Incorporating Tissue-Specific Gene Expression Data to Improve Chemical–Disease Inference of in Silico Toxicogenomics Methods
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene/Protein Expression Dataset
2.2. Thresholds for Defining Low, Medium, and High Expression Levels
2.3. Methodology of Chemical–Protein–Disease Inference
2.4. Dataset and Measurement for Evaluating Model Performance
3. Results and Discussion
3.1. Diseases Inference Augmented by Incorporating Tissue-Specific Gene Expression
3.2. Identification of Disease-Relevant Chemicals
3.3. Web-Based User Interface
3.4. Case Study: Melamine
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Z.; Huang, R.; Roberts, R.; Tong, W. Toxicogenomics: A 2020 Vision. Trends Pharmacol. Sci. 2019, 40, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-H.; Tung, C.-W. Identification of Consensus Biomarkers for Predicting Non-Genotoxic Hepatocarcinogens. Sci. Rep. 2017, 7, 41176. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-H.; Lin, Y.-C.; Tung, C.-W. Identification of Time-Invariant Biomarkers for Non-Genotoxic Hepatocarcinogen Assessment. Int. J. Environ. Res. Public Health 2020, 17, E4298. [Google Scholar] [CrossRef] [PubMed]
- Black, M.B.; Stern, A.; Efremenko, A.; Mallick, P.; Moreau, M.; Hartman, J.K.; McMullen, P.D. Biological System Considerations for Application of Toxicogenomics in Next-Generation Risk Assessment and Predictive Toxicology. Toxicol. In Vitro 2022, 80, 105311. [Google Scholar] [CrossRef] [PubMed]
- Tung, C.-W. ChemDIS: A Chemical-Disease Inference System Based on Chemical-Protein Interactions. J. Cheminform. 2015, 7, 25. [Google Scholar] [CrossRef]
- Tung, C.-W.; Wang, S.-S. ChemDIS 2: An Update of Chemical-Disease Inference System. Database 2018, 2018, bay077. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Wiegers, T.C.; King, B.L.; Wiegers, J.; Grondin, C.J.; Sciaky, D.; Johnson, R.J.; Mattingly, C.J. Generating Gene Ontology-Disease Inferences to Explore Mechanisms of Human Disease at the Comparative Toxicogenomics Database. PLoS ONE 2016, 11, e0155530. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2021. Nucleic Acids Res. 2021, 49, D1138–D1143. [Google Scholar] [CrossRef]
- Davis, A.P.; Wiegers, T.C.; Wiegers, J.; Johnson, R.J.; Sciaky, D.; Grondin, C.J.; Mattingly, C.J. Chemical-Induced Phenotypes at CTD Help Inform the Predisease State and Construct Adverse Outcome Pathways. Toxicol. Sci. 2018, 165, 145–156. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Santos, A.; von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting Protein-Chemical Interaction Networks with Tissue and Affinity Data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef]
- Jeong, J.; Choi, J. Advancing the Adverse Outcome Pathway for PPARγ Inactivation Leading to Pulmonary Fibrosis Using Bradford-Hill Consideration and the Comparative Toxicogenomics Database. Chem. Res. Toxicol. 2022, 35, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Qi, G.; Shou, Y.; Li, D.; Liu, Y.; Guan, H.; Zhang, Q.; Chen, S.; Luo, J.; Xu, L.; et al. High Throughput Data-Based, Toxicity Pathway-Oriented Development of a Quantitative Adverse Outcome Pathway Network Linking AHR Activation to Lung Damages. J. Hazard. Mater. 2022, 425, 128041. [Google Scholar] [CrossRef] [PubMed]
- Tung, C.-W.; Cheng, H.-J.; Wang, C.-C.; Wang, S.-S.; Lin, P. Leveraging Complementary Computational Models for Prioritizing Chemicals of Developmental and Reproductive Toxicity Concern: An Example of Food Contact Materials. Arch. Toxicol. 2020, 94, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-C.; Liang, Y.-C.; Wang, S.-S.; Lin, P.; Tung, C.-W. A Machine Learning-Driven Approach for Prioritizing Food Contact Chemicals of Carcinogenic Concern Based on Complementary in Silico Methods. Food Chem. Toxicol. 2022, 160, 112802. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Li, Y.; Be, X.; Howes, S.; Liu, W. Detecting and Profiling Tissue-Selective Genes. Physiol. Genom. 2006, 26, 158–162. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xiao, S.-J.; Zhang, C.; Zou, Q.; Ji, Z.-L. TiSGeD: A Database for Tissue-Specific Genes. Bioinformatics 2010, 26, 1273–1275. [Google Scholar] [CrossRef] [PubMed]
- Moreno, P.; Fexova, S.; George, N.; Manning, J.R.; Miao, Z.; Mohammed, S.; Muñoz-Pomer, A.; Fullgrabe, A.; Bi, Y.; Bush, N.; et al. Expression Atlas Update: Gene and Protein Expression in Multiple Species. Nucleic Acids Res. 2022, 50, D129–D140. [Google Scholar] [CrossRef] [PubMed]
- Asmann, Y.W.; Necela, B.M.; Kalari, K.R.; Hossain, A.; Baker, T.R.; Carr, J.M.; Davis, C.; Getz, J.E.; Hostetter, G.; Li, X.; et al. Detection of Redundant Fusion Transcripts as Biomarkers or Disease-Specific Therapeutic Targets in Breast Cancer. Cancer Res. 2012, 72, 1921–1928. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Barbosa-Morais, N.L.; Irimia, M.; Pan, Q.; Xiong, H.Y.; Gueroussov, S.; Lee, L.J.; Slobodeniuc, V.; Kutter, C.; Watt, S.; Colak, R.; et al. The Evolutionary Landscape of Alternative Splicing in Vertebrate Species. Science 2012, 338, 1587–1593. [Google Scholar] [CrossRef]
- GTEx Consortium Human Genomics. The Genotype-Tissue Expression (GTEx) Pilot Analysis: Multitissue Gene Regulation in Humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Habuka, M.; Fagerberg, L.; Hallström, B.M.; Pontén, F.; Yamamoto, T.; Uhlen, M. The Urinary Bladder Transcriptome and Proteome Defined by Transcriptomics and Antibody-Based Profiling. PLoS ONE 2015, 10, e0145301. [Google Scholar] [CrossRef] [PubMed]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the Human Tissue-Specific Expression by Genome-Wide Integration of Transcriptomics and Antibody-Based Proteomics. Mol. Cell Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-S.; Pinto, S.M.; Getnet, D.; Nirujogi, R.S.; Manda, S.S.; Chaerkady, R.; Madugundu, A.K.; Kelkar, D.S.; Isserlin, R.; Jain, S.; et al. A Draft Map of the Human Proteome. Nature 2014, 509, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Eraslan, B.; Wieland, T.; Hallström, B.; Hopf, T.; Zolg, D.P.; Zecha, J.; Asplund, A.; Li, L.-H.; Meng, C.; et al. A Deep Proteome and Transcriptome Abundance Atlas of 29 Healthy Human Tissues. Mol. Syst. Biol. 2019, 15, e8503. [Google Scholar] [CrossRef] [PubMed]
- Schriml, L.M.; Arze, C.; Nadendla, S.; Chang, Y.-W.W.; Mazaitis, M.; Felix, V.; Feng, G.; Kibbe, W.A. Disease Ontology: A Backbone for Disease Semantic Integration. Nucleic Acids Res. 2012, 40, D940–D946. [Google Scholar] [CrossRef] [PubMed]
- Schriml, L.M.; Mitraka, E.; Munro, J.; Tauber, B.; Schor, M.; Nickle, L.; Felix, V.; Jeng, L.; Bearer, C.; Lichenstein, R.; et al. Human Disease Ontology 2018 Update: Classification, Content and Workflow Expansion. Nucleic Acids Res. 2019, 47, D955–D962. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; François, R.; Henry, L.; Müller, K. Dplyr: A Grammar of Data Manipulation. R Package Version 1.0.5. 2021. Available online: https://CRAN.R-project.org/package=dplyr (accessed on 7 October 2021).
- Swaen, G.M. Urolithiasis in Children and Exposure to Melamine: A Review of the Epidemiological Literature. Toxicol. Res. Appl. 2019, 3, 2397847319861601. [Google Scholar] [CrossRef]
- Bolden, A.L.; Rochester, J.R.; Kwiatkowski, C.F. Melamine, beyond the Kidney: A Ubiquitous Endocrine Disruptor and Neurotoxicant? Toxicol. Lett. 2017, 280, 181–189. [Google Scholar] [CrossRef]
- Li, Q.; Song, P.; Wen, J. Melamine and Food Safety: A 10-Year Review. Curr. Opin. Food Sci. 2019, 30, 79–84. [Google Scholar] [CrossRef]
- Zheng, X.; Zhao, A.; Xie, G.; Chi, Y.; Zhao, L.; Li, H.; Wang, C.; Bao, Y.; Jia, W.; Luther, M.; et al. Melamine-Induced Renal Toxicity Is Mediated by the Gut Microbiota. Sci. Transl. Med. 2013, 5, 172ra22. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.-J.; Wu, P.-Y.; Huang, J.-C.; Chen, S.-C. Environmental Pollution and Chronic Kidney Disease. Int. J. Med. Sci. 2021, 18, 1121–1129. [Google Scholar] [CrossRef] [PubMed]
- Goyal, J.; Jain, P.; Jain, V.; Banerjee, D.; Bhattacharyya, R.; Dey, S.; Sharma, R.; Rai, N. Melamine Exacerbates Neurotoxicity in D-Galactose-Induced Neuronal SH-SY5Y Cells. J. Aging Res. 2023, 2023, 6635370. [Google Scholar] [CrossRef] [PubMed]
- Pavek, P.; Dvorak, Z. Xenobiotic-Induced Transcriptional Regulation of Xenobiotic Metabolizing Enzymes of the Cytochrome P450 Superfamily in Human Extrahepatic Tissues. Curr. Drug Metab. 2008, 9, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Reddy, J.K.; Rao, M.S. Xenobiotic-Induced Peroxisome Proliferation: Role of Tissue Specificity and Species Differences in Response in the Evaluation of the Implications for Human Health. Arch. Toxicol. Suppl. 1987, 10, 43–53. [Google Scholar] [CrossRef]
- Watanabe, H.; Suzuki, A.; Goto, M.; Lubahn, D.B.; Handa, H.; Iguchi, T. Tissue-Specific Estrogenic and Non-Estrogenic Effects of a Xenoestrogen, Nonylphenol. J. Mol. Endocrinol. 2004, 33, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, Y.; Sun, Y.; Wu, H.; Hou, J. Tissue-Specific Toxic Effects of Nano-Copper on Zebrafish. Environ. Res. 2024, 242, 117717. [Google Scholar] [CrossRef] [PubMed]
- Antony, B.; Johny, J.; Abdelazim, M.M.; Jakše, J.; Al-Saleh, M.A.; Pain, A. Global Transcriptome Profiling and Functional Analysis Reveal That Tissue-Specific Constitutive Overexpression of Cytochrome P450s Confers Tolerance to Imidacloprid in Palm Weevils in Date Palm Fields. BMC Genom. 2019, 20, 440. [Google Scholar] [CrossRef]
- Ju, Q.; Li, X.; Jiang, X.-J.; Qu, M.-J.; Guo, X.-Q.; Han, Z.-J.; Li, F. Transcriptome and Tissue-Specific Expression Analysis of Obp and Csp Genes in the Dark Black Chafer. Arch. Insect Biochem. Physiol. 2014, 87, 177–200. [Google Scholar] [CrossRef]
- Martelli, F.; Ravenscroft, T.A.; Hutchison, W.; Batterham, P. Tissue-Specific Transcriptome Analyses in Drosophila Provide Novel Insights into the Mode of Action of the Insecticide Spinosad and the Function of Its Target, nAChRα6. Pest. Manag. Sci. 2023, 79, 3913–3925. [Google Scholar] [CrossRef] [PubMed]
- Geist, J.; Werner, I.; Eder, K.J.; Leutenegger, C.M. Comparisons of Tissue-Specific Transcription of Stress Response Genes with Whole Animal Endpoints of Adverse Effect in Striped Bass (Morone saxatilis) Following Treatment with Copper and Esfenvalerate. Aquat. Toxicol. 2007, 85, 28–39. [Google Scholar] [CrossRef] [PubMed]


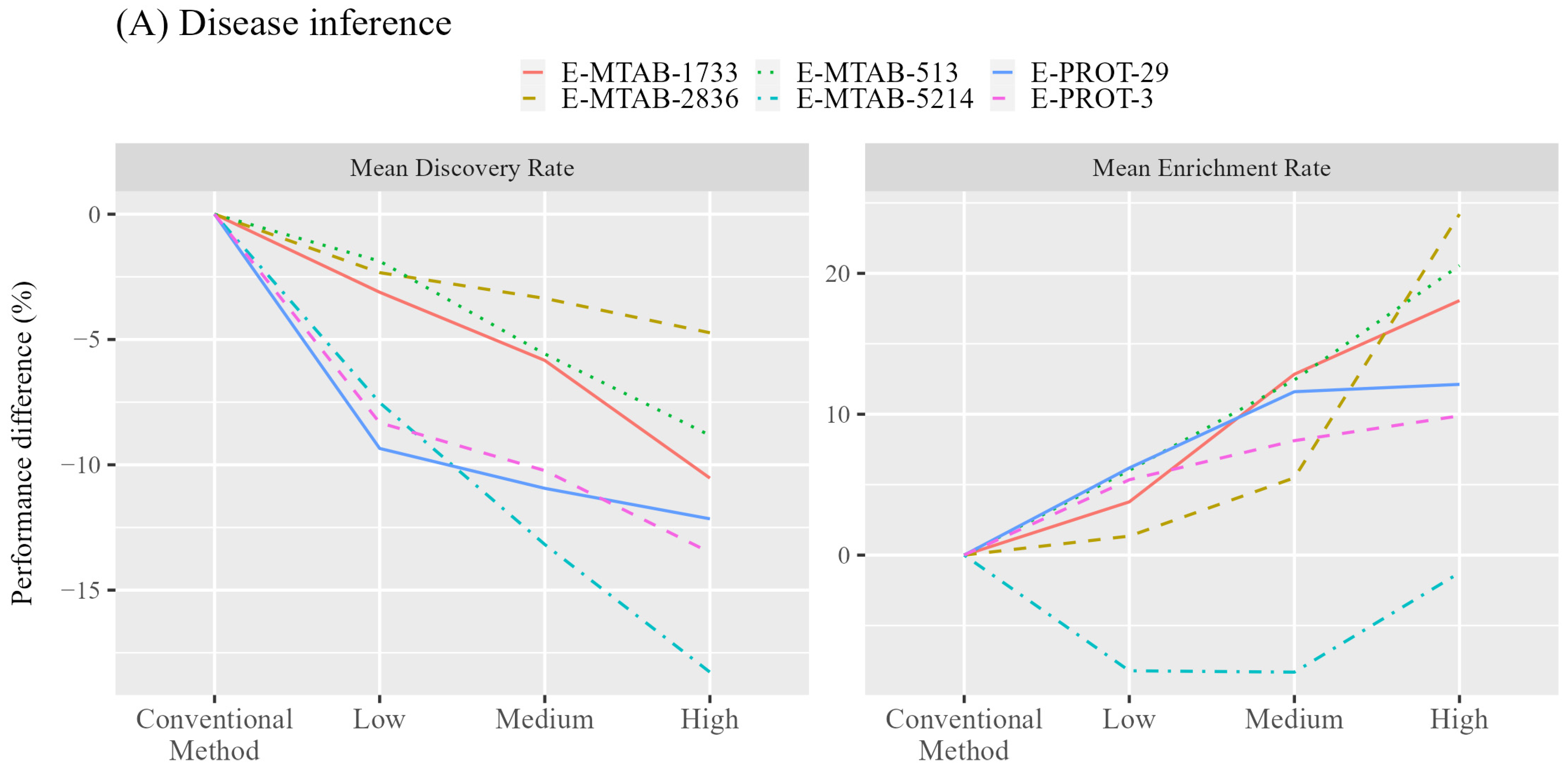
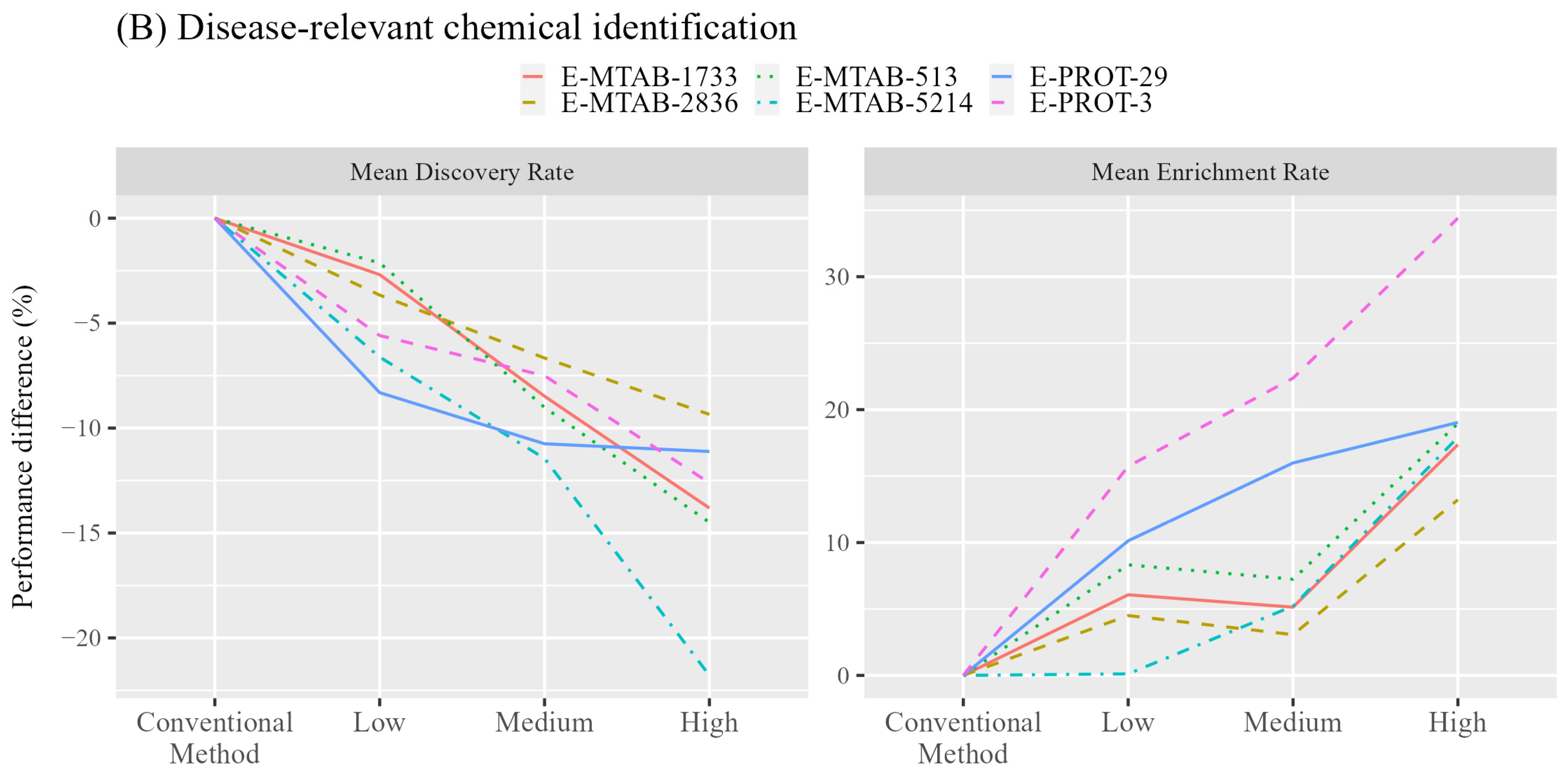

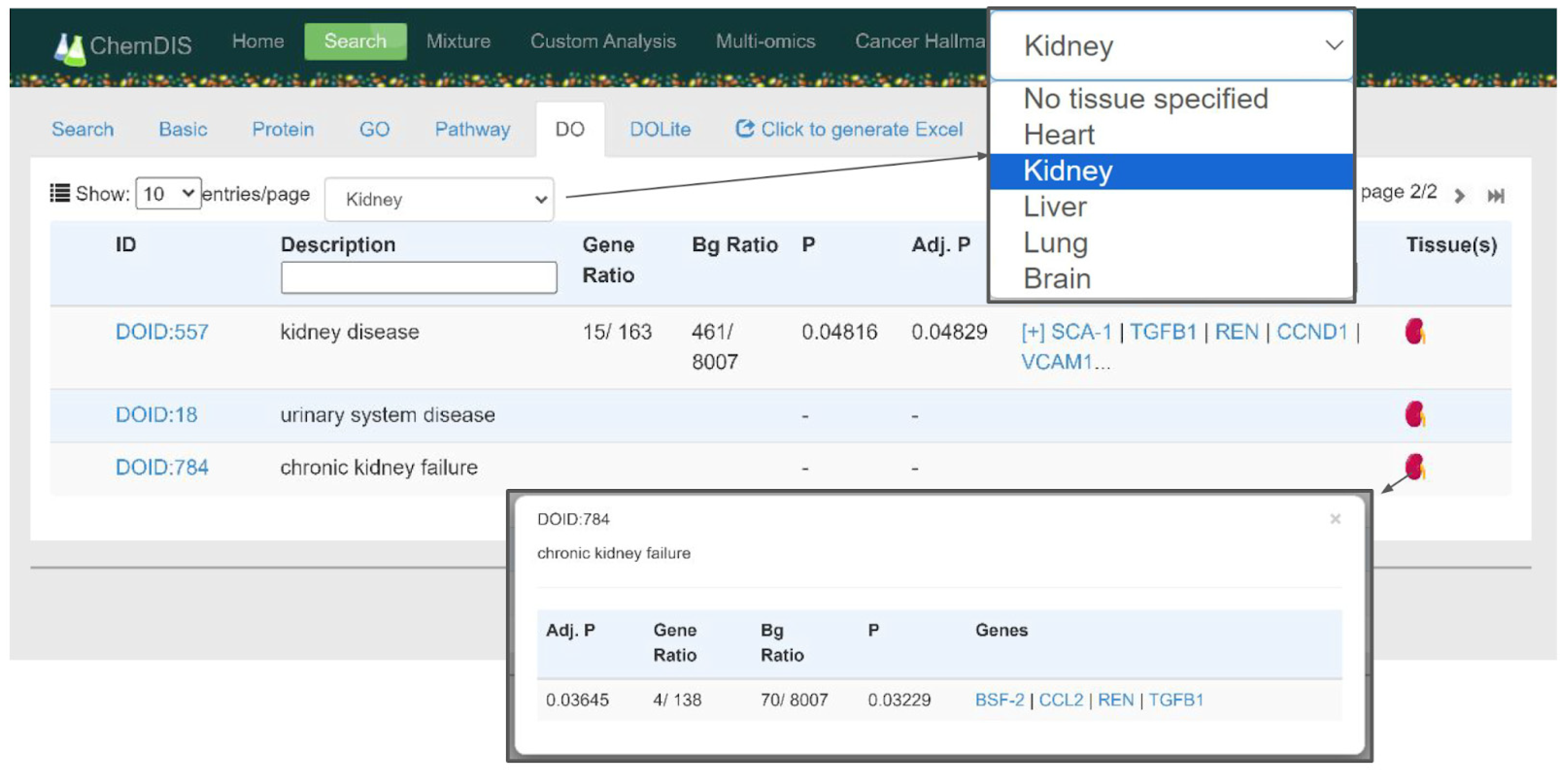
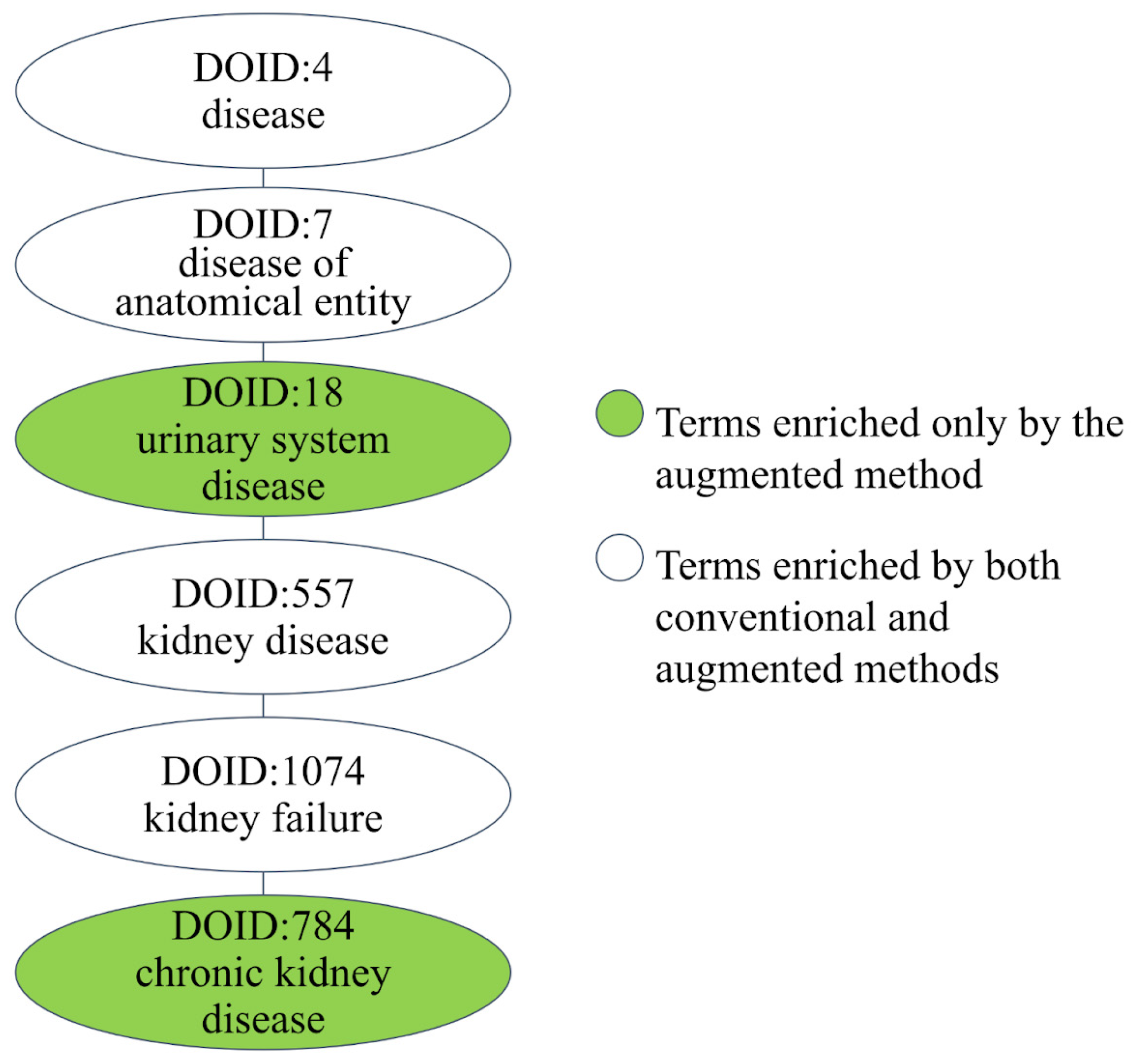
| Type | ID | Tissues | Expression Value | References |
|---|---|---|---|---|
| RNA-Seq mRNA | E-MTAB-513 | 16 | 0 to 75,295 TPM | [18,19,20] |
| E-MTAB-5214 | 53 | 0 to 315,499 TPM | [21] | |
| E-MTAB-2836 | 32 | 0 to 137,860 TPM | [22,23] | |
| E-MTAB-1733 | 27 | 0 to 137,868 TPM | [24] | |
| Proteomics | E-PROT-3 | 44 | 1 (low), 2 (medium), 3 (high) | [22,25] |
| E-PROT-29 | 31 | 0 to 51,851,041 ppb | [26] |
| ID | Blood (n = 128) | Skin (n = 98) | Brain (n = 73) | Lung (n = 62) | Heart (n = 50) | Kidney (n = 37) | Liver (n = 31) | Breast (n = 22) | Bone Marrow (n = 22) | Vagina (n = 18) |
|---|---|---|---|---|---|---|---|---|---|---|
| E-MTAB-513 | V | V | V | V | V | |||||
| E-MTAB-5214 | V | V | V | V | V | |||||
| E-MTAB-2836 | V | V | V | V | V | |||||
| E-MTAB-1733 | V | V | V | V | V | |||||
| E-PROT-3 | V | V | V | V | V | |||||
| E-PROT-29 | V | V | V | V | V |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.-S.; Wang, C.-C.; Wang, C.-L.; Lin, Y.-C.; Tung, C.-W. Incorporating Tissue-Specific Gene Expression Data to Improve Chemical–Disease Inference of in Silico Toxicogenomics Methods. J. Xenobiot. 2024, 14, 1023-1035. https://doi.org/10.3390/jox14030057
Wang S-S, Wang C-C, Wang C-L, Lin Y-C, Tung C-W. Incorporating Tissue-Specific Gene Expression Data to Improve Chemical–Disease Inference of in Silico Toxicogenomics Methods. Journal of Xenobiotics. 2024; 14(3):1023-1035. https://doi.org/10.3390/jox14030057
Chicago/Turabian StyleWang, Shan-Shan, Chia-Chi Wang, Chien-Lun Wang, Ying-Chi Lin, and Chun-Wei Tung. 2024. "Incorporating Tissue-Specific Gene Expression Data to Improve Chemical–Disease Inference of in Silico Toxicogenomics Methods" Journal of Xenobiotics 14, no. 3: 1023-1035. https://doi.org/10.3390/jox14030057
APA StyleWang, S.-S., Wang, C.-C., Wang, C.-L., Lin, Y.-C., & Tung, C.-W. (2024). Incorporating Tissue-Specific Gene Expression Data to Improve Chemical–Disease Inference of in Silico Toxicogenomics Methods. Journal of Xenobiotics, 14(3), 1023-1035. https://doi.org/10.3390/jox14030057








