The Unique Capability of Endolysin to Tackle Antibiotic Resistance: Cracking the Barrier
Abstract
:1. Introduction
2. Global Antibiotic Usage
3. Causes of the Crisis in Antibiotic Resistance
3.1. Misuse of Antibiotics
3.1.1. Bad Prescription Practices
3.1.2. Widespread Use in Agriculture
3.1.3. Limited Supply of New Antibiotics
3.1.4. Regulating Obstacles
4. What Is the Solution?
5. Bacteriophages
5.1. Phage Therapy
5.2. How About Using Phages Alone?
6. Endolysins
6.1. Endolysins and Associated Phage Proteins
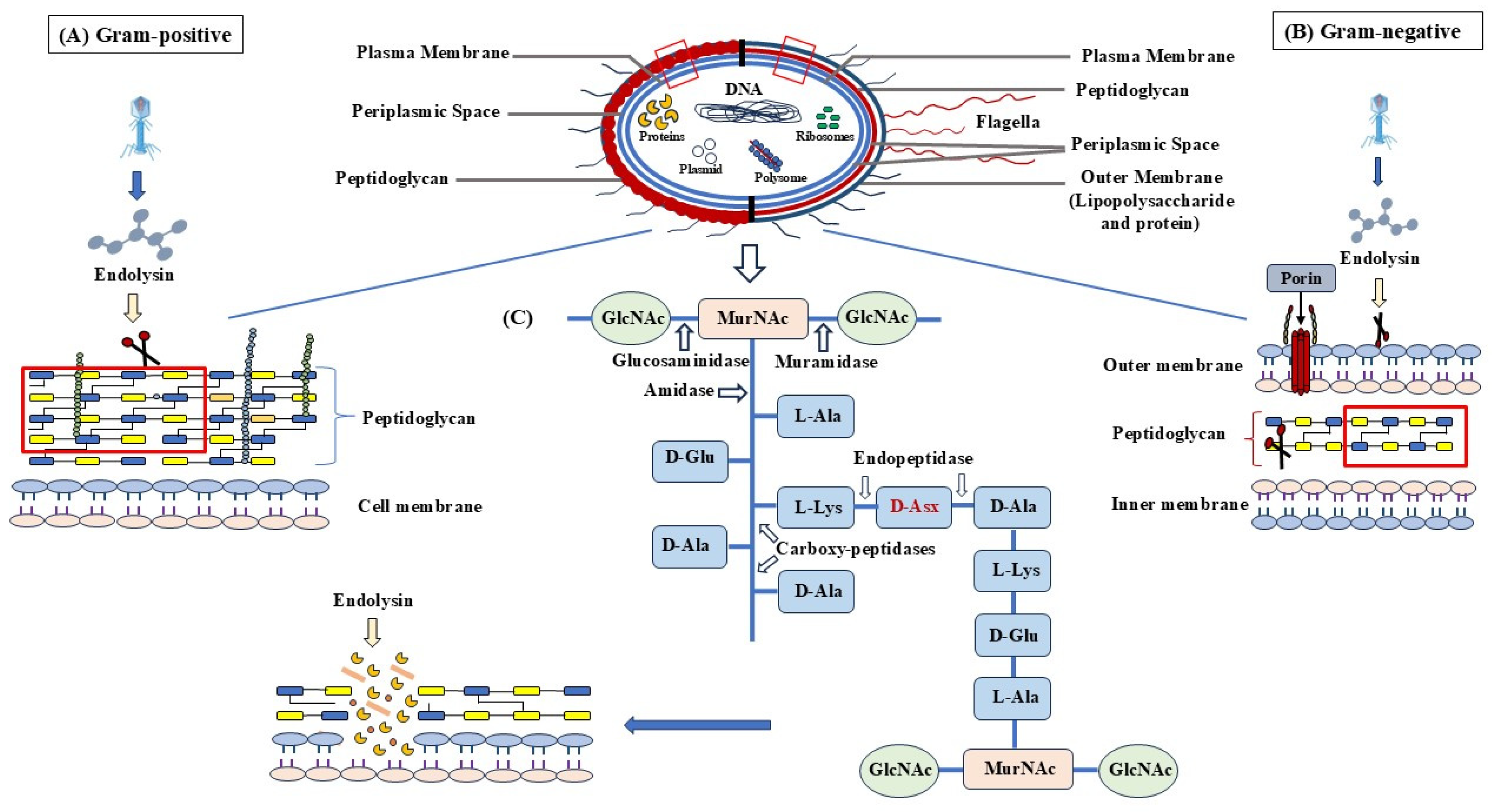
- Holins
- Signal Sequences
- Spanins
6.2. Classification of Phage Lysins
| Protein Name | Length | Gene | Source Organism | Antibacterial Activity | Effective Against G+ or G- Bacteria | Test Done In Vitro/In Vivo | Animal Model | MDR (YES/NO) | ATCC strain or Clinical Isolate | Expression Vector | Expression Host | pH | Temperature | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LysMR-5 | 495 | lysMR-5 | S. aureus phage MR-5 | S. aureus ATCC 43300 (MRSA), S. aureus ATCC 33,591 (MRSA), S. aureus ATCC 25923 (MSSA), and S. aureus ATCC 29,213 (MSSA) | Gram-positive | In Vitro | No | YES | ATCC | pET28a | E. coli BL21 | 7 | 37 °C | [51] |
| LYS_BPS13 | 91 | E | Enterobacteria phage S13 (Bacteriophage S13) | B. cereus | Gram-positive | In Vitro | No | Not available | ATCC | pET15b | E. coli BL21 | 9.5 | 42–45 °C | [52] |
| LysB4 | 262 | lysB4 | Bacillus phage B4 | B. cereus | Both | In vitro | No | Not available | ATCC | pET15b | E. coli BL21 | 8.0–10.0 | 50 °C | [53] |
| PlyB | 326 | plyB | Aspergillus nidulans | Bacillus | Not available | Both | Mouse | Not available | ATCC | pBAD24 (60) | Escherichia coli strain TOP10 | Not available | Not available | [54] |
| PlyC | 465 | orf11 | Streptococcus phage C1 | Staphylococcus aureus (MRSA), Enterococcus, E. coli, and Gram positive Lactococcus lactis | Both | In vitro | No | Not available | Clinical Isolate | pEX and pET-22b | E. coli strain BL21 (DE3) | Not available | 5 to 60 °C | [55] |
| Lys68 | 162 | Lys68 | Salmonella phage phi68 | Salmonella, Klebsiella, Pseudomonas etc. | Gram-positive | In Vitro | No | Not available | Not available | pET-28a | E. coli BL21(DE3) | 7 | 4 C to 40 °C | [56] |
| LysH5 | 481 | LysH5 | Staphylococcus phage phiH5 (Bacteriophage phiH5) | Staphylococcus aureus and Staphylococcus epidermidis | Gram-positive | In Vitro | No | Not available | Clinical Isolate | No | No | 7 | 37 °C | [57] |
| Endolysin | 185 | ABgp46 | Acinetobacter phage vB_AbaP_CEB1 | A. baumannii, S. typhimurium LT2, E.coli, etc. | Both | In Vitro | No | Not available | Both | pET15b-ABgp46 | Escherichia coli BL21(DE3) | 4.0–10.0 | Up to 50 °C | [58] |
| Putative phage lysin | 245 | phi7917_002 | Streptococcus phage phi7917 | E. coli, Salmonella, B. subtilis, S. aureus, S. suis | Both | Both | Mice | Not available | Both | pSJ2 | E. coli BL21 (DE3) | 6.0–9.0 | Up to 50 °C | [59] |
| Ribonucleoside-diphosphate reductase, 1.17.4.1 | 695 | PBC4_057 | Bacillus phage PBC4 | B. cereus | Gram-positive | In Vitro | No | Not available | ATCC | Not available | Not available | Not available | Not available | [60] |
| CHAP domain protein, Lysin | 238 AA | VD13_036, X878_0033 | Enterococcus phage VD13 | E. faecalis, Staphylococcus aureus, Escherichia coli DH5α | Both | In vitro | No | YES | Both | pET21a | E. coli BL21(DE3) | 4–10 (At 5 highest activity | 4–100 (At 50 highest activity | [61] |
| ST01 protein | 96 | st01 | Escherichia coli | P. aeruginosa, K. pneumoniae, E. coli | Gram negative | Both | Galleria mellonella larvae | YES | ATCC, KCTC, CCARM | pAS008 or pAS047 | BL21 (DE3) | Not available | Not available | [62] |
| ClyC/NocO | 434 | clyC/nocO | Nodularia sp. LEGE 06071 | S. aureus, Enterococcus faecalis, Bacillus cereus | Gram positive | Both | Mouse | YES | Both | pET28a | E. coli BL21(DE3) | Not available | 4−65 °C | [63] |
| lysozyme | 274 | phiCTP1_gp29 | Clostridium phage phiCTP1 | Clostridium species, lactic acid bacteria, Bacillus cereus. | Gram positive | In vitro | None | YES | NCIMB (Aberdeen, UK), ATCC (Manassas, VA, USA), CECT (Valencia, Spain), the BCCM/LMG (Ghent, Belgium) | pET15b | E. coli BL21 (DE3) | Not available | Not available | [64] |
| N-acetylmuramoyl-L-alanine amidase | 289 | PHIM1EF22_0110 | Enterococcus phage phiM1EF22 | E. faecalis | Gram positive | In vitro | None | Not available | Both | pDP2 | E. coli CG61 | 4–10 pH | 10–60 °C | [65] |
| N-acetylmuramoyl-L-alanine amidase | 233 | PlyG, GAMMALSU_0017, GAMMAUSAM_0017 | Bacillus phage Gamma | Bacillus anthracis | Gram positive | In vitro | None | YES | ATCC | pET-19b | Escherichia coli [BL21(DE3) | 7 | 40 | [66] |
| Portal protein | 602 AA | ORF17 | Helicobacter pylori bacteriophage KHP30 | H. pylori | Gram negative | In vitro | None | YES | ATCC | Not available | E. coli BL21(DE3) | 5–10 pH | 10–55 °C | [67] |
| L-alanyl-D-glutamate peptidase | 137 | lys | Escherichia phage T5 (Enterobacteria phage T5) | Escherichia coli | Gram negative | In vitro | None | YES | National Collection of Micro-organisms IBPM RAS | pT5lys | Escherichia coli BL21(DE3) | 3–10 pH | 10–60 °C | [68] |
| Endolysin | 133 | elyY | Yersinia enterocolitica (type O:9) | Yersinia enterocolitica, E. coli | Gram negative | In vitro | None | YES | Both | pET28-elyY | E. coli BL21(DE3) | 7 | 37 °C | [69] |
| N-acetylmuramoyl- L-alanine amidase | 289 | Thymidylate synthase | Enterococcus phage PBEF129 | E. faecalis | Gram positive | In vitro | None | YES | Culture Collection of Antibiotic-Resistant Microorganisms in Korea | pET21-a(+) | Escherichia coli BL21 (DE3)pLyss | pH 5–9 | 37 °C | [70] |
| dihydrofolate reductase, 1.5.1.3 | 169 | qdvp001_068 | Vibrio phage qdvp001 | Vibrio parahaemolyticus | Gram negative | In vitro | None | YES | ATCC | pET-30a | E. coli BL21 | 8 | 40 °C | [71] |
| Lysozyme, 3.2.1.17, CP-1 lysin, Endolysin, Muramidase | 339 | CPL1, 22 | Streptococcus phage Cp-1 (Bacteriophage Cp-1) | Streptococcus pneumoniae | Gram positive | In vitro | None | YES | ATCC | pT7–7 | E. coli BL21 (DE3) | 8 | 37 °C | [72] |
| PHIKZ144 | 260 | Transglycosylase gp144 | Pseudomonas phage phiKZ | Escherichia coli | Gram negative | In vitro | None | Not available | pQE-30 | Escherichia coli | 7 | 40 °C | [73] | |
| N-acetyl muramyl-L-alanine amidase | 308 | PlyPSA | Listeria phage PSU-VKH-LP041 | L. monocytogenes | Gram positive | In vitro | None | YES | Not available | pASK-IBA5 | E. coli K-12 | 7 | 45 °C | [74] |
| L-alanyl-D-glutamate peptidase | 289 | ply, ply500 | Listeria phage A500 (Bacteriophage A500) | Listeria species | Gram positive | In vitro | None | Not available | ATCC | pASK-IBA5 | E. coli K-12 | 7 | 45 °C | [75] |
| Endolysin | 266 | vB_BceM_AP3_0015 | Burkholderia phage AP3 | E. coli, K. pneumoniae, P. aeruginosa, B. cenocepacia, S. enterica, Staphylococcus aureus and S. epidermidis | Both | In vitro | None | YES | ATCC | pEXP5-CT/TOPO | Escherichia coli BL21-AI | pH 3–10 | 10–55 °C | [76] |
| Phage protein | 68 | SPN1S_0005 | Salmonella phage SPN1S | Salmonella typhimurium, Escherichia coli | Gram-negative | In vitro | None | YES | ATCC | pET-28a | E. coli BL21 (DE3) | pH 4–10 | 40 °C | [77] |
7. Applications of Endolysins
7.1. Application of Endolysins as Human Therapeutics
7.2. Application of Endolysins in the Veterinary Sector
7.3. Endolysins in Food and Other Sectors
| Application of Endolysins as Human Therapeutics | ||||
|---|---|---|---|---|
| Infection | Species | Antibiotics Resistance | Endolysin | Reference |
| Skin and respiratory infections | Staphylococcus aureus (MRSA) | Methicillin | LysK ClyS CF-301 MR-10 Staphefekt | [94,95,96,97,98,99,100,101] |
| Corneal infections | Staphylococcus simulans | Doxycycline, tetracycline | MV-L | [102,103] |
| Endocarditis, sepsis | Staphylococcus epidermidis | Rifamycin, fluoroquinolones, gentamicin, tetracycline, clindamycin | MV-L | [102,104] |
| Urinary tract infections, hemolytic–uremic syndrome, neonatal meningitis, hemorrhagic colitis | Escherichia coli | Penicillin, cephalosporins, cephamycins, carbapenems | MV-L | [102,105,106,107,108] |
| Nosocomial infections | Enterococcus faecalis | Vancomycin | PlyV12 EFAP-1, EFAL-1 IME-EF1 EF-P10 EC300 Lys170 LysEF-P10 | [109,110,111,112,113,114,115,116] |
| Strep throat, pneumonia, skin infections, and meningitis | S. pneumoniae | Penicillin, erythromycin, clarithromycin, ceftriaxone | Cpl-1 | [47,117,118,119,120,121] |
| Hospital-acquired pneumonia, community-acquired pneumonia, Community-acquired pneumonia, Bloodstream infections | Acinetobacter baumannii | Cephalosporin, carbapenem, ceftazidime, liprofloxacin | LysAB2 PlyF307 | [122,123,124,125,126,127,128,129,130] |
| Malignant external otitis, endophthalmitis, endocarditis, meningitis, pneumonia, and septicemia | P. aeruginosa | Carbapenem, aminoglycosides (gentamicin, tobramycin, amikacin, neomycin, plazomicin, streptomycin) | OBPgp279 | [126,130,131] |
| Recurrent urinary tract infections (rUTI), pneumonia, and bloodstream infections | Klebsiella pneumoniae | Carbapenem | LysPA26 | [79,132,133] |
| Application of Endolysins in the Veterinary Sector | ||||
| Necrotic enteritis and sub-clinical disease | Clostridium perfringens | Tetracycline, bacitracin | CP25L Psm | [134,135,136,137,138,139,140] |
| Anthrax | Bacillus anthracis | Streptomycin | PlyG | [141,142,143,144] |
| Equine strangles | Streptococcus equi. | Vancomycin | PlyC | [145,146,147,148] |
| Arthritis, meningitis, septicemia, and endocarditis | Streptococcus suis | Penicillin, ampicillin | LySMP | [149,150,151,152] |
| Bloodstream infection intra-abdominal infection bacteremia endocarditis | Enterococcus faecium E. faecalis | Vancomycin, lincomycin, bambermycin, bacitracin, tetracycline, ciprofloxacin, erythromycin, kanamycin, penicillin, tylosin, streptomycin, vancomycin, gentamycin, streptogramins, avilamycin | PlyV12 | [110,140] |
| Endolysins in Food and Other Sectors | ||||
| LysSA11 | After 15 min of endolysin treatment, viable MRSA levels decreased in experimentally contaminated ham and pasteurized products. | S. aureus | Milk Products | [153,154] |
| Gp110 | This endolysin, with a novel enzyme structure and N-acetylmuramidase lysis domain, exhibited exceptional in vitro activity against Salmonella and other Gram-negative pathogens. | Salmonella spp. | Sea Foods | [155,156] |
| LysCs4 SPN1S Lys68 | Peptidoglycan from Gram-positive and Gram-negative bacteria from six distinct genera could be broken down by the refined lysozymes, which could also lyse C. sakazakii that had an outer membrane permeabilized. | C. sakazakii | Milk powders, herbal teas, and other dried products. | [56,157,158,159] |
| PlyBa Ply12 Ply21 LysBPS13 LysB4 | Endolysins effectively combat 24 B. cereus and B. thuringiensis strains, contaminating food. Endopeptidase exhibits bactericidal activity against Gram-positive bacteria, including B. cereus, B. subtilis, and monocytogenes. | B. cereus | Dairy Products | [52,53,160,161] |
| CS74L CPT1l | It is also shown that these enzymes were active against Clostridium acetobutylicum and C. tyrobutyricum using the turbidity assay and fresh bacterial cells, indicating that they could be used as a potential bio preservative in cheese. Another endolysin that was recovered from a virulent phage was also described by the same family; however, this enzyme’s host range was more constrained. | Clostridium sporogenes, Clostridium acetobutylicum, Clostridium tyrobutyricum | In poultry, clostridial species are linked to food spoilage. Germinated Clostridium sporogenes and Clostridium tyrobutyricum have the potential to produce gases and acids in the dairy sector that alter the structural and sensory characteristics of cheeses. | [83,162,163] |
8. Administration Routes
| Target Pathogen | Phage | Enzyme (Endolysin) | Activity (Mode of Action) | Administration Route | References |
|---|---|---|---|---|---|
| MRSA | GH15 | LysGH15 | Amidase and endopeptidase | Intravenous and Intraperitoneal | [170,171] |
| Streptococcus pneumoniae | Cp1 | Cpl-1 | Muramidase | Intravenous, nasal, oral, aerosols, and Intraperitoneal | [120,172,173,174,175] |
| MRSA | MR11 | MV-L | Amidase and endopeptidase | Intraperitoneal, nasal | [102] |
| Streptococcus pyogenes | MGAS5005 prophage | PlyPy | Endopeptidase | Intraperitoneal | [176] |
| MRSA | phiSH2 prophage, phiP68, phiWMY, phi80α, phi11 2854, prophage K | phiSH2, P68, LysWMY, 80αLyt2, phi11, 2638A, LysK | Amidase and endopeptidase | Intraperitoneal | [177] |
| Pseudomonas aeruginosa | phage PVP-SE1 | Artilysin® Engineered Endolysin-Based (PVP-SE1gp146) | Muramidase | Oral and topical | [165] |
| Streptococcus agalactiae | NCTC11261 | PlyGBS | Endopeptidase and Muramidase | Intravaginal, oral and intranasal | [178] |
| Pseudomonas aeruginosa | P. aeruginosa phage | PlyPa03 | Muramidase | Topical | [179] |
| Streptococcus pneumoniae | CP-7 | Cpl-7 | Muramidase | Immersion | [180] |
| Acinetobacter baumannii | RL-2015 | PlyF307 | Muramidase | intraperitoneal and Topical | [129] |
| Enterococcus faecalis | E. faecalis phage IME-EF1 | LysIME-EF1 | Endopeptidase | Intraperitoneal | [181] |
| Acinetobacter baumannii | SS3e | LysSS | Muramidase | Intraperitoneal | [182] |
| Streptococcus pyogenes | C1 | PlyC | Amidase | Oral, nasal | [46] |
| Bacillus anthracis | γ-phage | PlyG | Amidase | Intraperitoneal | [100] |
9. Functional Improvements
| Endolysin | Improvements | Assets | Activity Against | References |
|---|---|---|---|---|
| CHAPk | Full-length enzyme truncation | Enhanced solubility and catalytic activity | Methicillin-resistant Staphylococcus aureus | [183] |
| ClyS | Combination of EADs (enzymatically-active domain) and CWBDs (cell wall-binding domain) from several endolysins | Improved solubility and catalytic potential | Methicillin-resistant S. aureus (MRSA) | [97] |
| Art-Bp7e6 | A random peptide was fused with the phage endolysin Bp7e | To create a chimeric endolysin library | ß-lactamase-resistant E. coli, Salmonella enterica serovar Enteritidis | [184] |
| EC300 | Combination of CWBD of endolysin with virion-associated lysin | Enhanced effectiveness | Vancomycin-resistant Enterococcus faecalis | [65] |
| SA2-E-Lyso-SH3b, SA2-E-LysK-SH3b | Proteins with switched specificity are produced when distinct-origin CWBDs and EADs are combined | Enhanced catalytic efficiency and expanded lytic range | Cephalosporins-resistant Listeria monocytogenes | [147] |
| OBPgp279, PVP-SE1g-146 | Combining endolysin and OMP (outer-membrane permeabilizer) | Improved capacity to combat Gram-negative bacteria. | Ceftazidime and tetracycline-resistant Pseudomonas aeruginosa and Acinetobacter baumannii | [165] |
| Art-175 | AMP (antimicrobial peptide)-mediated endolysin fusion | Enhanced ability to combat Gram-negative bacteria | Methicillin-resistant Staphylococcus aureus (MRSA) | [165] |
| PlyG | Combination of EADs and CWBDs from several endolysins | Ability to manage the temperature | Clindamycin-resistant C. perfringens | [138] |
| LysAB2 | Site-directed mutagenesis as well as truncation | Improvement of AMP | Colistin-resistant A. baumannii | [185] |
9.1. Domain Swapping and Shuffling
9.2. Mutagenesis
9.3. Lysin Translocation
10. Synergism with Antibiotics
11. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Woolhouse, M.; Farrar, J. Policy: An intergovernmental panel on antimicrobial resistance. Nature 2014, 509, 555–557. [Google Scholar] [CrossRef] [PubMed]
- Laxminarayan, R.; Matsoso, P.; Pant, S.; Brower, C.; Røttingen, J.A.; Klugman, K.; Davies, S. Access to effective antimicrobials: A worldwide challenge. Lancet 2016, 387, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Abat, C.; Rolain, J.M.; Dubourg, G.; Fournier, P.E.; Chaudet, H.; Raoult, D. Evaluating the clinical burden and mortality attributable to antibiotic resistance: The disparity of empirical data and simple model estimations. Clin. Infect. Dis. 2017, 65 (Suppl. S1), S58–S63. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Gandra, S.; Ashok, A.; Caudron, Q.; Grenfell, B.T.; Levin, S.A.; Laxminarayan, R. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect. Dis. 2014, 14, 742–750. [Google Scholar] [CrossRef]
- Orubu, E.S.F.; Samad, M.A.; Rahman, M.T.; Zaman, M.H.; Wirtz, V.J. Mapping the antimicrobial supply chain in Bangladesh: A scoping-review-based ecological assessment approach. Glob. Health Sci. Pract. 2021, 9, 532–547. [Google Scholar] [CrossRef]
- Zanichelli, V.; Sharland, M.; Cappello, B.; Moja, L.; Getahun, H.; Pessoa-Silva, C.; Sati, H.; van Weezenbeek, C.; Balkhy, H.; Simão, M.; et al. The WHO AWaRe (Access, Watch, Reserve) antibiotic book and prevention of antimicrobial resistance. Bull. World Health Organ. 2023, 101, 290. [Google Scholar] [CrossRef]
- Howard, P.; Pulcini, C.; Levy Hara, G.; West, R.M.; Gould, I.M.; Harbarth, S.; Nathwani, D. An international cross-sectional survey of antimicrobial stewardship programmes in hospitals. J. Antimicrob. Chemother. 2015, 70, 1245–1255. [Google Scholar] [CrossRef]
- World Health Organisation. Antimicrobial Resistance: Global Report on Surveillance. 2014. Available online: http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf?ua=1 (accessed on 20 May 2024).
- Spellberg, B.; Gilbert, D.N. The future of antibiotics and resistance: A tribute to a career of leadership by John Bartlett. Clin. Infect. Dis. 2014, 59 (Suppl. S2), S71–S75. [Google Scholar] [CrossRef]
- Bartlett, J.G.; Gilbert, D.N.; Spellberg, B. Seven ways to preserve the miracle of antibiotics. Clin. Infect. Dis. 2013, 56, 1445–1450. [Google Scholar] [CrossRef]
- Nature, E. The antibiotic alarm. Nature 2013, 495, 141. [Google Scholar]
- Read, A.F.; Woods, R.J. Antibiotic resistance management. Evol. Med. Public Health 2014, 2014, 147. [Google Scholar] [CrossRef] [PubMed]
- Gross, M. Antibiotics in crisis. Curr. Biol. 2013, 23, R1063–R1065. [Google Scholar] [CrossRef] [PubMed]
- Michael, C.A.; Dominey-Howes, D.; Labbate, M. The antimicrobial resistance crisis: Causes, consequences, and management. Front. Public Health 2014, 2, 110657. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention, Office of Infectious Disease. Antibiotic Resistance Threats in the United States. 2013. Available online: https://www.cdc.gov/drugresistance/threat-report-2013 (accessed on 21 May 2024).
- Luyt, C.E.; Bréchot, N.; Trouillet, J.L.; Chastre, J. Antibiotic stewardship in the intensive care unit. Crit. Care 2014, 18, 480. [Google Scholar] [CrossRef]
- Golkar, Z.; Bagasra, O.; Pace, D.G. Bacteriophage therapy: A potential solution for the antibiotic resistance crisis. J. Infect. Dev. Ctries. 2014, 8, 129–136. [Google Scholar] [CrossRef]
- Gould, I.M.; Bal, A.M. New antibiotic agents in the pipeline and how they can help overcome microbial resistance. Virulence 2013, 4, 185–191. [Google Scholar] [CrossRef]
- Wright, G.D. Something old, something new: Revisiting natural products in antibiotic drug discovery. Can. J. Microbiol. 2014, 60, 147–154. [Google Scholar] [CrossRef]
- Piddock, L.J. The crisis of no new antibiotics—What is the way forward? Lancet Infect. Dis. 2012, 12, 249–253. [Google Scholar] [CrossRef]
- Young, R.Y. Bacteriophage lysis: Mechanism and regulation. Microbiol. Rev. 1992, 56, 430–481. [Google Scholar] [CrossRef]
- Moak, M.; Molineux, I.J. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol. Microbiol. 2004, 51, 1169–1183. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; Martínez, B.; Donovan, D.M.; Rodríguez, A.; García, P. Bacteriophage virion-associated peptidoglycan hydrolases: Potential new enzybiotics. Crit. Rev. Microbiol. 2013, 39, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Wittebole, X.; De Roock, S.; Opal, S.M. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 2014, 5, 226–235. [Google Scholar] [CrossRef]
- Deresinski, S. Bacteriophage therapy: Exploiting smaller fleas. Clin. Infect. Dis. 2009, 48, 1096–1101. [Google Scholar] [CrossRef] [PubMed]
- Endersen, L.; O’Mahony, J.; Hill, C.; Ross, R.P.; McAuliffe, O.; Coffey, A. Phage therapy in the food industry. Annu. Rev. Food Sci. Technol. 2014, 5, 327–349. [Google Scholar] [CrossRef]
- O’Flaherty, S.; Ross, R.P.; Coffey, A. Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol. Rev. 2009, 33, 801–819. [Google Scholar] [CrossRef]
- Sulakvelidze, A.; Alavidze, Z.; Morris, J.G., Jr. Bacteriophage therapy. Antimicrob. Agents Chemother. 2001, 45, 649–659. [Google Scholar] [CrossRef]
- Fischetti, V.A. Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 2008, 11, 393–400. [Google Scholar] [CrossRef]
- Lai, W.C.B.; Chen, X.; Ho, M.K.Y.; Xia, J.; Leung, S.S.Y. Bacteriophage-derived endolysins to target gram-negative bacteria. Int. J. Pharm. 2020, 589, 119833. [Google Scholar] [CrossRef]
- São-José, C.; Parreira, R.; Vieira, G.; Santos, M.A. The N-terminal region of the Oenococcus oeni bacteriophage fOg44 lysin behaves as a bona fide signal peptide in Escherichia coli and as a cis-inhibitory element, preventing lytic activity on oenococcal cells. J. Bacteriol. 2000, 182, 5823–5831. [Google Scholar] [CrossRef]
- Kakikawa, M.; Yokoi, K.J.; Kimoto, H.; Nakano, M.; Kawasaki, K.I.; Taketo, A.; Kodaira, K.I. Molecular analysis of the lysis protein Lys encoded by Lactobacillus plantarum phage ϕg1e. Gene 2002, 299, 227–234. [Google Scholar] [CrossRef]
- Guo, T.; Zhang, C.; Liu, W.; Wang, S.; Kong, J. Functional analysis of the N-terminal region of endolysin Lyb5 encoded by Lactobacillus fermentum bacteriophage φPYB5. Int. J. Food Microbiol. 2015, 203, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Summer, E.J.; Berry, J.; Tran, T.A.T.; Niu, L.; Struck, D.K.; Young, R. Rz/Rz1 lysis gene equivalents in phages of Gram-negative hosts. J. Mol. Biol. 2007, 373, 1098–1112. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.; Rajaure, M.; Pang, T.; Young, R. The spanin complex is essential for lambda lysis. J. Bacteriol. 2012, 194, 5667–5674. [Google Scholar] [CrossRef]
- Ajuebor, J.; McAuliffe, O.; O’Mahony, J.; Ross, R.P.; Hill, C.; Coffey, A. Bacteriophage endolysins and their applications. Sci. Prog. 2016, 99, 183–199. [Google Scholar] [CrossRef]
- Dörr, T.; Moynihan, P.J.; Mayer, C. Bacterial cell wall structure and dynamics. Front. Microbiol. 2019, 10, 487209. [Google Scholar] [CrossRef]
- Fenton, M.; McAuliffe, O.; O’Mahony, J.; Coffey, A. Recombinant bacteriophage lysins as antibacterials. Bioeng. Bugs 2010, 1, 9–16. [Google Scholar] [CrossRef]
- Ragland, S.A.; Criss, A.K. From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog. 2017, 13, e1006512. [Google Scholar] [CrossRef]
- Twort, F.W. An investigation on the nature of ultra-microscopic viruses. Lancet 1915, 186, 1241–1243. Available online: https://scholar.archive.org/work/wuup4h47hbco7kysfzxn2tz4du/access/wayback/http://sgc.anlis.gob.ar/bitstream/123456789/752/2/ActaKravsi1961_2_37-40.pdf (accessed on 10 October 2024). [CrossRef]
- Reynals, F.D. Bacteriophage et microbes tues. C. R. Soc. Biol. 1926, 94, 242–243. [Google Scholar]
- Evans, A.C. Streptococcus bacteriophage: A study of four serological types. Public Health Rep. 1934, 49, 1386–1401. [Google Scholar] [CrossRef]
- Maxted, W.R. The active agent in nascent phage lysis of streptococci. Microbiology 1957, 16, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Gondil, V.S.; Harjai, K.; Chhibber, S. Endolysins as emerging alternative therapeutic agents to counter drug-resistant infections. Int. J. Antimicrob. Agents 2020, 55, 105844. [Google Scholar] [CrossRef] [PubMed]
- Fischetti, V.A. Development of phage lysins as novel therapeutics: A historical perspective. Viruses 2018, 10, 310. [Google Scholar] [CrossRef]
- Nelson, D.; Loomis, L.; Fischetti, V.A. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. USA 2001, 98, 4107–4112. [Google Scholar] [CrossRef]
- Loeffler, J.M.; Djurkovic, S.; Fischetti, V.A. Phage lytic enzyme Cpl-1 as a novel antimicrobial for pneumococcal bacteremia. Infect. Immun. 2003, 71, 6199–6204. [Google Scholar] [CrossRef]
- Ho, M.K.Y.; Zhang, P.; Chen, X.; Xia, J.; Leung, S.S.Y. Bacteriophage endolysins against gram-positive bacteria, an overview on the clinical development and recent advances on the delivery and formulation strategies. Crit. Rev. Microbiol. 2022, 48, 303–326. [Google Scholar] [CrossRef]
- Xu, Y. Phage and phage lysins: New era of bio-preservatives and food safety agents. J. Food Sci. 2021, 86, 3349–3373. [Google Scholar] [CrossRef]
- Abdelrahman, F.; Easwaran, M.; Daramola, O.I.; Ragab, S.; Lynch, S.; Oduselu, T.J.; Khan, F.M.; Ayobami, A.; Adnan, F.; Torrents, E.; et al. Phage-encoded endolysins. Antibiotics 2021, 10, 124. [Google Scholar] [CrossRef]
- Kaur, J.; Singh, P.; Sharma, D.; Harjai, K.; Chhibber, S. A potent enzybiotic against methicillin-resistant Staphylococcus aureus. Virus Genes 2020, 56, 480–497. [Google Scholar] [CrossRef]
- Park, J.; Yun, J.; Lim, J.A.; Kang, D.H.; Ryu, S. Characterization of an endolysin, LysBPS13, from a Bacillus cereus bacteriophage. FEMS Microbiol. Lett. 2012, 332, 76–83. [Google Scholar] [CrossRef]
- Son, B.; Yun, J.; Lim, J.A.; Shin, H.; Heu, S.; Ryu, S. Characterization of LysB4, an endolysin from the Bacillus cereus-infecting bacteriophage B4. BMC Microbiol. 2012, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Schuch, R.; Pelzek, A.J.; Nelson, D.C.; Fischetti, V.A. The PlyB endolysin of bacteriophage vB_BanS_Bcp1 exhibits broad-spectrum bactericidal activity against Bacillus cereus sensu lato isolates. Appl. Environ. Microbiol. 2019, 85, e00003-19. [Google Scholar] [CrossRef] [PubMed]
- Haddad Kashani, H.; Fahimi, H.; Dasteh Goli, Y.; Moniri, R. A novel chimeric endolysin with antibacterial activity against methicillin-resistant Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2017, 7, 290. [Google Scholar] [CrossRef]
- Oliveira, H.; Thiagarajan, V.; Walmagh, M.; Sillankorva, S.; Lavigne, R.; Neves-Petersen, M.T.; Kluskens, L.D.; Azeredo, J. A thermostable Salmonella phage endolysin, Lys68, with broad bactericidal properties against gram-negative pathogens in presence of weak acids. PLoS ONE 2014, 9, e108376. [Google Scholar] [CrossRef]
- Gutierrez, D.; Ruas-Madiedo, P.; Martinez, B.; Rodríguez, A.; Garcia, P. Effective removal of staphylococcal biofilms by the endolysin LysH5. PLoS ONE 2014, 9, e107307. [Google Scholar] [CrossRef]
- Oliveira, H.; Vilas Boas, D.; Mesnage, S.; Kluskens, L.D.; Lavigne, R.; Sillankorva, S.; Secundo, F.; Azeredo, J. Structural and enzymatic characterization of ABgp46, a novel phage endolysin with broad anti-gram-negative bacterial activity. Front. Microbiol. 2016, 7, 184317. [Google Scholar] [CrossRef]
- Ji, W.; Huang, Q.; Sun, L.; Wang, H.; Yan, Y.; Sun, J. A novel endolysin disrupts Streptococcus suis with high efficiency. FEMS Microbiol. Lett. 2015, 362, fnv205. [Google Scholar] [CrossRef]
- Na, H.; Kong, M.; Ryu, S. Characterization of LysPBC4, a novel Bacillus cereus-specific endolysin of bacteriophage PBC4. FEMS Microbiol. Lett. 2016, 363, fnw092. [Google Scholar] [CrossRef]
- Swift, S.M.; Rowley, D.T.; Young, C.; Franks, A.; Hyman, P.; Donovan, D.M. The endolysin from the Enterococcus faecalis bacteriophage VD13 and conditions stimulating its lytic activity. FEMS Microbiol. Lett. 2016, 363, fnw216. [Google Scholar] [CrossRef]
- Lim, J.; Hong, J.; Jung, Y.; Ha, J.; Kim, H.; Myung, H.; Song, M. Bactericidal effect of Cecropin A fused endolysin on drug-resistant gram-negative pathogens. J. Microbiol. Biotechnol. 2022, 32, 816. [Google Scholar] [CrossRef]
- Lee, C.; Kim, J.; Son, B.; Ryu, S. Development of advanced chimeric endolysin to control multidrug-resistant Staphylococcus aureus through domain shuffling. ACS Infect. Dis. 2021, 7, 2081–2092. [Google Scholar] [CrossRef]
- Gómez-Torres, N.; Dunne, M.; Garde, S.; Meijers, R.; Narbad, A.; Ávila, M.; Mayer, M.J. Development of a specific fluorescent phage endolysin for in situ detection of Clostridium species associated with cheese spoilage. Microb. Biotechnol. 2018, 11, 332–345. [Google Scholar] [CrossRef] [PubMed]
- Proença, D.; Velours, C.; Leandro, C.; Garcia, M.; Pimentel, M.; São-José, C. A two-component, multimeric endolysin encoded by a single gene. Mol. Microbiol. 2015, 95, 739–753. [Google Scholar] [CrossRef] [PubMed]
- Kikkawa, H.S.; Ueda, T.; Suzuki, S.I.; Yasuda, J. Characterization of the catalytic activity of the γ-phage lysin, PlyG, specific for Bacillus anthracis. FEMS Microbiol. Lett. 2008, 286, 236–240. [Google Scholar] [CrossRef]
- Xu, D.; Zhao, S.; Dou, J.; Xu, X.; Zhi, Y.; Wen, L. Engineered endolysin-based “artilysins” for controlling the gram-negative pathogen Helicobacter pylori. AMB Express 2021, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Shavrina, M.S.; Zimin, A.A.; Molochkov, N.V.; Chernyshov, S.V.; Machulin, A.V.; Mikoulinskaia, G.V. In vitro study of the antibacterial effect of the bacteriophage T5 thermostable endolysin on Escherichia coli cells. J. Appl. Microbiol. 2016, 121, 1282–1290. [Google Scholar] [CrossRef] [PubMed]
- Springer, K.; Reuter, S.; Knüpfer, M.; Schmauder, L.; Sänger, P.A.; Felsl, A.; Fuchs, T.M. Activity of a holin-endolysin system in the insecticidal pathogenicity island of Yersinia enterocolitica. J. Bacteriol. 2018, 200, e00180-18. [Google Scholar] [CrossRef]
- Oh, H.K.; Hwang, Y.J.; Hong, H.W.; Myung, H. Comparison of Enterococcus faecalis biofilm removal efficiency among bacteriophage PBEF129, its endolysin, and cefotaxime. Viruses 2021, 13, 426. [Google Scholar] [CrossRef]
- Wang, W.; Li, M.; Lin, H.; Wang, J.; Mao, X. The Vibrio parahaemolyticus-infecting bacteriophage qdvp001: Genome sequence and endolysin with a modular structure. Arch. Virol. 2016, 161, 2645–2652. [Google Scholar] [CrossRef]
- Hermoso, J.A.; Monterroso, B.; Albert, A.; Galán, B.; Ahrazem, O.; García, P.; Martínez-Ripoll, M.; García, J.L.; Menéndez, M. Structural basis for selective recognition of pneumococcal cell wall by modular endolysin from phage Cp-1. Structure 2003, 11, 1239–1249. [Google Scholar] [CrossRef]
- Fokine, A.; Miroshnikov, K.A.; Shneider, M.M.; Mesyanzhinov, V.V.; Rossmann, M.G. Structure of the bacteriophage φKZ lytic transglycosylase gp144. J. Biol. Chem. 2008, 283, 7242–7250. [Google Scholar] [CrossRef] [PubMed]
- Korndörfer, I.P.; Danzer, J.; Schmelcher, M.; Zimmer, M.; Skerra, A.; Loessner, M.J. The crystal structure of the bacteriophage PSA endolysin reveals a unique fold responsible for specific recognition of Listeria cell walls. J. Mol. Biol. 2006, 364, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Korndörfer, I.P.; Kanitz, A.; Danzer, J.; Zimmer, M.; Loessner, M.J.; Skerra, A. Structural analysis of the L-alanoyl-D-glutamate endopeptidase domain of Listeria bacteriophage endolysin Ply500 reveals a new member of the LAS peptidase family. Acta Crystallogr. Sect. D Biol. Crystallogr. 2008, 64, 644–650. [Google Scholar] [CrossRef] [PubMed]
- Maciejewska, B.; Źrubek, K.; Espaillat, A.; Wiśniewska, M.; Rembacz, K.P.; Cava, F.; Dubin, G.; Drulis-Kawa, Z. Modular endolysin of Burkholderia AP3 phage has the largest lysozyme-like catalytic subunit discovered to date and no catalytic aspartate residue. Sci. Rep. 2017, 7, 14501. [Google Scholar] [CrossRef]
- Park, Y.; Lim, J.A.; Kong, M.; Ryu, S.; Rhee, S. Structure of bacteriophage SPN 1 S endolysin reveals an unusual two-module fold for the peptidoglycan lytic and binding activity. Mol. Microbiol. 2014, 92, 316–325. [Google Scholar] [CrossRef]
- Jun, S.Y.; Jang, I.J.; Yoon, S.; Jang, K.; Yu, K.S.; Cho, J.Y.; Seong, M.-W.; Jung, G.M.; Yoon, S.J.; Kang, S.H. Pharmacokinetics and tolerance of the phage endolysin-based candidate drug SAL200 after a single intravenous administration among healthy volunteers. Antimicrob. Agents Chemother. 2017, 61, e02629-16. [Google Scholar] [CrossRef]
- Love, M.J.; Bhandari, D.; Dobson, R.C.; Billington, C. Potential for bacteriophage endolysins to supplement or replace antibiotics in food production and clinical care. Antibiotics 2018, 7, 17. [Google Scholar] [CrossRef]
- Wertheim, H.F.; Melles, D.C.; Vos, M.C.; Van Leeuwen, W.; Van Belkum, A.; Verbrugh, H.A.; Nouwen, J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005, 5, 751–762. [Google Scholar] [CrossRef]
- Golban, M.; Charostad, J.; Kazemian, H.; Heidari, H. Phage-Derived Endolysins Against Resistant Staphylococcus spp.: A Review of Features, Antibacterial Activities, and Recent Applications. Infect. Dis. Ther. 2024, 1–45. [Google Scholar] [CrossRef]
- Azzopardi, E.A.; Azzopardi, S.M.; Boyce, D.E.; Dickson, W.A. Emerging gram-negative infections in burn wounds. J. Burn Care Res. 2011, 32, 570–576. [Google Scholar] [CrossRef]
- Briers, Y.; Walmagh, M.; Van Puyenbroeck, V.; Cornelissen, A.; Cenens, W.; Aertsen, A.; Oliveira, H.; Azeredo, J.; Verween, G.; Pirnay, J.-P.; et al. Engineered endolysin-based “Artilysins” to combat multidrug-resistant gram-negative pathogens. mBio 2014, 5, e01379-14. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.E.; Ward, M.J. Sources of antimicrobial resistance. Science 2013, 341, 1460–1461. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; Gutiérrez, D.; Rodríguez, A.; García, P. Application of bacteriophages in the agro-food sector: A long way toward approval. Front. Cell. Infect. Microbiol. 2018, 8, 296. [Google Scholar] [CrossRef] [PubMed]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Garvey, M. Foodborne Clostridioides Species: Pathogenicity, Virulence and Biocontrol Options. Microorganisms 2023, 11, 2483. [Google Scholar] [CrossRef]
- Jończyk-Matysiak, E.; Kłak, M.; Weber-Dąbrowska, B.; Borysowski, J.; Górski, A. Possible use of bacteriophages active against Bacillus anthracis and other B. cereus group members in the face of a bioterrorism threat. BioMed Res. Int. 2014, 2014, 735413. [Google Scholar] [CrossRef]
- Brunel, A.S.; Guery, B. Multidrug resistant (or antimicrobial-resistant) pathogens-alternatives to new antibiotics? Swiss Med. Wkly. 2017, 147, w14553. [Google Scholar]
- Zhang, H.; Bao, H.; Billington, C.; Hudson, J.A.; Wang, R. Isolation and lytic activity of the Listeria bacteriophage endolysin LysZ5 against Listeria monocytogenes in soya milk. Food Microbiol. 2012, 31, 133–136. [Google Scholar] [CrossRef]
- Van Nassau, T.J.; Lenz, C.A.; Scherzinger, A.S.; Vogel, R.F. Combination of endolysins and high pressure to inactivate Listeria monocytogenes. Food Microbiol. 2017, 68, 81–88. [Google Scholar] [CrossRef]
- Linden, S.B.; Alreja, A.B.; Nelson, D.C. Application of bacteriophage-derived endolysins to combat streptococcal disease: Current state and perspectives. Curr. Opin. Biotechnol. 2021, 68, 213–220. [Google Scholar] [CrossRef]
- Roach, D.R.; Khatibi, P.A.; Bischoff, K.M.; Hughes, S.R.; Donovan, D.M. Bacteriophage-encoded lytic enzymes control growth of contaminating Lactobacillus found in fuel ethanol fermentations. Biotechnol. Biofuels 2013, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Bristol, N. The US Government and Antimicrobial Resistance; Center for Strategic and International Studies (CSIS): Washington, DC, USA, 2020. [Google Scholar]
- O’flaherty, S.; Coffey, A.; Meaney, W.; Fitzgerald, G.F.; Ross, R.P. The recombinant phage lysin LysK has a broad spectrum of lytic activity against clinically relevant staphylococci, including methicillin-resistant Staphylococcus aureus. J. Bacteriol. 2005, 187, 7161–7164. [Google Scholar] [CrossRef] [PubMed]
- Filatova, L.Y.; Becker, S.C.; Donovan, D.M.; Gladilin, A.K.; Klyachko, N.L. LysK, the enzyme lysing Staphylococcus aureus cells: Specific kinetic features and approaches towards stabilization. Biochimie 2010, 92, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Daniel, A.; Euler, C.; Collin, M.; Chahales, P.; Gorelick, K.J.; Fischetti, V.A. Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2010, 54, 1603–1612. [Google Scholar] [CrossRef]
- Schuch, R.; Lee, H.M.; Schneider, B.C.; Sauve, K.L.; Law, C.; Khan, B.K. Combination therapy with lysin cf-301 and antibiotic is superior to antibiotic alone for treating MRS-induced murine bacteremia. J. Infect. Dis. 2013, 209, 1469–1478. [Google Scholar] [CrossRef]
- Altoparlak, U.; Erol, S.; Akcay, M.N.; Celebi, F.; Kadanali, A. The time-related changes of antimicrobial resistance patterns and predominant bacterial profiles of burn wounds and body flora of burned patients. Burns 2004, 30, 660–664. [Google Scholar] [CrossRef]
- Chopra, S.; Harjai, K.; Chhibber, S. Potential of combination therapy of endolysin MR-10 and minocycline in treating MRSA induced systemic and localized burn wound infections in mice. Int. J. Med. Microbiol. 2016, 306, 707–716. [Google Scholar] [CrossRef]
- Totté, J.E.; van Doorn, M.B.; Pasmans, S.G. Successful treatment of chronic Staphylococcus aureus-related dermatoses with the topical endolysin Staphefekt SA. 100: A report of 3 cases. Case Rep. Dermatol. 2017, 9, 19–25. [Google Scholar] [CrossRef]
- Rashel, M.; Uchiyama, J.; Ujihara, T.; Uehara, Y.; Kuramoto, S.; Sugihara, S.; Yagyu, K.-I.; Muraoka, A.; Sugai, M.; Hiramatsu, K.; et al. Efficient elimination of multidrug-resistant Staphylococcus aureus by cloned lysin derived from bacteriophage ϕMR11. J. Infect. Dis. 2007, 196, 1237–1247. [Google Scholar] [CrossRef]
- Shields, B.E.; Tschetter, A.J.; Wanat, K.A. Staphylococcus simulans: An emerging cutaneous pathogen. JAAD Case Rep. 2016, 2, 428–429. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus epidermidis—The ‘accidental’ pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.Y.; Yoon, J.W.; Hovde, C.J. A brief overview of Escherichia coli O157: H7 and its plasmid O157. J. Microbiol. Biotechnol. 2010, 20, 5. [Google Scholar] [CrossRef]
- Chique, C.; Hynds, P.; Burke, L.P.; Morris, D.; Ryan, M.P.; O’Dwyer, J. Contamination of domestic groundwater systems by verotoxigenic Escherichia coli (VTEC), 2003–2019: A global scoping review. Water Res. 2021, 188, 116496. [Google Scholar] [CrossRef]
- Hilbert, D.W. Uropathogenic Escherichia coli: The pre-eminent urinary tract infection pathogen. In E. coli Infections: Causes, Treatment and Prevention; Nova Science Publishers: Hauppauge, NY, USA, 2011. [Google Scholar]
- Wu, D.; Ding, Y.; Yao, K.; Gao, W.; Wang, Y. Antimicrobial resistance analysis of clinical Escherichia coli isolates in neonatal ward. Front. Pediatr. 2021, 9, 670470. [Google Scholar] [CrossRef]
- Murray, B.E. The life and times of the Enterococcus. Clin. Microbiol. Rev. 1990, 3, 46–65. [Google Scholar] [CrossRef]
- Hammerum, A.M. Enterococci of animal origin and their significance for public health. Clin. Microbiol. Infect. 2012, 18, 619–625. [Google Scholar] [CrossRef]
- Courvalin, P. Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 2006, 42 (Suppl. S1), S25–S34. [Google Scholar] [CrossRef]
- Yoong, P.; Schuch, R.; Nelson, D.; Fischetti, V.A. Identification of a broadly active phage lytic enzyme with lethal activity against antibiotic-resistant Enterococcus faecalis and Enterococcus faecium. J. Bacteriol. 2004, 186, 4808–4812. [Google Scholar] [CrossRef]
- Son, J.; Jun, S.; Kim, E.; Park, J.; Paik, H.; Yoon, S.; Kang, S.; Choi, Y.-J. Complete genome sequence of a newly isolated lytic bacteriophage, EFAP-1 of Enterococcus faecalis, and antibacterial activity of its endolysin EFAL-1. J. Appl. Microbiol. 2010, 108, 1769–1779. [Google Scholar] [CrossRef]
- Zhang, W.; Mi, Z.; Yin, X.; Fan, H.; An, X.; Zhang, Z.; Chen, J.; Tong, Y. Characterization of Enterococcus faecalis phage IME-EF1 and its endolysin. PLoS ONE 2013, 8, e80435. [Google Scholar] [CrossRef]
- Cheng, M.; Zhang, Y.; Li, X.; Liang, J.; Hu, L.; Gong, P.; Zhang, L.; Cai, R.; Zhang, H.; Ge, J.; et al. Endolysin LysEF-P10 shows potential as an alternative treatment strategy for multidrug-resistant Enterococcus faecalis infections. Sci. Rep. 2017, 7, 10164. [Google Scholar] [CrossRef] [PubMed]
- Proença, D.; Leandro, C.; Garcia, M.; Pimentel, M.; São-José, C. EC300: A phage-based, bacteriolysin-like protein with enhanced antibacterial activity against Enterococcus faecalis. Appl. Microbiol. Biotechnol. 2015, 99, 5137–5149. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, M.R. Drug-resistant Streptococcus pneumoniae: Rational antibiotic choices. Am. J. Med. 1999, 106, 19–25. [Google Scholar] [CrossRef]
- Loeffler, J.M.; Nelson, D.; Fischetti, V.A. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 2001, 294, 2170–2172. [Google Scholar] [CrossRef]
- Jedrzejas, M.J. Pneumococcal virulence factors: Structure and function. Microbiol. Mol. Biol. Rev. 2001, 65, 187–207. [Google Scholar] [CrossRef]
- García, J.L.; García, E.; Arrarás, A.; García, P.; Ronda, C.; López, R. Cloning, purification, and biochemical characterization of the pneumococcal bacteriophage Cp-1 lysin. J. Virol. 1987, 61, 2573–2580. [Google Scholar] [CrossRef]
- Tran-Quang, K.; Nguyen-Thi-Dieu, T.; Tran-Xuan, B.; Larsson, M.; Duong-Quy, S. Antibiotic resistance of Streptococcus pneumoniae in Vietnamese children with severe pneumonia: A cross-sectional study. Front. Public Health 2023, 11, 1110903. [Google Scholar] [CrossRef]
- Howard, A.; O’Donoghue, M.; Feeney, A.; Sleator, R.D. Acinetobacter baumannii: An emerging opportunistic pathogen. Virulence 2012, 3, 243–250. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef]
- Bodey, G.P.; Bolivar, R.; Fainstein, V.; Jadeja, L. Infections caused by Pseudomonas aeruginosa. Rev. Infect. Dis. 1983, 5, 279–313. [Google Scholar] [CrossRef] [PubMed]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.J.; Lin, N.T.; Hu, A.; Soo, P.C.; Chen, L.K.; Chen, L.H.; Chang, K.C. Antibacterial activity of Acinetobacter baumannii phage ϕAB2 endolysin (LysAB2) against both gram-positive and gram-negative bacteria. Appl. Microbiol. Biotechnol. 2011, 90, 529–539. [Google Scholar] [CrossRef]
- Lood, R.; Winer, B.Y.; Pelzek, A.J.; Diez-Martinez, R.; Thandar, M.; Euler, C.W.; Schuch, R.; Fischetti, V.A. Novel phage lysin capable of killing the multidrug-resistant gram-negative bacterium Acinetobacter baumannii in a mouse bacteremia model. Antimicrob. Agents Chemother. 2015, 59, 1983–1991. [Google Scholar] [CrossRef] [PubMed]
- Walmagh, M.; Briers, Y.; Santos, S.B.D.; Azeredo, J.; Lavigne, R. Characterization of modular bacteriophage endolysins from Myoviridae phages OBP, 201φ2-1 and PVP-SE1. PLoS ONE 2012, 7, e36991. [Google Scholar] [CrossRef]
- Sato, H.; Feix, J.B. Peptide–membrane interactions and mechanisms of membrane destruction by amphipathic α-helical antimicrobial peptides. Biochim. Biophys. Acta (BBA)-Biomembr. 2006, 1758, 1245–1256. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Kalu, M.; Tan, K.; Algorri, M.; Jorth, P.; Wong-Beringer, A. In-Human Multiyear Evolution of Carbapenem-Resistant Klebsiella pneumoniae Causing Chronic Colonization and Intermittent Urinary Tract Infections: A Case Study. Msphere 2022, 7, e00190-22. [Google Scholar] [CrossRef]
- Lou, W.; Venkataraman, S.; Zhong, G.; Ding, B.; Tan, J.P.; Xu, L.; Fan, W.; Yang, Y.Y. Antimicrobial polymers as therapeutics for treatment of multidrug-resistant Klebsiella pneumoniae lung infection. Acta Biomater. 2018, 78, 78–88. [Google Scholar] [CrossRef]
- Cabello, F.C. Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and for the environment. Environ. Microbiol. 2006, 8, 1137–1144. [Google Scholar] [CrossRef]
- Timbermont, L.; Haesebrouck, F.; Ducatelle, R.; Van Immerseel, F. Necrotic enteritis in broilers: An updated review on the pathogenesis. Avian Pathol. 2011, 40, 341–347. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Goes, E.C.; Wakaruk, J.; Barreda, D.R.; Korver, D.R. A poultry subclinical Necrotic enteritis disease model based on natural Clostridium perfringens uptake. Front. Physiol. 2022, 13, 788592. [Google Scholar] [CrossRef] [PubMed]
- Swift, S.M.; Seal, B.S.; Garrish, J.K.; Oakley, B.B.; Hiett, K.; Yeh, H.Y.; Woolsey, R.; Schegg, K.M.; Line, J.E.; Donovan, D.M. A thermophilic phage endolysin fusion to a Clostridium perfringens-specific cell wall binding domain creates an anti-Clostridium antimicrobial with improved thermostability. Viruses 2015, 7, 3019–3034. [Google Scholar] [CrossRef]
- Tamai, E.; Yoshida, H.; Sekiya, H.; Nariya, H.; Miyata, S.; Okabe, A.; Kuwahara, T.; Maki, J.; Kamitori, S. X-ray structure of a novel endolysin encoded by episomal phage phiSM 101 of Clostridium perfringens. Mol. Microbiol. 2014, 92, 326–337. [Google Scholar] [CrossRef]
- Nariya, H.; Miyata, S.; Tamai, E.; Sekiya, H.; Maki, J.; Okabe, A. Identification and characterization of a putative endolysin encoded by episomal phage phiSM101 of Clostridium perfringens. Appl. Microbiol. Biotechnol. 2011, 90, 1973–1979. [Google Scholar] [CrossRef]
- Mak, P.H.; Rehman, M.A.; Kiarie, E.G.; Topp, E.; Diarra, M.S. Production systems and important antimicrobial resistant-pathogenic bacteria in poultry: A review. J. Anim. Sci. Biotechnol. 2022, 13, 148. [Google Scholar] [CrossRef]
- Cantas, L.; Shah, S.Q.; Cavaco, L.M.; Walsh, F.; Popowska, M.; Garelick, H.; Bürgmann, H. A brief multi-disciplinary review on antimicrobial resistance in medicine and its linkage to the global environmental microbiota. Front. Microbiol. 2013, 4, 42415. [Google Scholar] [CrossRef]
- Fasanella, A.; Galante, D.; Garofolo, G.; Jones, M.H. Anthrax undervalued zoonosis. Vet. Microbiol. 2010, 140, 318–331. [Google Scholar] [CrossRef]
- Inglesby, T.V.; O’Toole, T.; Henderson, D.A.; Bartlett, J.G.; Ascher, M.S.; Eitzen, E.; Friedlander, A.M.; Hauer, J.; McDade, J.; Osterholm, M.T. Working Group on Civilian Biodefense. Anthrax as a biological weapon, 2002: Updated recommendations for management. JAMA 2002, 287, 2236–2252. [Google Scholar] [CrossRef]
- Schuch, R.; Nelson, D.; Fischetti, V.A. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 2002, 418, 884–889. [Google Scholar] [CrossRef]
- Arafa, A.A.; Hedia, R.H.; Ata, N.S.; Ibrahim, E.S. Vancomycin resistant Streptococcus equi subsp. equi isolated from equines suffering from respiratory manifestation in Egypt. Vet. World 2021, 14, 1808. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Tchang, V.S.; Loessner, M.J. Domain shuffling and module engineering of Listeria phage endolysins for enhanced lytic activity and binding affinity. Microb. Biotechnol. 2011, 4, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, K.; Egberink, H.; Pennisi, M.G.; Lloret, A.; Addie, D.; Belák, S.; Boucraut-Baralon, C.; Frymus, T.; Gruffydd-Jones, T.; Hosie, M.J. Leptospira species infection in cats: ABCD guidelines on prevention and management. J. Feline Med. Surg. 2013, 15, 576–581. [Google Scholar] [CrossRef]
- Hoopes, J.T.; Stark, C.J.; Kim, H.A.; Sussman, D.J.; Donovan, D.M.; Nelson, D.C. Use of a bacteriophage lysin, PlyC, as an enzyme disinfectant against Streptococcus equi. Appl. Environ. Microbiol. 2009, 75, 1388–1394. [Google Scholar] [CrossRef]
- McEwen, S.A. Antibiotic use in animal agriculture: What have we learned and where are we going? Anim. Biotechnol. 2006, 17, 239–250. [Google Scholar] [CrossRef]
- Silbergeld, E.K.; Graham, J.; Price, L.B. Industrial food animal production, antimicrobial resistance, and human health. Annu. Rev. Public Health 2008, 29, 151–169. [Google Scholar] [CrossRef]
- Yi, L.; Jin, M.; Li, J.; Grenier, D.; Wang, Y. Antibiotic resistance related to biofilm formation in Streptococcus suis. Appl. Microbiol. Biotechnol. 2020, 104, 8649–8660. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, J.H.; Lu, C.P. Purified recombinant phage lysin LySMP: An extensive spectrum of lytic activity for swine streptococci. Curr. Microbiol. 2009, 58, 609–615. [Google Scholar] [CrossRef]
- Hennekinne, J.A.; De Buyser, M.L.; Dragacci, S. Staphylococcus aureus and its food poisoning toxins: Characterization and outbreak investigation. FEMS Microbiol. Rev. 2012, 36, 815–836. [Google Scholar] [CrossRef]
- Chang, Y.; Kim, M.; Ryu, S. Characterization of a novel endolysin LysSA11 and its utility as a potent biocontrol agent against Staphylococcus aureus on food and utensils. Food Microbiol. 2017, 68, 112–120. [Google Scholar] [CrossRef]
- Sahu, B.; Singh, S.D.; Behera, B.K.; Panda, S.K.; Das, A.; Parida, P.K. Rapid detection of Salmonella contamination in seafoods using multiplex PCR. Braz. J. Microbiol. 2019, 50, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rubio, L.; Gerstmans, H.; Thorpe, S.; Mesnage, S.; Lavigne, R.; Briers, Y. DUF3380 domain from a Salmonella phage endolysin shows potent N-acetylmuramidase activity. Appl. Environ. Microbiol. 2016, 82, 4975–4981. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.A.; Shin, H.; Kang, D.H.; Ryu, S. Characterization of endolysin from a Salmonella typhimurium-infecting bacteriophage SPN1S. Res. Microbiol. 2012, 163, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Endersen, L.; Guinane, C.M.; Johnston, C.; Neve, H.; Coffey, A.; Ross, R.P.; McAuliffe, O.; O’Mahony, J. Genome analysis of Cronobacter phage vB_CsaP_Ss1 reveals an endolysin with potential for biocontrol of Gram-negative bacterial pathogens. J. Gen. Virol. 2015, 96, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Endersen, L.; Coffey, A.; Ross, R.P.; McAuliffe, O.; Hill, C.; O’Mahony, J. Characterisation of the antibacterial properties of a bacterial derived peptidoglycan hydrolase (LysCs4), active against C. sakazakii and other Gram-negative food-related pathogens. Int. J. Food Microbiol. 2015, 215, 79–85. [Google Scholar] [CrossRef]
- Vidic, J.; Chaix, C.; Manzano, M.; Heyndrickx, M. Food sensing: Detection of Bacillus cereus spores in dairy products. Biosensors 2020, 10, 15. [Google Scholar] [CrossRef]
- Loessner, M.J.; Maier, S.K.; Daubek-Puza, H.; Wendlinger, G.; Scherer, S. Three Bacillus cereus bacteriophage endolysins are unrelated but reveal high homology to cell wall hydrolases from different bacilli. J. Bacteriol. 1997, 179, 2845–2851. [Google Scholar] [CrossRef]
- Mayer, M.J.; Payne, J.; Gasson, M.J.; Narbad, A. Genomic sequence and characterization of the virulent bacteriophage ΦCTP1 from Clostridium tyrobutyricum and heterologous expression of its endolysin. Appl. Environ. Microbiol. 2010, 76, 5415–5422. [Google Scholar] [CrossRef]
- Mayer, M.J.; Gasson, M.J.; Narbad, A. Genomic sequence of bacteriophage ATCC 8074-B1 and activity of its endolysin and engineered variants against Clostridium sporogenes. Appl. Environ. Microbiol. 2012, 78, 3685–3692. [Google Scholar] [CrossRef]
- Hathaway, H.; Ajuebor, J.; Stephens, L.; Coffey, A.; Potter, U.; Sutton, J.M.; Jenkins, A.T.A. Thermally triggered release of the bacteriophage endolysin CHAPK and the bacteriocin lysostaphin for the control of methicillin resistant Staphylococcus aureus (MRSA). J. Control. Release 2017, 245, 108–115. [Google Scholar] [CrossRef]
- Maciejewska, B.; Olszak, T.; Drulis-Kawa, Z. Applications of bacteriophages versus phage enzymes to combat and cure bacterial infections: An ambitious and also a realistic application? Appl. Microbiol. Biotechnol. 2018, 102, 2563–2581. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.M.; Valencia, P.M.; Zhang, L.; Langer, R.; Farokhzad, O.C. Polymeric nanoparticles for drug delivery. Cancer Nanotechnol. Methods Protoc. 2010, 624, 163–175. [Google Scholar]
- Matamp, N.; Bhat, S.G. Phage endolysins as potential antimicrobials against multidrug resistant Vibrio alginolyticus and Vibrio parahaemolyticus: Current status of research and challenges ahead. Microorganisms 2019, 7, 84. [Google Scholar] [CrossRef]
- Jun, S.Y.; Jung, G.M.; Yoon, S.J.; Oh, M.D.; Choi, Y.J.; Lee, W.J.; Kong, J.-C.; Seol, J.G.; Kang, S.H. Antibacterial properties of a pre-formulated recombinant phage endolysin, SAL-1. Int. J. Antimicrob. Agents 2013, 41, 156–161. [Google Scholar] [CrossRef]
- Jun, S.Y.; Jung, G.M.; Son, J.S.; Yoon, S.J.; Choi, Y.J.; Kang, S.H. Comparison of the antibacterial properties of phage endolysins SAL-1 and LysK. Antimicrob. Agents Chemother. 2011, 55, 1764–1767. [Google Scholar] [CrossRef]
- Zhang, L.; Li, D.; Li, X.; Hu, L.; Cheng, M.; Xia, F.; Gong, P.; Wang, B.; Ge, J.; Zhang, H.; et al. LysGH15 kills Staphylococcus aureus without being affected by the humoral immune response or inducing inflammation. Sci. Rep. 2016, 6, 29344. [Google Scholar] [CrossRef]
- Jingmin, G.; Zuo, J.; Lei, L.; Zhao, H.; Sun, C.; Feng, X.; Du, C.; Li, X.; Yang, Y.; Han, W. LysGH15 reduces the inflammation caused by lethal methicillin-resistant Staphylococcus aureus infection in mice. Bioeng. Bugs 2011, 2, 96–99. [Google Scholar]
- Entenza, J.M.; Loeffler, J.M.; Grandgirard, D.; Fischetti, V.A.; Moreillon, P. Therapeutic effects of bacteriophage Cpl-1 lysin against Streptococcus pneumoniae endocarditis in rats. Antimicrob. Agents Chemother. 2005, 49, 4789–4792. [Google Scholar] [CrossRef]
- McCullers, J.A.; Karlström, Å.; Iverson, A.R.; Loeffler, J.M.; Fischetti, V.A. Novel strategy to prevent otitis media caused by colonizing Streptococcus pneumoniae. PLOS Pathog. 2007, 3, e28. [Google Scholar] [CrossRef]
- Grandgirard, D.; Loeffler, J.M.; Fischetti, V.A.; Leib, S.L. Phage lytic enzyme Cpl-1 for antibacterial therapy in experimental Pneumococcal meningitis. J. Infect. Dis. 2008, 197, 1519–1522. [Google Scholar] [CrossRef]
- Witzenrath, M.; Schmeck, B.; Doehn, J.M.; Tschernig, T.; Zahlten, J.; Loeffler, J.M.; Zemlin, M.; Müller, H.; Gutbier, B.; Schütte, H.; et al. Systemic use of the endolysin Cpl-1 rescues mice with fatal Pneumococcal pneumonia. Crit. Care Med. 2009, 37, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Lood, R.; Raz, A.; Molina, H.; Euler, C.W.; Fischetti, V.A. A highly active and negatively charged Streptococcus pyogenes lysin with a rare D-alanyl-L-alanine endopeptidase activity protects mice against streptococcal bacteremia. Antimicrob. Agents Chemother. 2014, 58, 3073–3084. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Shen, Y.; Nelson, D.C.; Eugster, M.R.; Eichenseher, F.; Hanke, D.C.; Loessner, M.J.; Dong, S.; Pritchard, D.G.; Lee, J.C.; et al. Evolutionarily distinct bacteriophage endolysins featuring conserved peptidoglycan cleavage sites protect mice from MRSA infection. J. Antimicrob. Chemother. 2015, 70, 1453–1465. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Nelson, D.; Zhu, S.; Fischetti, V.A. Removal of group B streptococci colonizing the vagina and oropharynx of mice with a bacteriophage lytic enzyme. Antimicrob. Agents Chemother. 2005, 49, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Raz, A.; Serrano, A.; Hernandez, A.; Euler, C.W.; Fischetti, V.A. Isolation of phage lysins that effectively kill Pseudomonas aeruginosa in mouse models of lung and skin infection. Antimicrob. Agents Chemother. 2019, 63, e00024-19. [Google Scholar] [CrossRef]
- Doehn, J.M.; Fischer, K.; Reppe, K.; Gutbier, B.; Tschernig, T.; Hocke, A.C.; Fischetti, V.A.; Löffler, J.; Suttorp, N.; Hippenstiel, S.; et al. Delivery of the endolysin Cpl-1 by inhalation rescues mice with fatal Pneumococcal pneumonia. J. Antimicrob. Chemother. 2013, 68, 2111–2117. [Google Scholar] [CrossRef]
- Zhou, B.; Zhen, X.; Zhou, H.; Zhao, F.; Fan, C.; Perčulija, V.; Tong, Y.; Mi, Z.; Ouyang, S. Structural and functional insights into a novel two-component endolysin encoded by a single gene in Enterococcus faecalis phage. PLoS Pathog. 2020, 16, e1008394. [Google Scholar] [CrossRef]
- Kim, S.; Lee, D.W.; Jin, J.S.; Kim, J. Antimicrobial activity of LysSS, a novel phage endolysin, against Acinetobacter baumannii and Pseudomonas aeruginosa. J. Glob. Antimicrob. Resist. 2020, 22, 32–39. [Google Scholar] [CrossRef]
- Horgan, M.; O’Flynn, G.; Garry, J.; Cooney, J.; Coffey, A.; Fitzgerald, G.F.; Ross, R.P.; McAuliffe, O. Phage lysin LysK can be truncated to its CHAP domain and retain lytic activity against live antibiotic-resistant staphylococci. Appl. Environ. Microbiol. 2009, 75, 872–874. [Google Scholar] [CrossRef]
- Sui, B.; Wang, X.; Zhao, T.; Zhen, J.; Ren, H.; Liu, W.; Zhang, X.; Zhang, C. Design, screening, and characterization of engineered phage endolysins with extracellular antibacterial activity against Gram-negative bacteria. Appl. Environ. Microbiol. 2023, 89, e00581-23. [Google Scholar] [CrossRef]
- Peng, S.Y.; You, R.I.; Lai, M.J.; Lin, N.T.; Chen, L.K.; Chang, K.C. Highly potent antimicrobial modified peptides derived from the Acinetobacter baumannii phage endolysin LysAB2. Sci. Rep. 2017, 7, 11477. [Google Scholar] [CrossRef] [PubMed]
- Díez-Martínez, R.; de Paz, H.; Bustamante, N.; García, E.; Menéndez, M.; García, P. Improving the lethal effect of Cpl-7, a pneumococcal phage lysozyme with broad bactericidal activity, by inverting the net charge of its cell wall-binding module. Antimicrob. Agents Chemother. 2013, 57, 5355–5365. [Google Scholar] [CrossRef] [PubMed]
- Díez-Martínez, R.; De Paz, H.D.; García-Fernández, E.; Bustamante, N.; Euler, C.W.; Fischetti, V.A.; Menendez, M.; García, P. A novel chimeric phage lysin with high in vitro and in vivo bactericidal activity against Streptococcus pneumoniae. J. Antimicrob. Chemother. 2015, 70, 1763–1773. [Google Scholar] [CrossRef] [PubMed]
- Proença, D.; Fernandes, S.; Leandro, C.; Silva, F.A.; Santos, S.; Lopes, F.; Mato, R.; Cavaco-Silva, P.; Pimentel, M.; São-José, C. Phage endolysins with broad antimicrobial activity against Enterococcus faecalis clinical strains. Microb. Drug Resist. 2012, 18, 322–332. [Google Scholar] [CrossRef]
- Fernandes, S.; Proença, D.; Cantante, C.; Silva, F.A.; Leandro, C.; Lourenço, S.; Milheiriço, C.; de Lencastre, H.; Cavaco-Silva, P.; Pimentel, M.; et al. Novel chimerical endolysins with broad antimicrobial activity against methicillin-resistant Staphylococcus aureus. Microb. Drug Resist. 2012, 18, 333–343. [Google Scholar] [CrossRef]
- Oyston, P.C.F.; Fox, M.A.; Richards, S.J.; Clark, G.C. Novel peptide therapeutics for treatment of infections. J. Med. Microbiol. 2009, 58, 977–987. [Google Scholar] [CrossRef]
- Auclair, S.M.; Bhanu, M.K.; Kendall, D.A. Signal peptidase I: Cleaving the way to mature proteins. Protein Sci. 2012, 21, 13–25. [Google Scholar] [CrossRef]
- Gaeng, S.; Scherer, S.; Neve, H.; Loessner, M.J. Gene cloning and expression and secretion of Listeria monocytogenes bacteriophage-lytic enzymes in Lactococcus lactis. Appl. Environ. Microbiol. 2000, 66, 2951–2958. [Google Scholar] [CrossRef]
- Gervasi, T.; Horn, N.; Wegmann, U.; Dugo, G.; Narbad, A.; Mayer, M.J. Expression and delivery of an endolysin to combat Clostridium perfringens. Appl. Microbiol. Biotechnol. 2014, 98, 2495–2505. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; Gutiérrez, D.; Martínez, B.; Rodríguez, A.; García, P. Lytic activity of LysH5 endolysin secreted by Lactococcus lactis using the secretion signal sequence of bacteriocin Lcn972. Appl. Environ. Microbiol. 2012, 78, 3469–3472. [Google Scholar] [CrossRef]
- Kim, N.H.; Park, W.B.; Cho, J.E.; Choi, Y.J.; Choi, S.J.; Jun, S.Y.; Kang, C.K.; Song, K.-H.; Choe, P.G.; Bang, J.-H.; et al. Effects of phage endolysin SAL200 combined with antibiotics on Staphylococcus aureus infection. Antimicrob. Agents Chemother. 2018, 62, e00731-18. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabur, A.; Khan, A.; Borphukan, B.; Razzak, A.; Salimullah, M.; Khatun, M. The Unique Capability of Endolysin to Tackle Antibiotic Resistance: Cracking the Barrier. J. Xenobiot. 2025, 15, 19. https://doi.org/10.3390/jox15010019
Sabur A, Khan A, Borphukan B, Razzak A, Salimullah M, Khatun M. The Unique Capability of Endolysin to Tackle Antibiotic Resistance: Cracking the Barrier. Journal of Xenobiotics. 2025; 15(1):19. https://doi.org/10.3390/jox15010019
Chicago/Turabian StyleSabur, Abdus, Angkan Khan, B. Borphukan, Abdur Razzak, M. Salimullah, and Muslima Khatun. 2025. "The Unique Capability of Endolysin to Tackle Antibiotic Resistance: Cracking the Barrier" Journal of Xenobiotics 15, no. 1: 19. https://doi.org/10.3390/jox15010019
APA StyleSabur, A., Khan, A., Borphukan, B., Razzak, A., Salimullah, M., & Khatun, M. (2025). The Unique Capability of Endolysin to Tackle Antibiotic Resistance: Cracking the Barrier. Journal of Xenobiotics, 15(1), 19. https://doi.org/10.3390/jox15010019









