An Amplicon-Based Sequencing Approach for the Study of Aeromycology †
Introduction
Materials and Methods
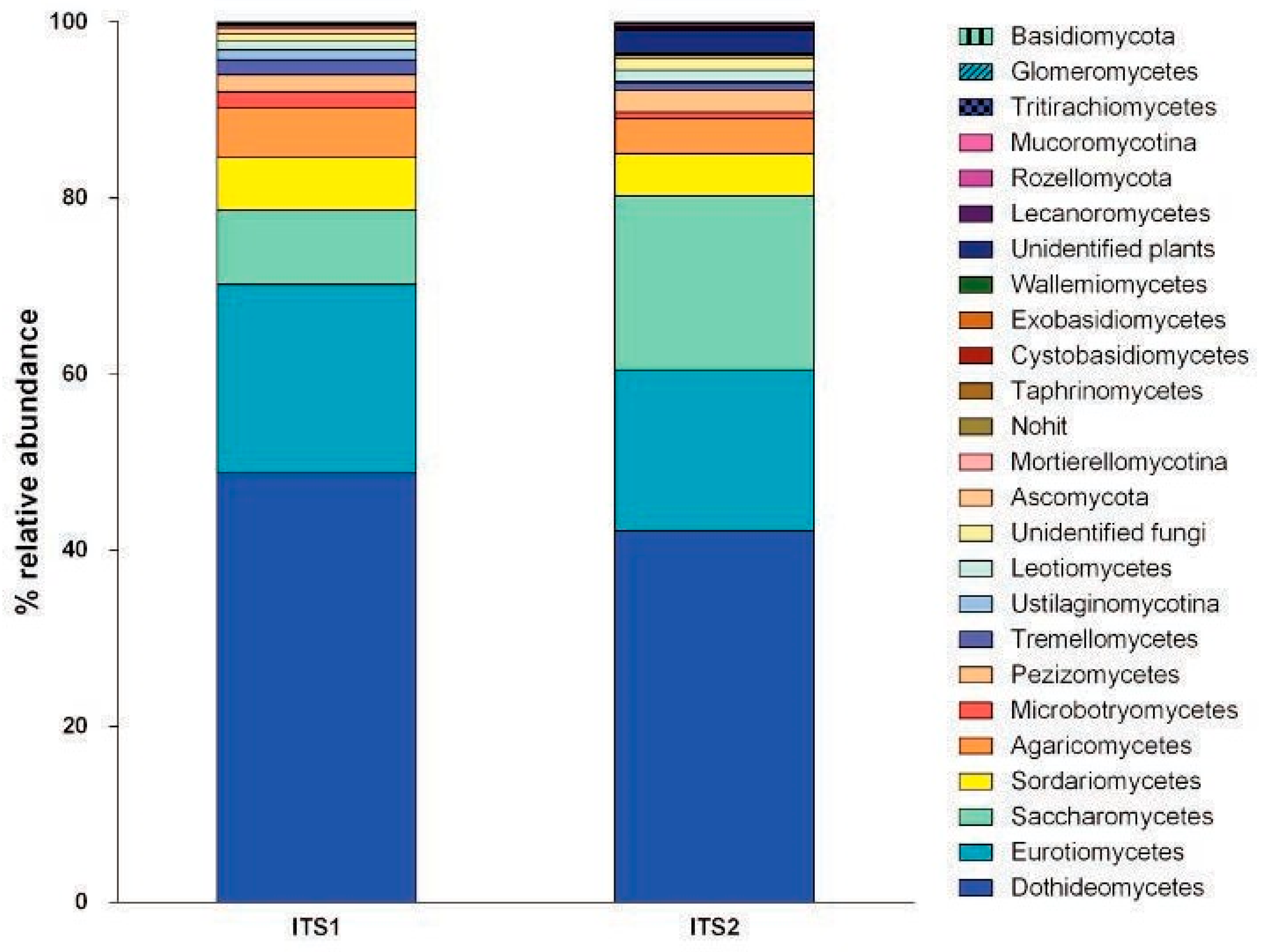
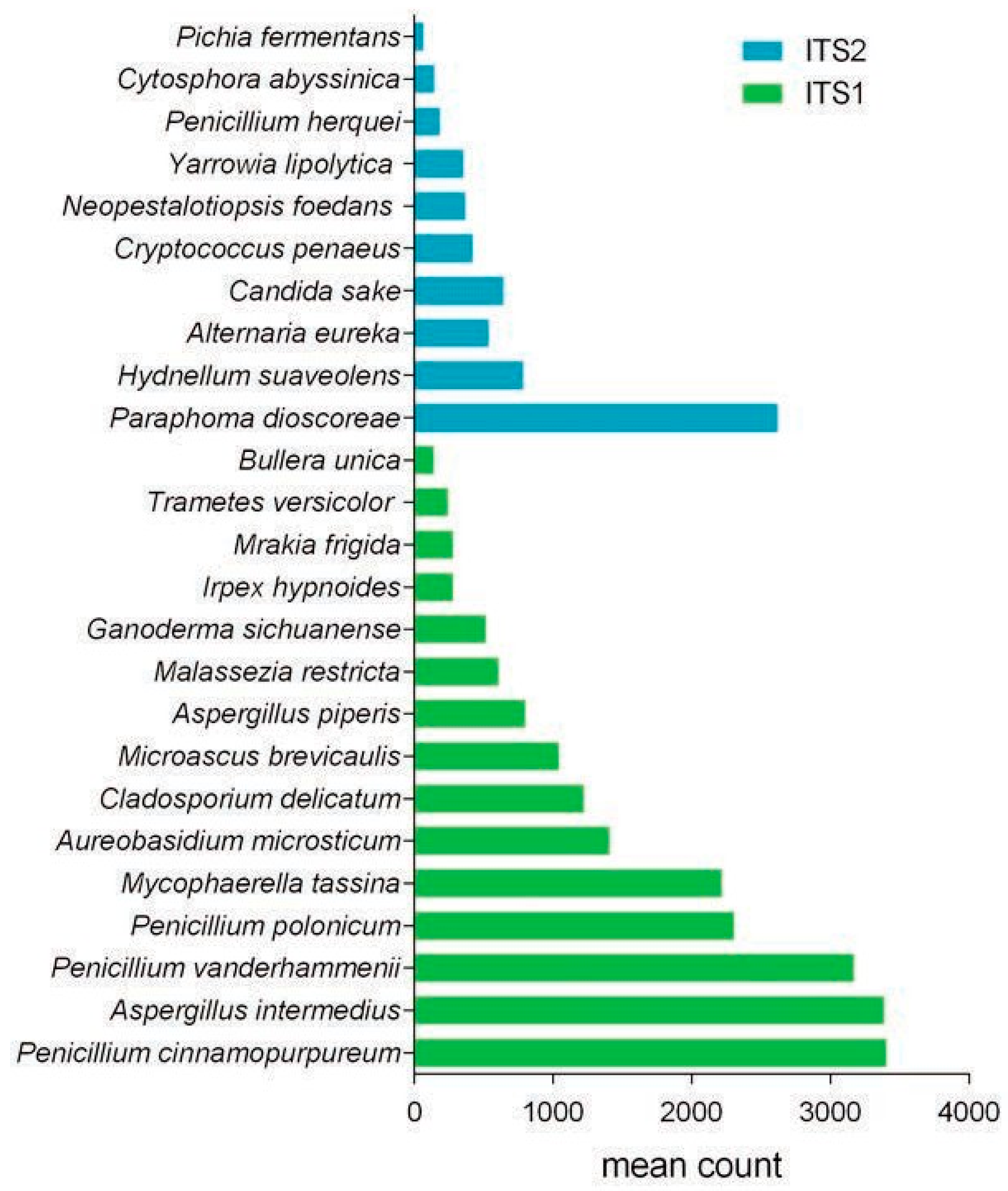
Results and Discussion
Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ruzer, L.S.; Harley, N.H. Aerosols hand-book: measurement, dosimetry and health effects; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Kakde, U.B. Fungal bioaerosols: global diversity, distribution and its impact on human beings and agricultural crops. Bionano Front 2012, 5, 323–9. [Google Scholar]
- Gilbert, Y.; Duchaine, C. Bioaerosols in industrial environments: a review. Canad J Civil Engin 2009, 36, 1873–86. [Google Scholar] [CrossRef]
- Mbareche, H.; Veillette, M.; Bonifait, L.; Dubuis, M.E.; Bernard, Y.; Marchand, G.; et al. A next generation sequencing approach with a suitable bioinformatics workflow to study fungal diversity in bioaerosols released from two different types of compost plants. Sci Total Environ 2017, 601–2. [Google Scholar] [CrossRef] [PubMed]
- Mbareche, H.; Veillette, M.; Dubuis, M.E.; Bakhiyi, B.; Zayed, J.; Lavoie, J.; et al. Fungal bioaerosols in biomethanization facilities. J Air Waste Manag Assoc 2018, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mbareche, H.; Veillette, M.; Bilodeau, G.J.; Duchaine, C. Fungal bioaerosols at five dairy farms: a novel approach to describe workers’ exposure. 2018. Available online: https://www.biorxiv.org/content/early/2 018/04/26/308825.full.pdf+html.
- Tischer, C.G.; Heinrich, J. Exposure assessment of residential mould, Fungi and microbial components in relation to children’s health: Achievements and challenges. Int J Hyg Environ Health 2013, 216, 109–14. [Google Scholar] [CrossRef] [PubMed]
- Roe, A.D.; Rice, A.V.; Bromilow, S.E.; Cooke, J.E.; Sperling, F.A. Multilocus species identification and fungal DNA barcoding: insights from blue stain fungal symbionts of the mountain pine beetle. Molec Ecol Resour 2010, 10, 946–59. [Google Scholar] [CrossRef] [PubMed]
- Dentinger, B.T.; Didukh, M.Y.; Moncalvo, J.M. Comparing COI and ITS as DNA barcode markers for mushrooms and allies (Agaricomycotina). PLoS ONE 2011, 6, e25081. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Nation Acad Sci Unit States Am 2012, 109, 6241–6. [Google Scholar]
- Tedersoo, L.; Anslan, S.; Bahram, M.; Polme, S.; Riit, T.; Liiv, I.; et al. Shotgun metagenomes and multiple pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 2015, 10, 1–43. [Google Scholar] [CrossRef]
- Perotto, S.; Nepote-Fus, P.; Saletta, L.; Bandi, C.; Young, J.P.W. W. A diverse population of introns in the nuclear ribosomal genes of ericoid mycorrhizal fungi includes elements with sequence similarity to endonuclease-coding genes. Mol Biol Evol 2000, 17, 44–59. [Google Scholar] [PubMed]
- Bhattacharya, D.; Lutzoni, F.; Reeb, V.; Simon, D.; Nason, J.; Fernandez, F. Widespread occurrence of spliceosomal introns in the rDNA genes of Ascomycetes. Molec Biol Evol 2000, 17, 1971–84. [Google Scholar] [CrossRef] [PubMed]
- Vrålstad, T.; Myhre, E.; Schumacher, T. ; Schumacher, T. Molecular diversity and phylogenetic affinities of symbiotic root-associated ascomycetes of the Helotiales in burnt and metal polluted habitats. New Phytol 2002, 155, 131–48. [Google Scholar] [PubMed]
- Bellemain, E.; Carlsen, T.; Brochmann, C.; Coissac, E.; Taberlet, P.; Kauserud, H. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiology 2010, 10, 189. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Zhu, Y.; Chen, X.; Liao, B.; Yao, H.; Song, J.; et al. The short ITS2 sequence serves as an efficient taxonomic sequence tag in comparison with the full-length, I.T.S. T.S. BioMed Res Int 2013, 741476. [Google Scholar] [CrossRef] [PubMed]
- McDowell, D.G.; Burns, N.A.; Parkes, H.C. Localised sequence regions possessing high melting temperatures prevent the amplification of a DNA mimic in competitive PCR. Nucl Acid Res 1998, 26, 3340–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Liu, C.; Huang, L.; Bengtsson-Palme, J.; Chen, H.; Zhang, J.H.; et al. ITS1: a DNA barcode better than ITS2 in eukaryotes? Molec Ecol Resour 2015, 15, 573–86. [Google Scholar] [CrossRef] [PubMed]
© Copyright H. Mbareche et al., 2018. Licensee PAGEPress, Italy. This work is licensed under a Creative Commons Attribution NonCommercial 4.0 License (CC BY-NC 4.0).
Share and Cite
Mbareche, H.; Veillette, M.; Bilodeau, G.J.; Duchaine, C. An Amplicon-Based Sequencing Approach for the Study of Aeromycology. J. Xenobiot. 2018, 8, 7810. https://doi.org/10.4081/xeno.2018.7810
Mbareche H, Veillette M, Bilodeau GJ, Duchaine C. An Amplicon-Based Sequencing Approach for the Study of Aeromycology. Journal of Xenobiotics. 2018; 8(1):7810. https://doi.org/10.4081/xeno.2018.7810
Chicago/Turabian StyleMbareche, Hamza, Marc Veillette, Guillaume J. Bilodeau, and Caroline Duchaine. 2018. "An Amplicon-Based Sequencing Approach for the Study of Aeromycology" Journal of Xenobiotics 8, no. 1: 7810. https://doi.org/10.4081/xeno.2018.7810
APA StyleMbareche, H., Veillette, M., Bilodeau, G. J., & Duchaine, C. (2018). An Amplicon-Based Sequencing Approach for the Study of Aeromycology. Journal of Xenobiotics, 8(1), 7810. https://doi.org/10.4081/xeno.2018.7810



