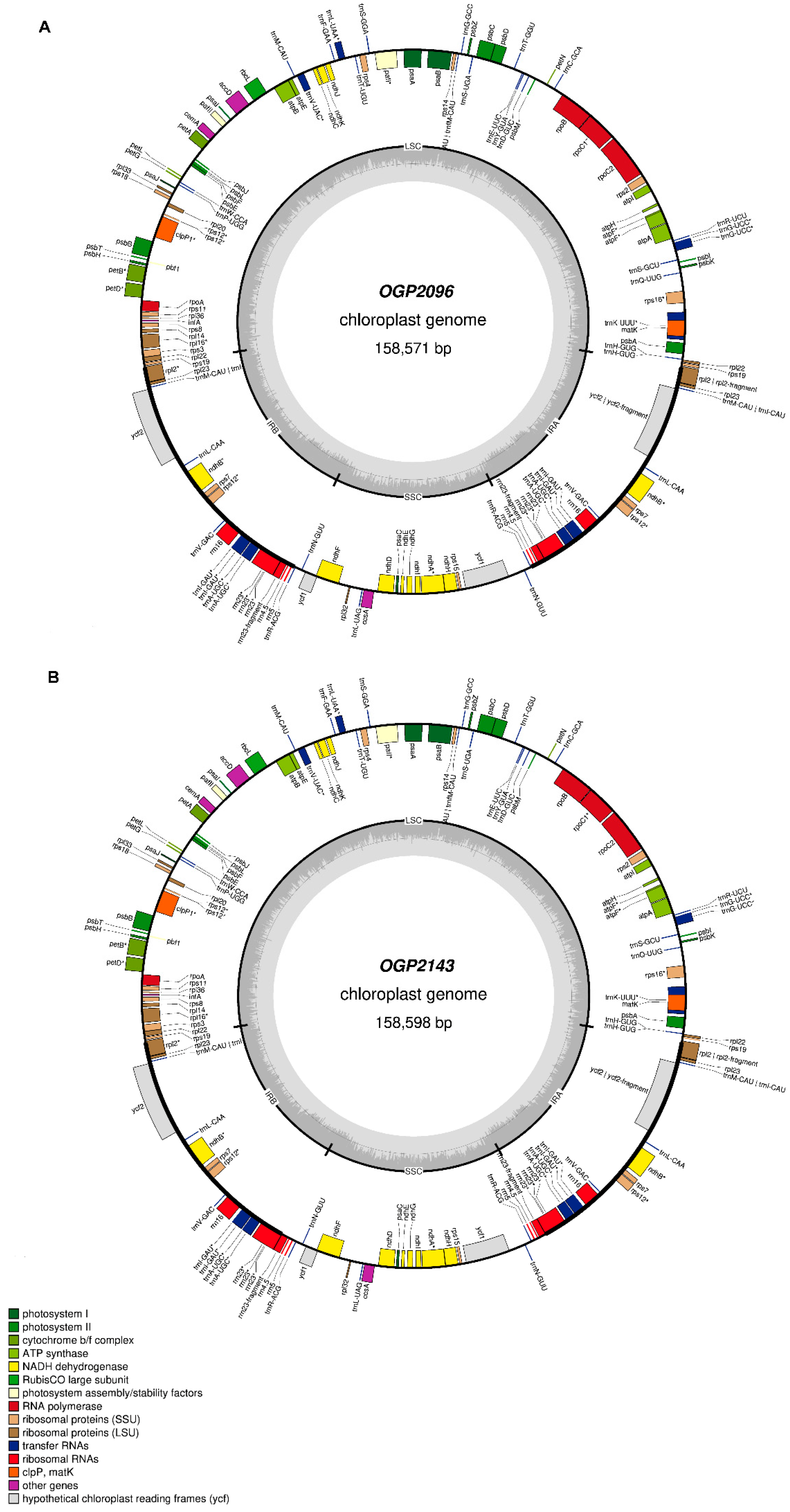
| CP Genomic Feature | Refseq (MG724915.1) | OGP2096 (OP750006) | OGP2143 (OP750007) |
|---|---|---|---|
| Genome size (kb) | 158.55 kb | 158.57 kb | 158.59 kb |
| GC content | 37.9% | 37.9% | 37.9% |
| Predicted genes | 132 | 132 | 132 |
| Long single copy (LSC) | 86.3 kb | 87.7 kb | 87.9 kb |
| Short single copy (SSC) | 18.3 kb | 22.7 kb | 25.5 kb |
| Inverted repeat (IR-A) | 26.89 kb | 24.03 kb | 22.56 kb |
| Inverted repeat (IR-B) | 26.89 kb | 24.03 kb | 22.56 kb |
| Individual | SNPs | INS | DEL | MNP | Total | |
|---|---|---|---|---|---|---|
| Unique | Total | |||||
| CEAN143 | 32 | 249 | 23 | 39 | 5 | 348 |
| CEFI211 | 64 | 277 | 21 | 45 | 11 | 418 |
| CEMO50 | 15 | 221 | 13 | 28 | 6 | 283 |
| CESA102 | 20 | 270 | 20 | 44 | 9 | 363 |
| COD10 | 1 | 221 | 21 | 34 | 9 | 286 |
| COD52 | 2 | 239 | 18 | 36 | 11 | 306 |
| COD162 | 1 | 238 | 17 | 37 | 8 | 301 |
| COD185 | 9 | 240 | 19 | 31 | 7 | 306 |
| COD202 | 20 | 267 | 21 | 32 | 10 | 350 |
| COD222 | 6 | 228 | 19 | 28 | 7 | 288 |
| COD277 | 1 | 227 | 16 | 35 | 9 | 288 |
| OGP2096 | 14 | 70 | 0 | 2 | 3 | 89 |
| OGP2143 | 2 | 46 | 0 | 2 | 1 | 51 |
| Total | 187 | 2793 | 208 | 393 | 96 | 3677 |
| POS (nt) | MG724915.1 | OGP2096 | OGP2143 | Type | Locus |
|---|---|---|---|---|---|
| 6727 | G | A | G | variant | intergenic |
| 9279 | G | A | G | variant | intergenic |
| 16,056 | C | T | C | variant | intergenic |
| 21,511 | C | T | C | missense | rpoC2 |
| 40,324 | T | A | T | missense | psaB |
| 44,137 | G | A | G | variant | intergenic |
| 66,416 | C | C | A | variant | intergenic |
| 75,039 | G | A | G | variant | intergenic |
| 77,654 | T | G | T | intron variant | psbH |
| 80,024 | C | T | C | intron variant | petD |
| 84,403 | G | A | G | intron variant * | Rpl16 |
| 87,863 | G | G | A | intron variant | rpl2 |
| 114,009 | C | T | C | missense | NdhF |
| 116,740 | C | T | C | intron variant | Rpl32 |
| 124,608 | G | A | G | intron variant | ndhA |
| 126,930 | T | G | T | missense | rps15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simbaqueba, J.; Garzón-Martínez, G.A.; Castano, N. New Variants in the Chloroplast Genome Sequence of Two Colombian Individuals of the Cedar Timber Species (Cedrela odorata L.), Using Long-Read Oxford Nanopore Technology. Int. J. Plant Biol. 2024, 15, 865-877. https://doi.org/10.3390/ijpb15030062
Simbaqueba J, Garzón-Martínez GA, Castano N. New Variants in the Chloroplast Genome Sequence of Two Colombian Individuals of the Cedar Timber Species (Cedrela odorata L.), Using Long-Read Oxford Nanopore Technology. International Journal of Plant Biology. 2024; 15(3):865-877. https://doi.org/10.3390/ijpb15030062
Chicago/Turabian StyleSimbaqueba, Jaime, Gina A. Garzón-Martínez, and Nicolas Castano. 2024. "New Variants in the Chloroplast Genome Sequence of Two Colombian Individuals of the Cedar Timber Species (Cedrela odorata L.), Using Long-Read Oxford Nanopore Technology" International Journal of Plant Biology 15, no. 3: 865-877. https://doi.org/10.3390/ijpb15030062
APA StyleSimbaqueba, J., Garzón-Martínez, G. A., & Castano, N. (2024). New Variants in the Chloroplast Genome Sequence of Two Colombian Individuals of the Cedar Timber Species (Cedrela odorata L.), Using Long-Read Oxford Nanopore Technology. International Journal of Plant Biology, 15(3), 865-877. https://doi.org/10.3390/ijpb15030062





