Phage Adsorption to Gram-Positive Bacteria
Abstract
:1. Introduction
| Phage | Morpho | Host | Receptor | Reference |
|---|---|---|---|---|
| Gram-positive hosts | ||||
| Gamma | S | Bacillus anthracis | Membrane surface-anchored protein (GamR) | [22] |
| PBP1 | S | Bacillus pumilus | Flagella (reversible) | [23] |
| SPP1 | S | Bacillus subtilis | Glc residues on WTA (reversible) and membrane protein YueB (irreversible) | [24] |
| PBS1 | S | B. subtilis | Flagella | [25] |
| CPS1 | P | Clostridium perfringens | Capsular polysaccharides (glucosamine and galactosamine) | [26] |
| VPE25 | S | Enterococcus faecalis | Membrane protein (PIPEF) as secondary receptor | [27] |
| ΦNPV1 | S | E. faecalis | Exopolysaccharides as primary receptor | [28] |
| LL-H | S | Lactobacillus delbrueckii | Glu moiety of LTA (reversible) and negatively charged GroP group of the LTA (irreversible) | [4] |
| B1 | S | Lactobacillus plantarum | Galactose of CW polysaccharides | [29] |
| c2 | S | Lactobacillus lactis | Rha in PG (reversible) and membrane protein (PIP) (irreversible) | [30] |
| p2 | S | L. lactis | CW pellicle | [31] |
| CHPC971 | S | L. lactis | CW pellicle | [32] |
| A511 | M | Listeria monocytogenes | GlcNAc and Rha on WTA and PG | [33] |
| A118 | S | L. monocytogenes | Rha on WTA | [34] |
| P35 | S | L. monocytogenes | GlcNAc and Rha on WTA | [35] |
| Φ11 | S | Staphylococcus aureus | GlcNAc residue in RboP WTA, O-acetylated PG | [36] |
| 187 | S | S. aureus | GalNAc residue in GroP WTA | [37] |
| P68 | P | S. aureus | β-GlcNAc residue in RboP WTA | [38] |
| SA012 | M | S. aureus | α-GlcNAc residue and backbone of RboP WTA | [39] |
| SA039 | M | S. aureus | β-GlcNAc residue and backbone of RboP WTA | [9] |
| K | M | S. aureus | Backbone of WTA | [36] |
| Dp-1 | S | Streptococcus pneumoniae | Choline containing teichoic acids | [37] |
| 9871 | S | Streptococcus thermophilus | Exopolysaccharides | [40] |
| CHPC951 | S | S. thermophilus | Rha-Glc CW polysaccharides | [41] |
| Gram-negative | ||||
| 7-7-1 | M | Agrobacterium sp. | Flagella and LPS | [42] |
| F336 | M | Campylobacter jejuni | Capsular polysaccharides | [43] |
| φCb13 | S | Caulobacter crescentus | Flagellum and pilus portals on the cell pole | [44] |
| T4 | M | Escherichia coli | OmpC and LPS | [45] |
| T5 | S | E. coli | Polymannose sequence in LPS and FhuA | [5,6] |
| λ | S | E. coli | LamB protein | [46] |
| SRD2021 | S | Klebsiella pneumoniae | Capsular polysaccharide | [47] |
| MPK7 | P | Pseudomonas aeruginosa | Type IV pili | [48] |
| JG004 | M | P. aeruginosa | LPS | [49] |
| P22 | P | Salmonella enterica | α-Rhamnosyl 1–3 galactose linkage of LPS O-chain | [50] |
| iEPS5 | S | S. enterica | Flagella molecular ruler protein FliK | [51] |
| SPC35 | S | S. enterica | BtuB protein | [52] |
| L-413C | M | Yersinia pestis | Terminal GlcNAc LPS outer core. HepII/HepIII and HepI/Glc residues also involved | [53] |
| YepE2 | P | Y. pestis | HepII/HepIII LPS inner core | [54] |
2. The Bacterial Side
2.1. The Gram-Negative Cell Surface
2.2. The Gram-Positive Cell Surface
2.2.1. The Peptidoglycan Layer and Associated Proteins
2.2.2. Gram-Positive Secondary Cell Wall Polysaccharides
2.3. Other Associated Envelope Components
2.3.1. S-Layer
2.3.2. Capsules and Exopolysaccharides (EPS)
2.3.3. Cell Appendages
3. The Phage Side
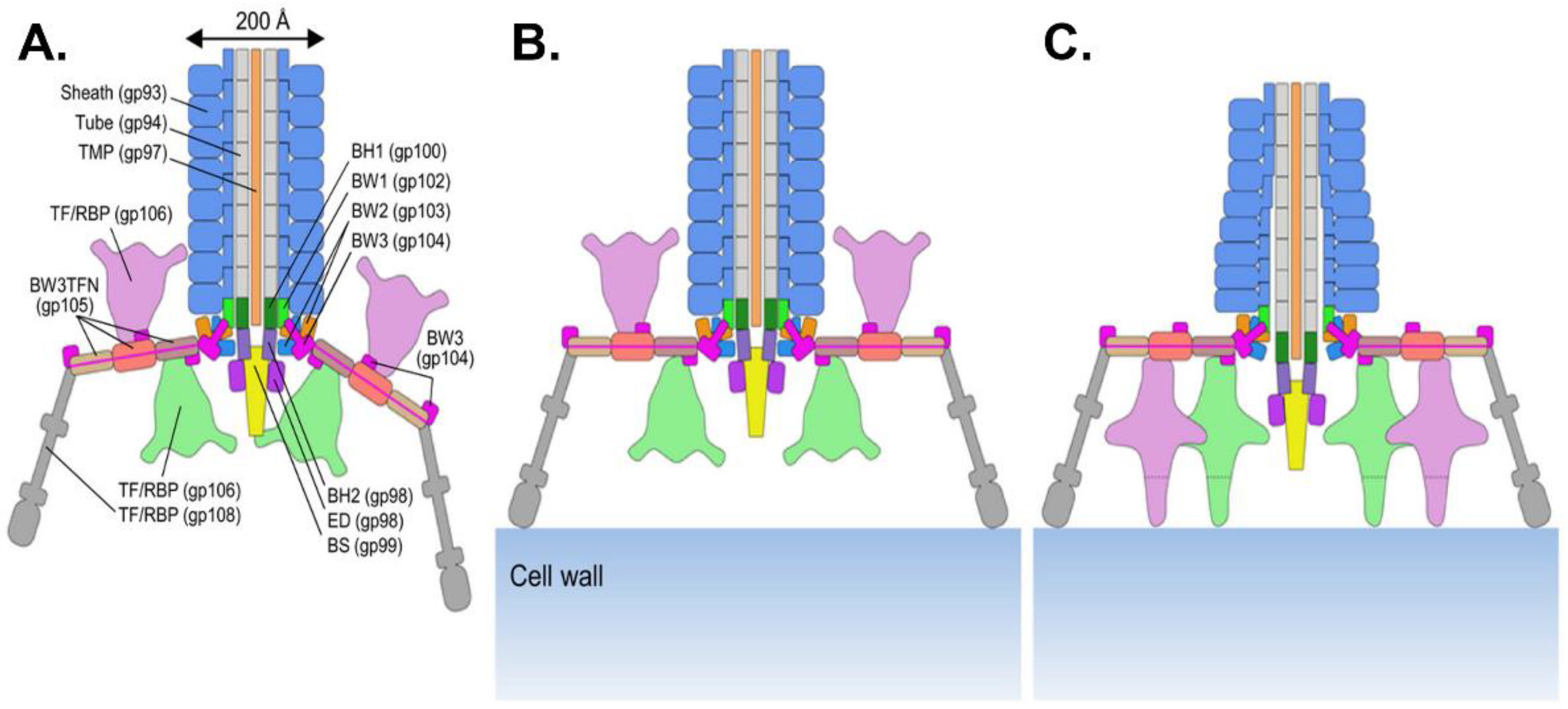
3.1. Myoviruses
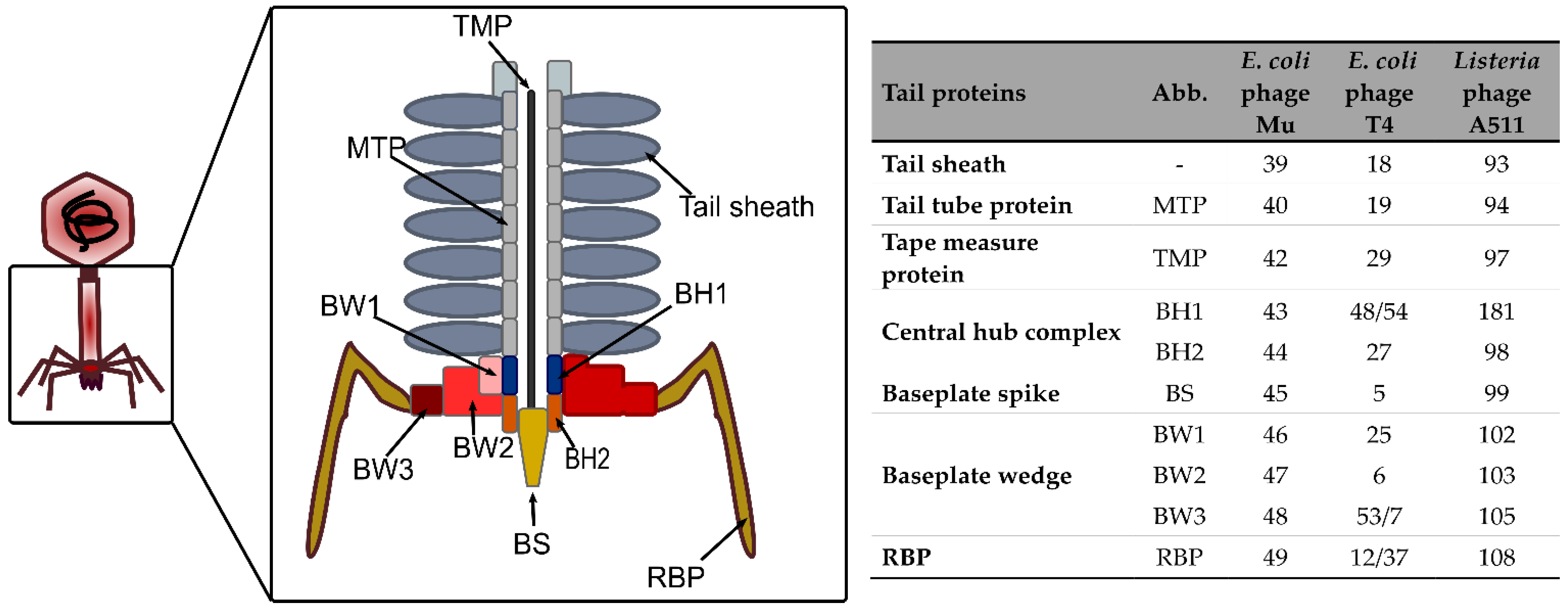
3.1.1. The Tail Tube
3.1.2. The Baseplate
3.1.3. Tail Proteins Associated with Enzymatic Activity
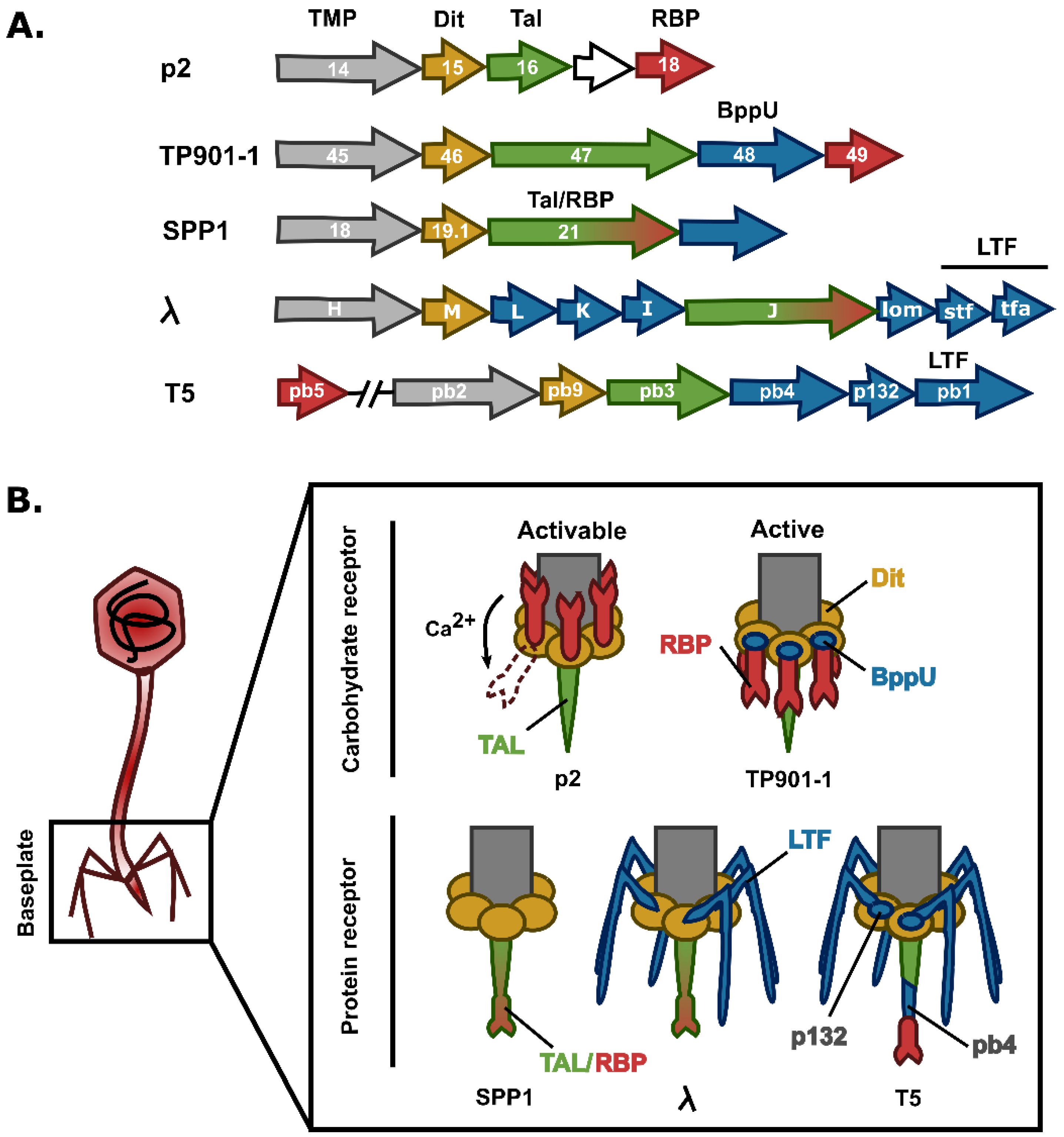
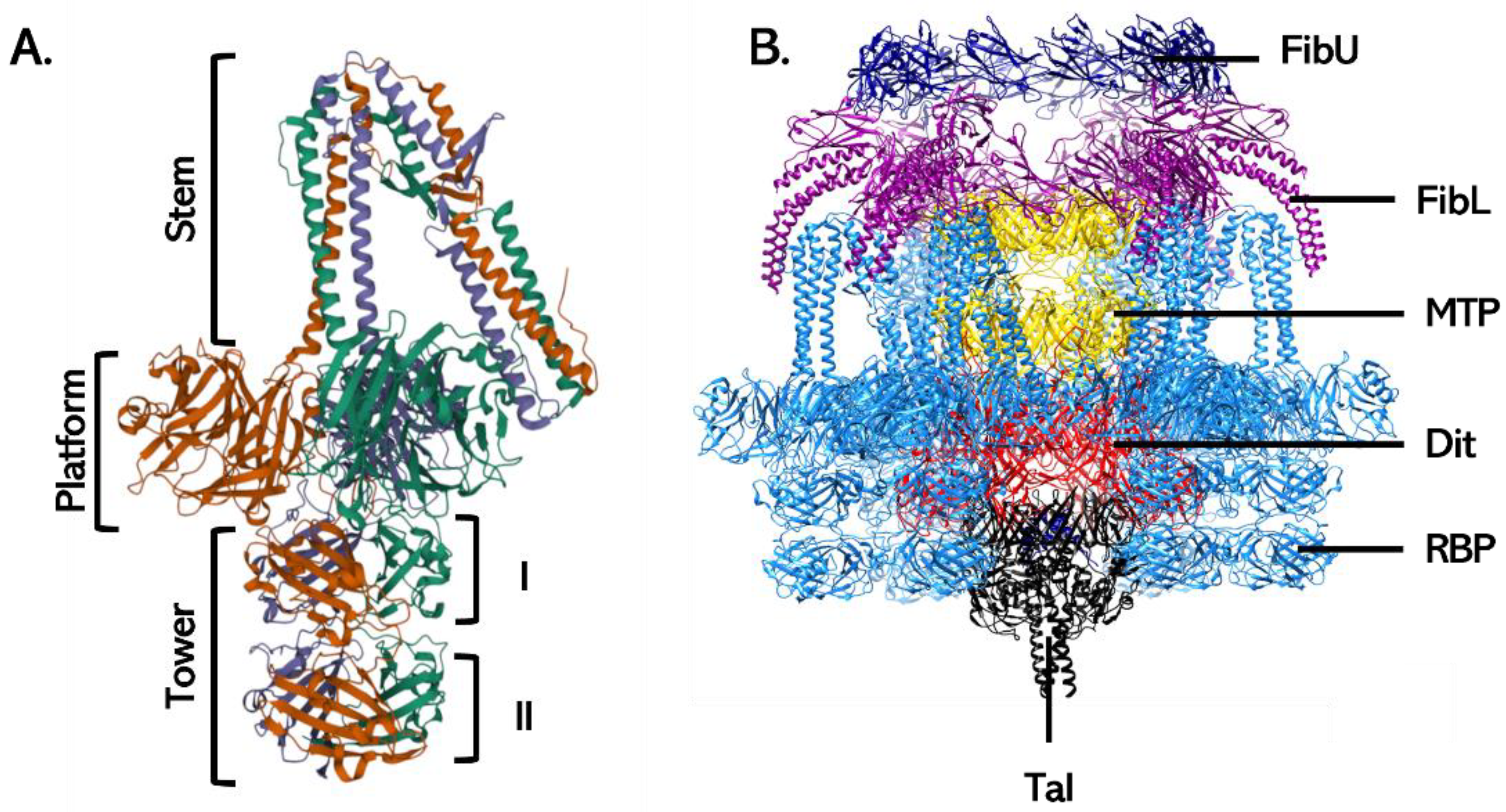
3.2. Siphoviruses
3.2.1. The Tail Tube
3.2.2. Baseplate Proteins
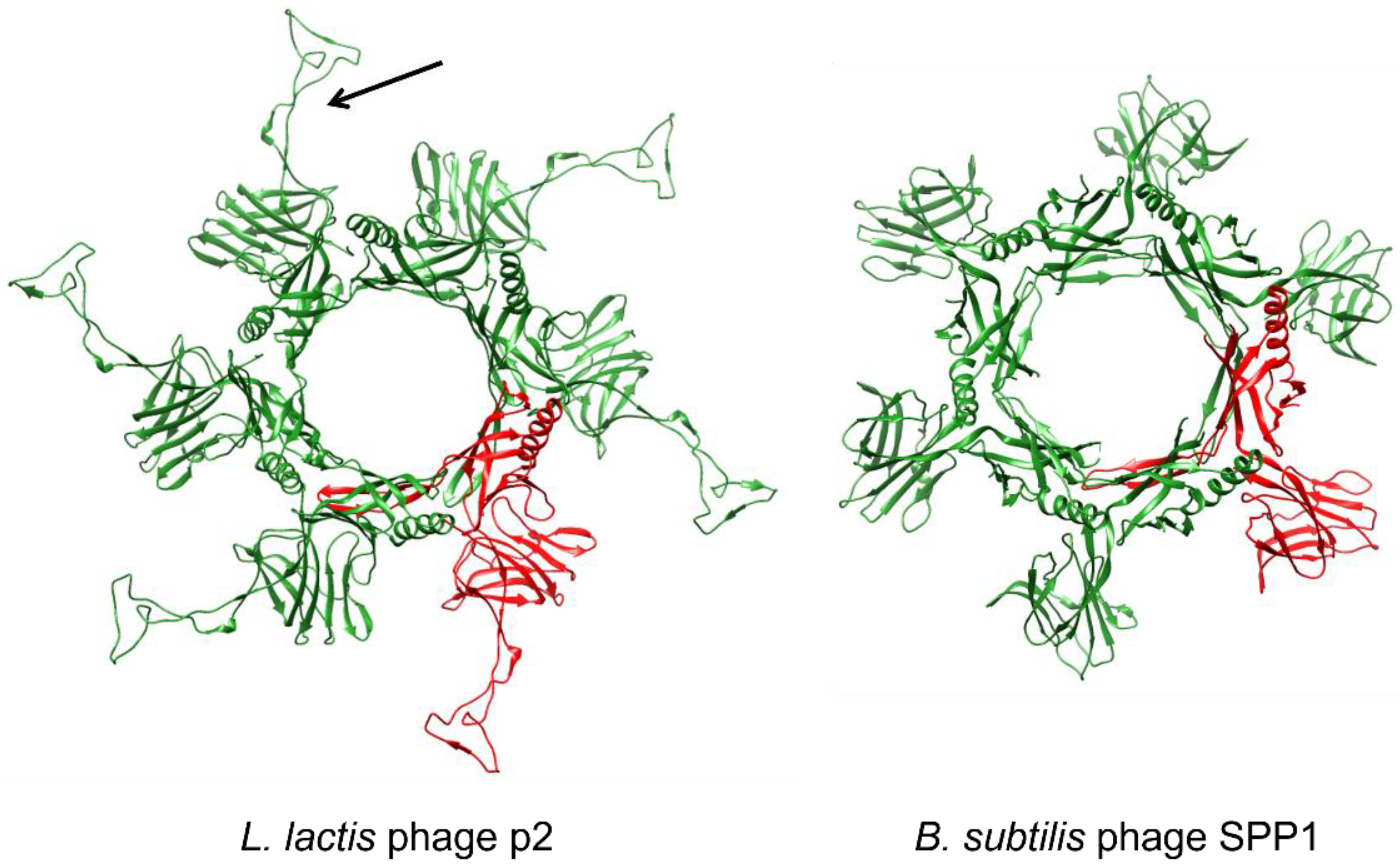
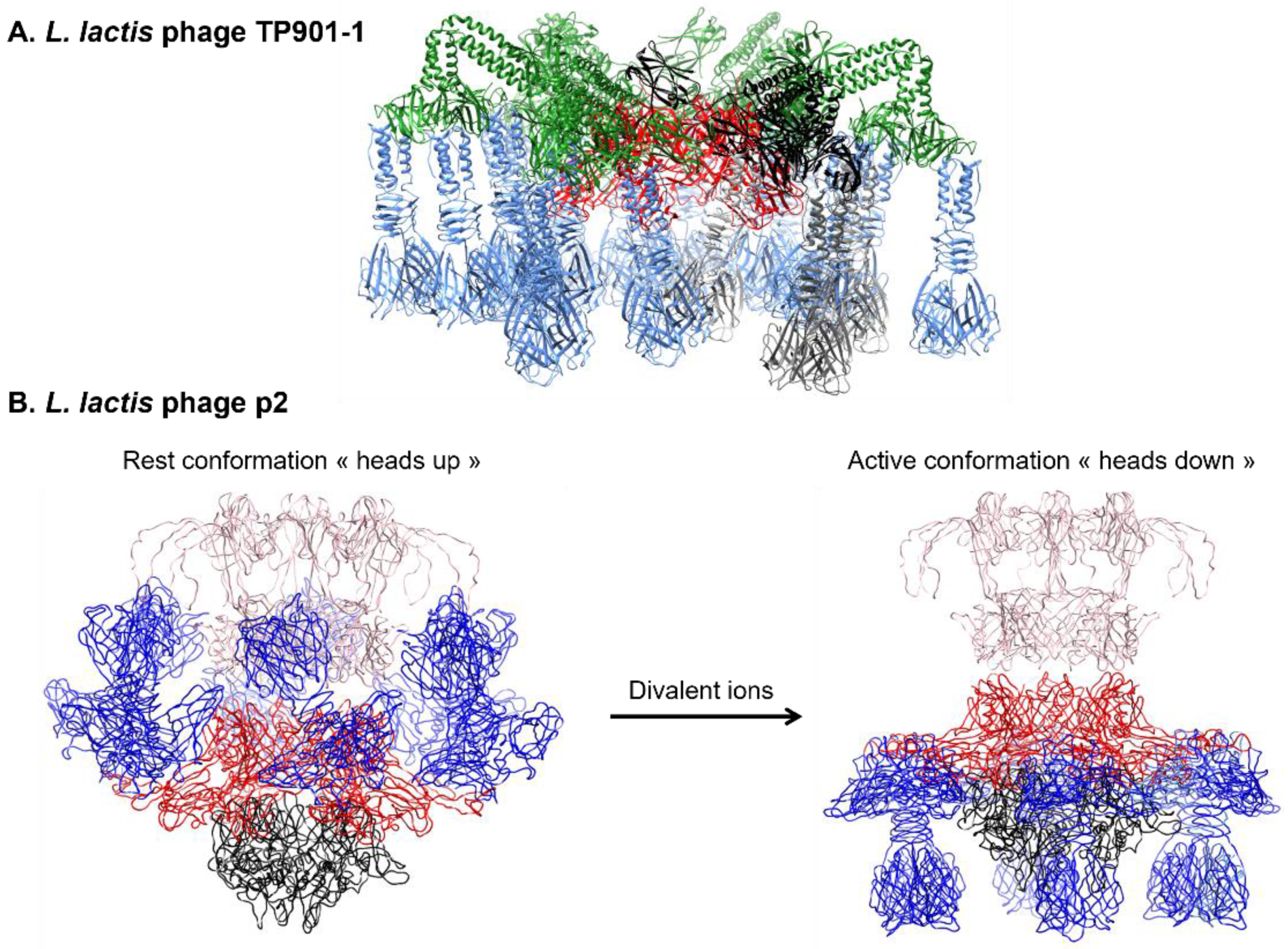
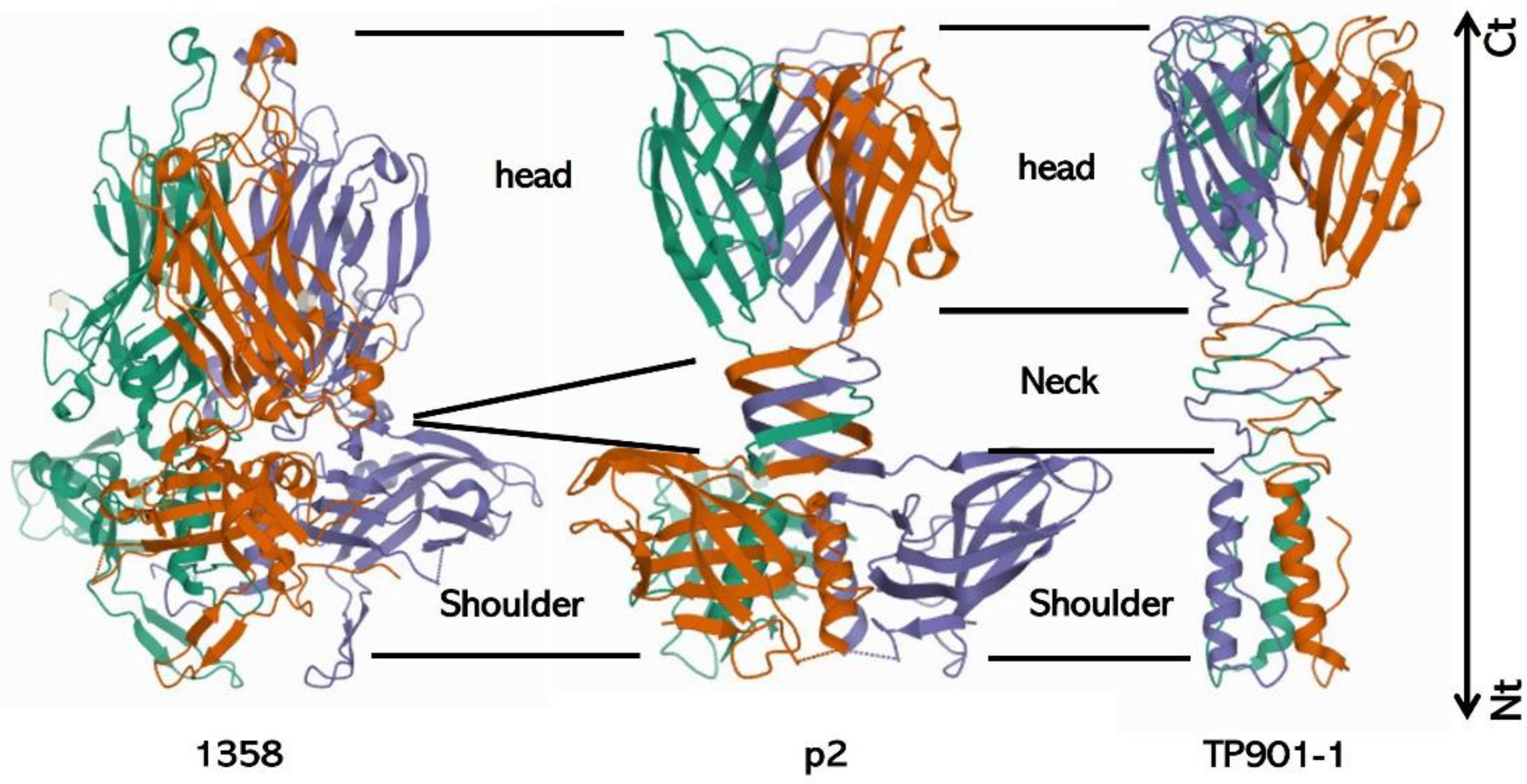
3.2.3. Baseplate Architecture
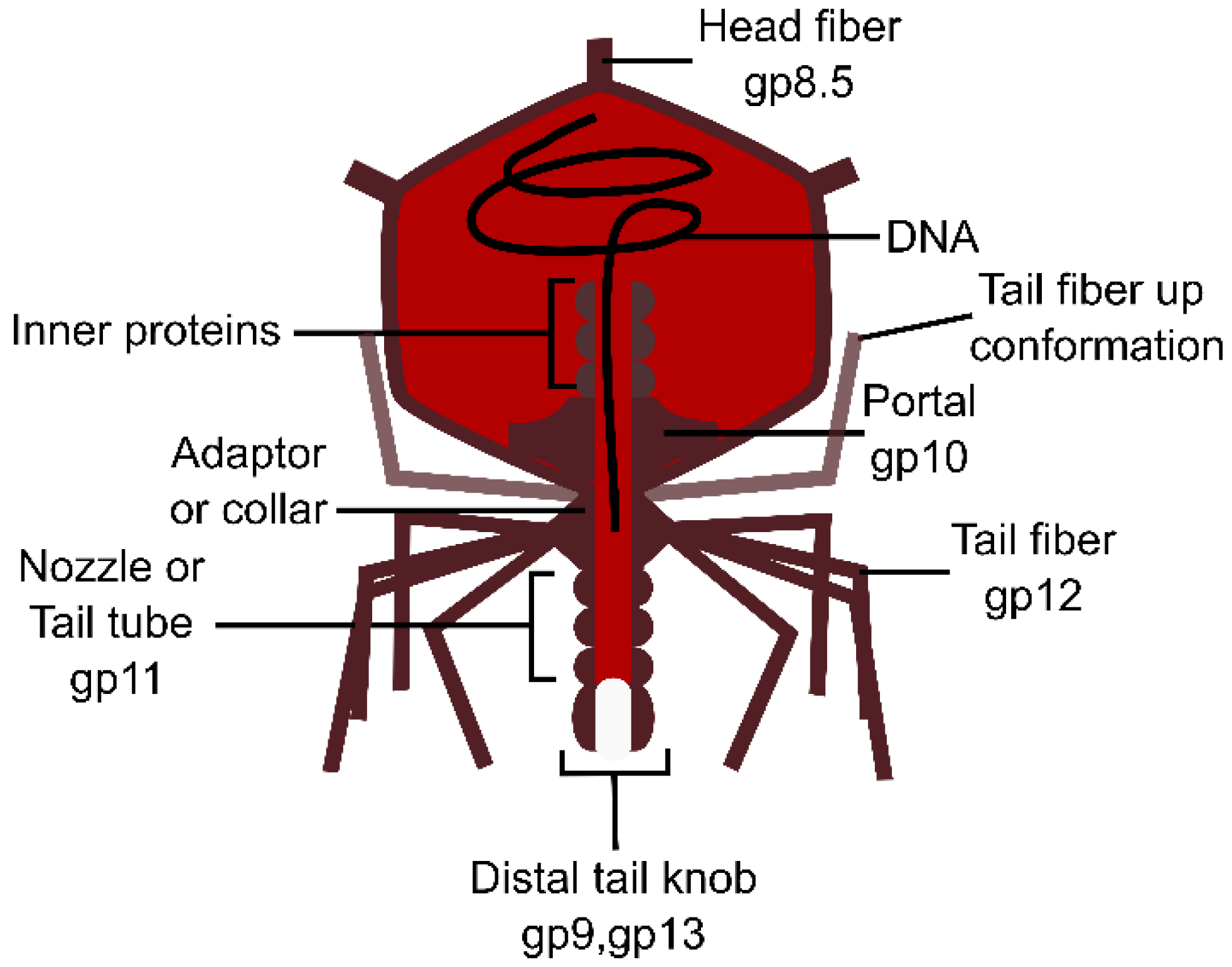
3.3. Podoviruses
4. Adsorption of Phages Infecting Gram-Positive Bacteria
4.1. Phages Infecting Lactic Acid Bacteria
4.1.1. Lactococcal Phages
4.1.2. Streptococcal Phages
4.2. Listeria spp. Phages
4.3. Staphylococcal Phages
4.4. Bacillus spp. Phages
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nobrega, F.L.; Vlot, M.; de Jonge, P.A.; Dreesens, L.L.; Beaumont, H.J.E.; Lavigne, R.; Dutilh, B.E.; Brouns, S.J.J. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 2018, 16, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Munsch-Alatossava, P.; Alatossava, T. The extracellular phage-host interactions involved in the bacteriophage ll-h infection of Lactobacillus Delbrueckii sp. Lactis ATCC 15808. Front. Microbiol. 2013, 4, 408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Degroux, S.; Effantin, G.; Linares, R.; Schoehn, G.; Breyton, C. Deciphering bacteriophage T5 host recognition mechanism and infection trigger. BioRxiv 2022. [Google Scholar] [CrossRef]
- Van den Berg, B.; Silale, A.; Baslé, A.; Brandner, A.F.; Mader, S.L.; Khalid, S. Structural basis for host recognition and superinfection exclusion by bacteriophage T5. Proc. Natl. Acad. Sci. USA 2022, 119, e2211672119. [Google Scholar] [CrossRef]
- Hampton, H.G.; Watson, B.N.J.; Fineran, P.C. The arms race between bacteria and their phage foes. Nature 2020, 577, 327–336. [Google Scholar] [CrossRef]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
- Azam, A.H.; Hoshiga, F.; Takeuchi, I.; Miyanaga, K.; Tanji, Y. Analysis of phage resistance in Staphylococcus aureus SA003 reveals different binding mechanisms for the closely related Twort-like phages ΦSA012 and ΦSA039. Appl. Microbiol. Biotechnol. 2018, 102, 8963–8977. [Google Scholar] [CrossRef]
- Scholl, D.; Adhya, S.; Merril, C. Escherichia coli K1′s capsule is a barrier to bacteriophage T7. Appl. Environ. Microbiol. 2005, 71, 4872–4874. [Google Scholar] [CrossRef]
- Gordillo Altamirano, F.; Forsyth, J.H.; Patwa, R.; Kostoulias, X.; Trim, M.; Subedi, D.; Archer, S.K.; Morris, F.C.; Oliveira, C.; Kielty, L.; et al. Bacteriophage-resistant Acinetobacter Baumannii are resensitized to antimicrobials. Nat. Microbiol. 2021, 6, 157–161. [Google Scholar] [CrossRef]
- Bondy-Denomy, J.; Qian, J.; Westra, E.R.; Buckling, A.; Guttman, D.S.; Davidson, A.R.; Maxwell, K.L. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 2016, 10, 2854–2866. [Google Scholar] [CrossRef] [Green Version]
- Destoumieux-Garzón, D.; Duquesne, S.; Peduzzi, J.; Goulard, C.; Desmadril, M.; Letellier, L.; Rebuffat, S.; Boulanger, P. The iron–siderophore transporter FhuA is the receptor for the antimicrobial peptide microcin J25: Role of the microcin Val11–Pro16 β-hairpin region in the recognition mechanism. Biochem. J. 2005, 389, 869–876. [Google Scholar] [CrossRef] [Green Version]
- Seed, K.D.; Faruque, S.M.; Mekalanos, J.J.; Calderwood, S.B.; Qadri, F.; Camilli, A. Phase variable O antigen biosynthetic genes control expression of the major protective antigen and bacteriophage receptor in Vibrio cholerae O1. PLOS Pathog. 2012, 8, e1002917. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Deora, R.; Doulatov, S.R.; Gingery, M.; Eiserling, F.A.; Preston, A.; Maskell, D.J.; Simons, R.W.; Cotter, P.A.; Parkhill, J.; et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 2002, 295, 2091–2094. [Google Scholar] [CrossRef]
- Hoque, M.M.; Naser, I.B.; Bari, S.M.N.; Zhu, J.; Mekalanos, J.J.; Faruque, S.M. Quorum regulated resistance of Vibrio cholerae against environmental bacteriophages. Sci. Rep. 2016, 6, 37956. [Google Scholar] [CrossRef] [Green Version]
- León-Félix, J.; Villicaña, C. The impact of quorum Sensing on the modulation of phage-host interactions. J. Bacteriol. 2021, 203, e00687-20. [Google Scholar] [CrossRef]
- Samson, J.E.; Magadán, A.H.; Sabri, M.; Moineau, S. Revenge of the phages: Defeating bacterial defences. Nat. Rev. Microbiol. 2013, 11, 675–687. [Google Scholar] [CrossRef]
- Dennehy, J.J.; Abedon, S.T. Adsorption: Phage Acquisition of Bacteria. In Bacteriophages; Harper, D.R., Abedon, S.T., Burrowes, B.H., McConville, M.L., Eds.; Springer: Cham, Switzerland, 2021. [Google Scholar] [CrossRef]
- Fernandes, S.; São-José, C. Enzymes and mechanisms employed by tailed bacteriophages to breach the bacterial cell barriers. Viruses 2018, 10, 396. [Google Scholar] [CrossRef]
- Knecht, L.E.; Veljkovic, M.; Fieseler, L. Diversity and function of phage encoded depolymerases. Front. Microbiol. 2020, 10, 2949. [Google Scholar] [CrossRef]
- Davison, S.; Couture-Tosi, E.; Candela, T.; Mock, M.; Fouet, A. Identification of the Bacillus anthracis γ phage receptor. J. Bacteriol. 2005, 187, 6742–6749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lovett, P.S. PBP1: A flagella specific bacteriophage mediating transduction in Bacillus pumilus. Virology 1972, 47, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Baptista, C.; Santos, M.A.; São-José, C. Phage SPP1 reversible adsorption to Bacillus subtilis cell wall teichoic acids accelerates virus recognition of membrane receptor YueB. J. Bacteriol. 2008, 190, 4989–4996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raimondo, L.M.; Lundh, N.P.; Martinez, R.J. Primary adsorption site of phage PBS1: The flagellum of Bacillus subtilis. J. Virol. 1968, 2, 256–264. [Google Scholar] [CrossRef] [Green Version]
- Ha, E.; Chun, J.; Kim, M.; Ryu, S. Capsular polysaccharide is a receptor of a Clostridium perfringens bacteriophage CPS1. Viruses 2019, 11, 1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duerkop, B.A.; Huo, W.; Bhardwaj, P.; Palmer, K.L.; Hooper, L.V. Molecular basis for lytic bacteriophage resistance in Enterococci. mBio 2016, 7, e01304-16. [Google Scholar] [CrossRef] [Green Version]
- Ho, K.; Huo, W.; Pas, S.; Dao, R.; Palmer, K.L. Loss-of-function mutations in EpaR confer resistance to ΦNPV1 infection in Enterococcus faecalis OG1RF. Antimicrob. Agents Chemother. 2018, 62, e00758-18. [Google Scholar] [CrossRef] [Green Version]
- Douglas, L.J.; Wolin, M.J. Cell wall polymers and phage lysis of Lactobacillus plantarum. Biochemistry 1971, 10, 1551–1555. [Google Scholar] [CrossRef]
- Mooney, D.T.; Jann, M.; Geller, B.L. Subcellular location of Phage Infection Protein (Pip) in Lactococcus lactis. Can. J. Microbiol. 2006, 52, 664–672. [Google Scholar] [CrossRef]
- Bebeacua, C.; Tremblay, D.; Farenc, C.; Chapot-Chartier, M.-P.; Sadovskaya, I.; van Heel, M.; Veesler, D.; Moineau, S.; Cambillau, C. Structure, adsorption to host, and infection mechanism of virulent lactococcal phage P2. J. Virol. 2013, 87, 12302–12312. [Google Scholar] [CrossRef] [Green Version]
- Marcelli, B.; de Jong, A.; Karsens, H.; Janzen, T.; Kok, J.; Kuipers, O.P. A specific sugar moiety in the Lactococcus lactis cell wall pellicle is required for infection by CHPC971, a member of the rare 1706 phage species. Appl. Environ. Microbiol. 2019, 85, e01224-19. [Google Scholar] [CrossRef] [Green Version]
- Habann, M.; Leiman, P.G.; Vandersteegen, K.; Van den Bossche, A.; Lavigne, R.; Shneider, M.M.; Bielmann, R.; Eugster, M.R.; Loessner, M.J.; Klumpp, J. Listeria Phage A511, a model for the contractile tail machineries of SPO1-related bacteriophages. Mol. Microbiol. 2014, 92, 84–99. [Google Scholar] [CrossRef] [Green Version]
- Bielmann, R.; Habann, M.; Eugster, M.R.; Lurz, R.; Calendar, R.; Klumpp, J.; Loessner, M.J. Receptor binding proteins of Listeria monocytogenes bacteriophages A118 and P35 recognize serovar-specific teichoic acids. Virology 2015, 477, 110–118. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Koç, C.; Kühner, P.; Stierhof, Y.-D.; Krismer, B.; Enright, M.C.; Penadés, J.R.; Wolz, C.; Stehle, T.; Cambillau, C.; et al. An essential role for the baseplate protein Gp45 in phage adsorption to Staphylococcus aureus. Sci. Rep. 2016, 6, 26455. [Google Scholar] [CrossRef] [Green Version]
- Winstel, V.; Sanchez-Carballo, P.; Holst, O.; Xia, G.; Peschel, A. Biosynthesis of the unique wall teichoic acid of Staphylococcus aureus lineage ST395. mBio 2014, 5, e00869-14. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Gerlach, D.; Du, X.; Larsen, J.; Stegger, M.; Kühner, P.; Peschel, A.; Xia, G.; Winstel, V. An Accessory wall teichoic acid glycosyltransferase protects Staphylococcus aureus from the lytic activity of Podoviridae. Sci. Rep. 2015, 5, 17219. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, I.; Osada, K.; Azam, A.H.; Asakawa, H.; Miyanaga, K.; Tanji, Y. The presence of two receptor-binding proteins contributes to the wide host range of Staphylococcal Twort-Like phages. Appl. Environ. Microbiol. 2016, 82, 5763–5774. [Google Scholar] [CrossRef] [Green Version]
- Lopez, R.; Garcia, E.; Garcia, P.; Ronda, C.; Tomasz, A. Choline-containing bacteriophage receptors in Streptococcus pneumoniae. J. Bacteriol. 1982, 151, 1581–1590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonnell, B.; Hanemaaijer, L.; Bottacini, F.; Kelleher, P.; Lavelle, K.; Sadovskaya, I.; Vinogradov, E.; Ver Loren van Themaat, E.; Kouwen, T.; Mahony, J.; et al. A cell wall-associated polysaccharide is required for bacteriophage adsorption to the Streptococcus thermophilus cell surface. Mol. Microbiol. 2020, 114, 31–45. [Google Scholar] [CrossRef]
- Szymczak, P.; Filipe, S.R.; Covas, G.; Vogensen, F.K.; Neves, A.R.; Janzen, T. Cell wall glycans mediate recognition of the dairy bacterium Streptococcus thermophilus by bacteriophages. Appl. Environ. Microbiol. 2018, 84, e01847-18. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, F.; Helm, R.F.; Broadway, K.M.; Scharf, B.E. More than rotating flagella: Lipopolysaccharide as a secondary receptor for flagellotropic phage 7-7-1. J. Bacteriol. 2018, 200, e00363-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sørensen, M.C.H.; van Alphen, L.B.; Harboe, A.; Li, J.; Christensen, B.B.; Szymanski, C.M.; Brøndsted, L. Bacteriophage F336 recognizes the capsular phosphoramidate modification of Campylobacter jejuni NCTC11168. J. Bacteriol. 2011, 193, 6742–6749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerrero-Ferreira, R.C.; Viollier, P.H.; Ely, B.; Poindexter, J.S.; Georgieva, M.; Jensen, G.J.; Wright, E.R. Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 2011, 108, 9963–9968. [Google Scholar] [CrossRef] [Green Version]
- Washizaki, A.; Yonesaki, T.; Otsuka, Y. Characterization of the interactions between Escherichia coli receptors, LPS and ompC, and bacteriophage T4 long tail fibers. MicrobiologyOpen 2016, 5, 1003–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gehring, K.; Charbit, A.; Brissaud, E.; Hofnung, M. Bacteriophage Lambda receptor site on the Escherichia coli K-12 LamB protein. J. Bacteriol. 1987, 169, 2103–2106. [Google Scholar] [CrossRef] [Green Version]
- Hao, G.; Shu, R.; Ding, L.; Chen, X.; Miao, Y.; Wu, J.; Zhou, H.; Wang, H. Bacteriophage SRD2021 recognizing capsular polysaccharide shows therapeutic potential in serotype K47 Klebsiella pneumoniae infections. Antibiotics 2021, 10, 894. [Google Scholar] [CrossRef]
- Bae, H.-W.; Cho, Y.-H. Complete genome sequence of Pseudomonas Aeruginosa podophage MPK7, which requires Type IV pili for infection. Genome Announc. 2013, 1, e00744-13. [Google Scholar] [CrossRef] [Green Version]
- Garbe, J.; Bunk, B.; Rohde, M.; Schobert, M. Sequencing and characterization of Pseudomonas aeruginosa phage JG004. BMC Microbiol. 2011, 11, 102. [Google Scholar] [CrossRef] [Green Version]
- Iwashita, S.; Kanegasaki, S. Smooth specific phage adsorption: Endorhamnosidase activity of tail parts of P22. Biochem. Biophys. Res. Commun. 1973, 55, 403–409. [Google Scholar] [CrossRef]
- Choi, Y.; Shin, H.; Lee, J.-H.; Ryu, S. Identification and characterization of a novel flagellum-dependent salmonella-infecting bacteriophage, IEPS5. Appl. Environ. Microbiol. 2013, 79, 4829–4837. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Ryu, S. Characterization of a T5-like Coliphage, SPC35, and differential development of resistance to SPC35 in Salmonella enterica serovar Typhimurium and Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 2042–2050. [Google Scholar] [CrossRef] [Green Version]
- Filippov, A.A.; Sergueev, K.V.; He, Y.; Huang, X.-Z.; Gnade, B.T.; Mueller, A.J.; Fernandez-Prada, C.M.; Nikolich, M.P. Bacteriophage-resistant mutants in Yersinia pestis: Identification of phage receptors and attenuation for mice. PLoS ONE 2011, 6, e25486. [Google Scholar] [CrossRef] [Green Version]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Weidenmaier, C.; Peschel, A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 2008, 6, 276–287. [Google Scholar] [CrossRef]
- Maffei, E.; Shaidullina, A.; Burkolter, M.; Heyer, Y.; Estermann, F.; Druelle, V.; Sauer, P.; Willi, L.; Michaelis, S.; Hilbi, H.; et al. Systematic exploration of Escherichia coli phage–host interactions with the BASEL phage collection. PLOS Biol. 2021, 19, e3001424. [Google Scholar] [CrossRef]
- Rakhuba, D.V.; Kolomiets, E.I.; Dey, E.S.; Novik, G.I. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 2010, 59, 145–155. [Google Scholar] [CrossRef]
- Kim, M.; Kim, S.; Park, B.; Ryu, S. Core lipopolysaccharide-specific phage SSU5 as an auxiliary component of a phage cocktail for Salmonella biocontrol. Appl. Environ. Microbiol. 2014, 80, 1026–1034. [Google Scholar] [CrossRef] [Green Version]
- Eriksson, U.; Svenson, S.B.; Lönngren, J.; Lindberg, A.A. Salmonella phage glycanases: Substrate specificity of the phage P22 endo-rhamnosidase. J. Gen. Virol. 1979, 43, 503–511. [Google Scholar] [CrossRef]
- Ricci, V.; Piddock, L.J.V. Exploiting the Role of TolC in Pathogenicity: Identification of a bacteriophage for eradication of Salmonella serovars from poultry. Appl. Environ. Microbiol. 2010, 76, 1704–1706. [Google Scholar] [CrossRef]
- Vollmer, W.; Blanot, D.; De Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef] [Green Version]
- Takumi, K.; Takeoka, A.; Kinouchi, T.; Kawata, T. Solubilization and partial properties of receptor substance for bacteriophage A2 induced from Clostridium botulinum Type A 190L. Microbiol. Immunol. 1985, 29, 1185–1195. [Google Scholar] [CrossRef] [PubMed]
- Wendlinger, G.; Loessner, M.J.; Scherer, S. Bacteriophage receptors on Listeria monocytogenes cells are the N-acetylglucosamine and rhamnose substituents of teichoic acids or the peptidoglycan itself. Microbiology 1996, 142 (Pt 4), 985–992. [Google Scholar] [CrossRef] [Green Version]
- São-José, C.; Baptista, C.; Santos, M.A. Bacillus subtilis operon encoding a membrane receptor for bacteriophage SPP1. J. Bacteriol. 2004, 186, 8337–8346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schäffer, C.; Messner, P. 2005 the structure of secondary cell wall polymers: How Gram-positive bacteria stick their cell walls together. Microbiology 2005, 151, 643–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, S.; Santa Maria, J.P.; Walker, S. Wall teichoic acids of Gram-positive bacteria. Ann. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef] [Green Version]
- Percy, M.G.; Gründling, A. Lipoteichoic acid synthesis and function in Gram-positive bacteria. Ann. Rev. Microbiol. 2014, 68, 81–100. [Google Scholar] [CrossRef] [Green Version]
- Weidenmaier, C.; Kokai-Kun, J.F.; Kristian, S.A.; Chanturiya, T.; Kalbacher, H.; Gross, M.; Nicholson, G.; Neumeister, B.; Mond, J.J.; Peschel, A. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 2004, 10, 243–245. [Google Scholar] [CrossRef]
- Bergmann, S.; Hammerschmidt, S. 2006 Versatility of pneumococcal surface proteins. Microbiology 2006, 152, 295–303. [Google Scholar] [CrossRef] [Green Version]
- Azam, A.H.; Tanji, Y. Peculiarities of Staphylococcus aureus phages and their possible application in phage therapy. Appl. Microbiol. Biotechnol. 2019, 103, 4279–4289. [Google Scholar] [CrossRef]
- Sumrall, E.T.; Keller, A.P.; Shen, Y.; Loessner, M.J. Structure and function of Listeria teichoic acids and their implications. Mol. Microbiol. 2020, 113, 627–637. [Google Scholar] [CrossRef] [Green Version]
- Mahony, J.; Frantzen, C.; Vinogradov, E.; Sadovskaya, I.; Theodorou, I.; Kelleher, P.; Chapot-Chartier, M.-P.; Cambillau, C.; Holo, H.; van Sinderen, D. The CWPS Rubik’s Cube: Linking diversity of cell wall polysaccharide structures with the encoded biosynthetic machinery of selected Lactococcus lactis strains. Mol. Microbiol. 2020, 114, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Missiakas, D.; Schneewind, O. Assembly and function of the Bacillus anthracis S-layer. Ann. Rev. Microbiol. 2017, 71, 79–98. [Google Scholar] [CrossRef]
- Mistou, M.-Y.; Sutcliffe, I.C.; van Sorge, N.M. Bacterial glycobiology: Rhamnose-containing cell wall polysaccharides in Gram-positive bacteria. FEMS Microbiol. Rev. 2016, 40, 464–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rismondo, J.; Gillis, A.; Gründling, A. Modifications of cell wall polymers in Gram-positive bacteria by multi-component transmembrane glycosylation systems. Curr. Opin. Microbiol. 2021, 60, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Poxton, I.R. Chapter 5—Teichoic acids, lipoteichoic acids and other secondary cell wall and membrane polysaccharides of Gram-positive bacteria. In Molecular Medical Microbiology, 2nd ed.; Tang, Y.-W., Sussman, M., Liu, D., Poxton, I., Schwartzman, J., Eds.; Academic Press: Boston, UK, 2015; pp. 91–103. ISBN 978-0-12-397169-2. [Google Scholar]
- Dunne, M.; Hupfeld, M.; Klumpp, J.; Loessner, M.J. Molecular basis of bacterial host interactions by Gram-positive targeting bacteriophages. Viruses 2018, 10, 397. [Google Scholar] [CrossRef] [Green Version]
- Sadovskaya, I.; Vinogradov, E.; Courtin, P.; Armalyte, J.; Meyrand, M.; Giaouris, E.; Palussière, S.; Furlan, S.; Péchoux, C.; Ainsworth, S.; et al. Another brick in the wall: A rhamnan polysaccharide trapped inside peptidoglycan of Lactococcus lactis. mBio 2017, 8, e01303-17. [Google Scholar] [CrossRef] [Green Version]
- Chapot-Chartier, M.-P.; Vinogradov, E.; Sadovskaya, I.; Andre, G.; Mistou, M.-Y.; Trieu-Cuot, P.; Furlan, S.; Bidnenko, E.; Courtin, P.; Péchoux, C.; et al. Cell surface of Lactococcus lactis is covered by a protective polysaccharide pellicle*. J. Bio. Chem. 2010, 285, 10464–10471. [Google Scholar] [CrossRef] [Green Version]
- Gerbino, E.; Carasi, P.; Mobili, P.; Serradell, M.A.; Gómez-Zavaglia, A. Role of S-Layer Proteins in Bacteria. World J. Microbiol. Biotechnol. 2015, 31, 1877–1887. [Google Scholar] [CrossRef]
- Plaut, R.D.; Beaber, J.W.; Zemansky, J.; Kaur, A.P.; George, M.; Biswas, B.; Henry, M.; Bishop-Lilly, K.A.; Mokashi, V.; Hannah, R.M.; et al. Genetice for the involvement of the S-Layer protein gene sap and the sporulation genes spo0A, spo0B, and spo0F in phage AP50c infection of Bacillus anthracis. J. Bacteriol. 2014, 196, 1143–1154. [Google Scholar] [CrossRef]
- Latka, A.; Leiman, P.G.; Drulis-Kawa, Z.; Briers, Y. Modeling the architecture of depolymerase-containing receptor binding proteins in Klebsiella phages. Front Microbiol 2019, 10, 2649. [Google Scholar] [CrossRef]
- Oliveira, H.; Costa, A.R.; Konstantinides, N.; Ferreira, A.; Akturk, E.; Sillankorva, S.; Nemec, A.; Shneider, M.; Dötsch, A.; Azeredo, J. Ability of Phages to infect Acinetobacter calcoaceticus-Acinetobacter baumannii complex species through acquisition of different pectate lyase depolymerase domains. Env. Microbiol. 2017, 19, 5060–5077. [Google Scholar] [CrossRef] [Green Version]
- Pickard, D.; Toribio, A.L.; Petty, N.K.; van Tonder, A.; Yu, L.; Goulding, D.; Barrell, B.; Rance, R.; Harris, D.; Wetter, M.; et al. A conserved acetyl esterase domain targets diverse bacteriophages to the VI capsular receptor of Salmonella enterica serovar Typhi. J. Bacteriol. 2010, 192, 5746–5754. [Google Scholar] [CrossRef] [Green Version]
- Ozaki, T.; Abe, N.; Kimura, K.; Suzuki, A.; Kaneko, J. Genomic analysis of Bacillus subtilis lytic bacteriophage ΦNIT1 capable of obstructing natto fermentation carrying genes for the capsule-lytic soluble enzymes poly-γ-glutamate hydrolase and levanase. Biosc. Biotechnol. Biochem. 2017, 81, 135–146. [Google Scholar] [CrossRef] [Green Version]
- Mohd Nadzir, M.; Nurhayati, R.W.; Idris, F.N.; Nguyen, M.H. Biomedical applications of bacterial exopolysaccharides: A review. Polymers 2021, 13, 530. [Google Scholar] [CrossRef]
- Whitfield, C.; Wear, S.S.; Sande, C. Assembly of bacterial capsular polysaccharides and exopolysaccharides. Ann. Rev. Microbiol. 2020, 74, 521–543. [Google Scholar] [CrossRef]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The Biofilm life cycle: Expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef]
- Chibeu, A.; Ceyssens, P.-J.; Hertveldt, K.; Volckaert, G.; Cornelis, P.; Matthijs, S.; Lavigne, R. The adsorption of Pseudomonas aeruginosa bacteriophage ΦKMV is dependent on expression regulation of Type IV pili genes. FEMS Microbiol. Lett. 2009, 296, 210–218. [Google Scholar] [CrossRef] [Green Version]
- Costa, T.R.D.; Ilangovan, A.; Ukleja, M.; Redzej, A.; Santini, J.M.; Smith, T.K.; Egelman, E.H.; Waksman, G. Structure of the bacterial sex F pilus reveals an assembly of a stoichiometric protein-phospholipid complex. Cell 2016, 166, 1436–1444.e10. [Google Scholar] [CrossRef] [Green Version]
- Veesler, D.; Spinelli, S.; Mahony, J.; Lichière, J.; Blangy, S.; Bricogne, G.; Legrand, P.; Ortiz-Lombardia, M.; Campanacci, V.; van Sinderen, D.; et al. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc. Natl. Acad. Sci. USA 2012, 109, 8954–8958. [Google Scholar] [CrossRef]
- Dunne, M.; Rupf, B.; Tala, M.; Qabrati, X.; Ernst, P.; Shen, Y.; Sumrall, E.; Heeb, L.; Plückthun, A.; Loessner, M.J.; et al. Reprogramming bacteriophage host range through structure-guided design of chimeric receptor binding proteins. Cell Rep. 2019, 29, 1336–1350.e4. [Google Scholar] [CrossRef] [Green Version]
- Huss, P.; Meger, A.; Leander, M.; Nishikawa, K.; Raman, S. Mapping the functional landscape of the receptor binding domain of T7 bacteriophage by Deep mutational scanning. eLife 2021, 10, e63775. [Google Scholar] [CrossRef] [PubMed]
- Büttner, C.R.; Wu, Y.; Maxwell, K.L.; Davidson, A.R. Baseplate assembly of phage Mu: Defining the conserved core components of contractile-tailed phages and related bacterial systems. Proc. Natl. Acad. Sci. USA 2016, 113, 10174–10179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.; Xiang, Y. Structures of the tailed bacteriophages that infect Gram-positive bacteria. Curr. Op. Virol. 2020, 45, 65–74. [Google Scholar] [CrossRef]
- Sanz-Gaitero, M.; Seoane-Blanco, M.; van Raaij, M.J. Structure and function of bacteriophages. In Bacteriophages; Harper, D.R., Abedon, S.T., Burrowes, B.H., McConville, M.L., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 19–91. ISBN 978-3-319-41985-5. [Google Scholar]
- Leiman, P.G.; Shneider, M.M. Contractile tail machines of bacteriophages. In Viral Molecular Machines; Rossmann, M.G., Rao, V.B., Eds.; Advances in Experimental Medicine and Biology; Springer US: Boston, MA, USA, 2012; pp. 93–114. ISBN 978-1-4614-0980-9. [Google Scholar]
- Yap, M.L.; Rossmann, M.G. Structure and function of bacteriophage T4. Future Microbiol. 2014, 9, 1319–1327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerrero-Ferreira, R.C.; Hupfeld, M.; Nazarov, S.; Taylor, N.M.; Shneider, M.M.; Obbineni, J.M.; Loessner, M.J.; Ishikawa, T.; Klumpp, J.; Leiman, P.G. Structure and transformation of bacteriophage A511 baseplate and tail upon infection of Listeria cells. EMBO J. 2019, 38, e99455. [Google Scholar] [CrossRef]
- Arisaka, F.; Yap, M.L.; Kanamaru, S.; Rossmann, M.G. Molecular assembly and structure of the bacteriophage T4 Tail. Biophys. Rev. 2016, 8, 385–396. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.Z.; Fokine, A.; Mahalingam, M.; Zhang, Z.; Garcia-Doval, C.; van Raaij, M.J.; Rossmann, M.G.; Rao, V.B. Molecular anatomy of the receptor binding module of a bacteriophage long tail fiber. PLOS Pathog. 2019, 15, e1008193. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, H.; Costa, A.R.; Konstantinidis, N.; Nemec, A.; Shneider, M.; Dötsch, A.; Sillankorva, S.; Azeredo, J. Capsule Depolymerase Activity of Phages Infecting the Acinetobacter baumannii-calcoaceticus complex; 2016; ISBN 978-5-9905908-2-3. Available online: https://repositorium.sdum.uminho.pt/handle/1822/45332 (accessed on 20 August 2021).
- Squeglia, F.; Maciejewska, B.; Łątka, A.; Ruggiero, A.; Briers, Y.; Drulis-Kawa, Z.; Berisio, R. Structural and functional studies of a Klebsiella phage capsule depolymerase tailspike: Mechanistic insights into capsular degradation. Structure 2020, 28, 613–624.e4. [Google Scholar] [CrossRef]
- Oliveira, H.; Costa, A.R.; Ferreira, A.; Konstantinides, N.; Santos, S.B.; Boon, M.; Noben, J.-P.; Lavigne, R.; Azeredo, J. Functional analysis and antivirulence properties of a new depolymerase from a myovirus that infects Acinetobacter baumannii capsule K45. J. Virol. 2019, 93, e01163-18. [Google Scholar] [CrossRef]
- Park, D.-W.; Park, J.-H. Characterization of a novel phage depolymerase specific to Escherichia coli O157:H7 and biofilm control on abiotic surfaces. J. Microbiol. 2021, 59, 1002–1009. [Google Scholar] [CrossRef]
- Shahed-Al-Mahmud, M.; Roy, R.; Sugiokto, F.G.; Islam, M.N.; Lin, M.-D.; Lin, L.-C.; Lin, N.-T. Phage ΦAB6-Borne Depolymerase combats Acinetobacter baumannii biofilm formation and infection. Antibiotics 2021, 10, 279. [Google Scholar] [CrossRef]
- Myers, C.L.; Ireland, R.G.; Garrett, T.A.; Brown, E.D. Characterization of wall teichoic acid degradation by the bacteriophage ϕ29 appendage protein GP12 using synthetic substrate analogs. J. Biol. Chem. 2015, 290, 19133–19145. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez, D.; Briers, Y.; Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; Lavigne, R.; García, P. Role of the pre-neck appendage protein (Dpo7) from phage vB_SepiS-phiIPLA7 as an anti-biofilm agent in Staphylococcal species. Front. Microbiol. 2015, 6, 1315. [Google Scholar] [CrossRef] [Green Version]
- Pires, D.P.; Oliveira, H.; Melo, L.D.R.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef] [Green Version]
- Majkowska-Skrobek, G.; Latka, A.; Berisio, R.; Squeglia, F.; Maciejewska, B.; Briers, Y.; Drulis-Kawa, Z. Phage-borne depolymerases decrease Klebsiella pneumoniae resistance to innate defense mechanisms. Front. Microbiol. 2018, 9, 2517. [Google Scholar] [CrossRef] [Green Version]
- Abedon, S.T. Lysis from without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; Martínez, B.; Donovan, D.M.; Rodríguez, A.; García, P. Bacteriophage virion-associated peptidoglycan hydrolases: Potential new enzybiotics. Crit. Rev. Microbiol. 2013, 39, 427–434. [Google Scholar] [CrossRef] [Green Version]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [Green Version]
- Goulet, A.; Spinelli, S.; Mahony, J.; Cambillau, C. Conserved and diverse traits of adhesion devices from siphoviridae recognizing proteinaceous or saccharidic receptors. Viruses 2020, 12, 512. [Google Scholar] [CrossRef]
- Mahony, J.; Alqarni, M.; Stockdale, S.; Spinelli, S.; Feyereisen, M.; Cambillau, C.; van Sinderen, D. Functional and structural dissection of the tape measure protein of lactococcal phage TP901-1. Sci. Rep. 2016, 6, 36667. [Google Scholar] [CrossRef] [Green Version]
- Hayes, S.; Vincentelli, R.; Mahony, J.; Nauta, A.; Ramond, L.; Lugli, G.A.; Ventura, M.; van Sinderen, D.; Cambillau, C. Functional carbohydrate binding modules identified in evolved Dits from siphophages infecting various Gram-positive bacteria. Mol. Microbiol. 2018, 110, 777–795. [Google Scholar] [CrossRef] [PubMed]
- Flayhan, A.; Vellieux, F.M.D.; Lurz, R.; Maury, O.; Contreras-Martel, C.; Girard, E.; Boulanger, P.; Breyton, C. Crystal structure of Pb9, the distal tail protein of bacteriophage T5: A conserved structural motif among all siphophages. J. Virol. 2014, 88, 820–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veesler, D.; Robin, G.; Lichière, J.; Auzat, I.; Tavares, P.; Bron, P.; Campanacci, V.; Cambillau, C. Crystal structure of bacteriophage SPP1 distal tail protein (Gp19.1): A baseplate hub paradigm in Gram-positive infecting phages. J. Bio. Chem. 2010, 285, 36666–36673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sciara, G.; Bebeacua, C.; Bron, P.; Tremblay, D.; Ortiz-Lombardia, M.; Lichière, J.; van Heel, M.; Campanacci, V.; Moineau, S.; Cambillau, C. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc. Natl. Acad. Sci. USA 2010, 107, 6852–6857. [Google Scholar] [CrossRef] [Green Version]
- Spinelli, S.; Tremblay, D.; Moineau, S.; Cambillau, C.; Goulet, A. Structural insights into lactococcal siphophage p2 baseplate activation mechanism. Viruses 2020, 12, 878. [Google Scholar] [CrossRef]
- Dieterle, M.-E.; Spinelli, S.; Sadovskaya, I.; Piuri, M.; Cambillau, C. Evolved distal tail carbohydrate binding modules of Lactobacillus phage J-1: A novel type of anti-receptor widespread among lactic acid bacteria phages. Mol. Microbiol. 2017, 104, 608–620. [Google Scholar] [CrossRef] [Green Version]
- Leprince, A.; Nuytten, M.; Mahillon, J. Viral proteins involved in the adsorption process of Deep-Purple, a siphovirus infecting members of the Bacillus cereus group. Appl. Env. Microbiol. 2022, 88, e02478-21. [Google Scholar] [CrossRef]
- Dieterle, M.E.; Bowman, C.; Batthyany, C.; Lanzarotti, E.; Turjanski, A.; Hatfull, G.; Piuri, M. Exposing the secrets of two well-known Lactobacillus casei phages, J-1 and Pl-1, by genomic and structural analysis. Appl. Env. Microbiol. 2014, 80, 7107–7121. [Google Scholar] [CrossRef] [Green Version]
- Lavelle, K.; Goulet, A.; McDonnell, B.; Spinelli, S.; van Sinderen, D.; Mahony, J.; Cambillau, C. Revisiting the host adhesion determinants of Streptococcus thermophilus siphophages. Microb. Biotechnol. 2020, 13, 1765–1779. [Google Scholar] [CrossRef]
- Stockdale, S.R.; Mahony, J.; Courtin, P.; Chapot-Chartier, M.-P.; van Pijkeren, J.-P.; Britton, R.A.; Neve, H.; Heller, K.J.; Aideh, B.; Vogensen, F.K.; et al. The lactococcal phages Tuc2009 and TP901-1 incorporate two alternate forms of their tail fiber into their virions for infection specialization. J. Biol. Chem. 2013, 288, 5581–5590. [Google Scholar] [CrossRef] [Green Version]
- Siponen, M.; Spinelli, S.; Blangy, S.; Moineau, S.; Cambillau, C.; Campanacci, V. Crystal structure of a chimeric receptor binding protein constructed from two lactococcal phages. J. Bacteriol. 2009, 191, 3220–3225. [Google Scholar] [CrossRef] [Green Version]
- Farenc, C.; Spinelli, S.; Vinogradov, E.; Tremblay, D.; Blangy, S.; Sadovskaya, I.; Moineau, S.; Cambillau, C. Molecular insights on the recognition of a Lactococcus lactis cell wall pellicle by the phage 1358 receptor binding protein. J. Virol. 2014, 88, 7005–7015. [Google Scholar] [CrossRef] [Green Version]
- Kizziah, J.L.; Manning, K.A.; Dearborn, A.D.; Dokland, T. Structure of the host cell recognition and penetration machinery of a Staphylococcus aureus bacteriophage. PLOS Pathog. 2020, 16, e1008314. [Google Scholar] [CrossRef] [Green Version]
- Koç, C.; Xia, G.; Kühner, P.; Spinelli, S.; Roussel, A.; Cambillau, C.; Stehle, T. Structure of the host-recognition device of Staphylococcus aureus phage Φ11. Sci. Rep. 2016, 6, 27581. [Google Scholar] [CrossRef] [Green Version]
- Spinelli, S.; Campanacci, V.; Blangy, S.; Moineau, S.; Tegoni, M.; Cambillau, C. Modular structure of the receptor binding proteins of Lactococcus lactis phages: The RBP structure of the temperate phage TP901-1. J. Bio. Chem. 2006, 281, 14256–14262. [Google Scholar] [CrossRef] [Green Version]
- Tremblay, D.M.; Tegoni, M.; Spinelli, S.; Campanacci, V.; Blangy, S.; Huyghe, C.; Desmyter, A.; Labrie, S.; Moineau, S.; Cambillau, C. Receptor-binding protein of Lactococcus lactis phages: Identification and characterization of the saccharide receptor-binding site. J. Bacteriol. 2006, 188, 2400–2410. [Google Scholar] [CrossRef] [Green Version]
- Guan, J.; Ibarra, D.; Zeng, L. The role of side tail fibers during the infection cycle of phage Lambda. Virology 2019, 527, 57–63. [Google Scholar] [CrossRef]
- Zivanovic, Y.; Confalonieri, F.; Ponchon, L.; Lurz, R.; Chami, M.; Flayhan, A.; Renouard, M.; Huet, A.; Decottignies, P.; Davidson, A.R.; et al. Insights into bacteriophage T5 structure from analysis of its morphogenesis genes and protein components. J. Virol. 2014, 88, 1162–1174. [Google Scholar] [CrossRef] [Green Version]
- Bebeacua, C.; Bron, P.; Lai, L.; Vegge, C.S.; Brøndsted, L.; Spinelli, S.; Campanacci, V.; Veesler, D.; van Heel, M.; Cambillau, C. Structure and molecular assignment of lactococcal phage TP901-1 baseplate. J. Bio. Chem. 2010, 285, 39079–39086. [Google Scholar] [CrossRef]
- Collins, B.; Bebeacua, C.; Mahony, J.; Blangy, S.; Douillard, F.P.; Veesler, D.; Cambillau, C.; van Sinderen, D. Structure and functional analysis of the host recognition device of lactococcal phage Tuc2009. J. Virol. 2013, 87, 8429–8440. [Google Scholar] [CrossRef] [Green Version]
- Hayes, S.; Duhoo, Y.; Neve, H.; Murphy, J.; Noben, J.-P.; Franz, C.M.A.P.; Cambillau, C.; Mahony, J.; Nauta, A.; Van Sinderen, D. Identification of dual receptor binding protein systems in lactococcal 936 group phages. Viruses 2018, 10, 668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casjens, S.R.; Molineux, I.J. Short Noncontractile Tail Machines: Adsorption and DNA delivery by podoviruses. In Viral Molecular Machines; Rossmann, M.G., Rao, V.B., Eds.; Advances in Experimental Medicine and Biology; Springer US: Boston, MA, USA, 2012; pp. 143–179. ISBN 978-1-4614-0980-9. [Google Scholar]
- Leiman, P.G.; Battisti, A.J.; Bowman, V.D.; Stummeyer, K.; Mühlenhoff, M.; Gerardy-Schahn, R.; Scholl, D.; Molineux, I.J. The Structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities. J. Mol. Biol. 2007, 371, 836–849. [Google Scholar] [CrossRef]
- Swanson, N.A.; Lokareddy, R.K.; Li, F.; Hou, C.-F.D.; Leptihn, S.; Pavlenok, M.; Niederweis, M.; Pumroy, R.A.; Moiseenkova-Bell, V.Y.; Cingolani, G. Cryo-EM structure of the periplasmic tunnel of T7 DNA-ejectosome at 2.7 Å resolution. Mol. Cell 2021, 81, 3145–3159.e7. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, D.; Gui, M.; Xiang, Y. Structural assembly of the tailed bacteriophage Φ29. Nat. Comm. 2019, 10, 2366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, Y.; Morais, M.C.; Cohen, D.N.; Bowman, V.D.; Anderson, D.L.; Rossmann, M.G. Crystal and CryoEM structural studies of a cell wall degrading enzyme in the bacteriophage Φ29 tail. Proc. Natl. Acad. Sci. USA 2008, 105, 9552–9557. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Gui, M.; Wang, D.; Xiang, Y. The bacteriophage Φ29 tail possesses a pore-forming loop for cell membrane penetration. Nature 2016, 534, 544–547. [Google Scholar] [CrossRef]
- Xiang, Y.; Rossmann, M.G. Structure of bacteriophage Φ29 head fibers has a supercoiled triple repeating helix-turn-helix motif. Proc. Natl. Acad. Sci. USA 2011, 108, 4806–4810. [Google Scholar] [CrossRef] [Green Version]
- Marcó, M.B.; Moineau, S.; Quiberoni, A. Bacteriophages and dairy fermentations. Bacteriophage 2012, 2, 149–158. [Google Scholar] [CrossRef] [Green Version]
- Deveau, H.; Labrie, S.J.; Chopin, M.-C.; Moineau, S. Biodiversity and classification of lactococcal phages. Appl. Env. Microbiol. 2006, 72, 4338–4346. [Google Scholar] [CrossRef]
- Spinelli, S.; Veesler, D.; Bebeacua, C.; Cambillau, C. Structures and host-adhesion mechanisms of lactococcal siphophages. Front Microbiol. 2014, 5, 3. [Google Scholar] [CrossRef] [Green Version]
- Ainsworth, S.; Sadovskaya, I.; Vinogradov, E.; Courtin, P.; Guerardel, Y.; Mahony, J.; Grard, T.; Cambillau, C.; Chapot-Chartier, M.-P.; van Sinderen, D. Differences in lactococcal cell wall polysaccharide structure are major determining factors in bacteriophage sensitivity. mBio 2014, 5, e00880-14. [Google Scholar] [CrossRef] [Green Version]
- Mahony, J.; Kot, W.; Murphy, J.; Ainsworth, S.; Neve, H.; Hansen, L.H.; Heller, K.J.; Sorensen, S.J.; Hammer, K.; Cambillau, C.; et al. Investigation of the relationship between lactococcal host cell wall polysaccharide genotype and 936 phage receptor binding protein phylogeny. Appl. Env. Microbiol. 2013, 79, 4385–4392. [Google Scholar] [CrossRef] [Green Version]
- Murphy, J.; Bottacini, F.; Mahony, J.; Kelleher, P.; Neve, H.; Zomer, A.; Nauta, A.; van Sinderen, D. Comparative genomics and functional analysis of the 936 group of lactococcal Siphoviridae phages. Sci. Rep. 2016, 6, 21345. [Google Scholar] [CrossRef] [Green Version]
- Mahony, J.; Randazzo, W.; Neve, H.; Settanni, L.; van Sinderen, D. Lactococcal 949 group phages recognize a carbohydrate receptor on the host cell surface. Appl. Env. Microbiol. 2015, 81, 3299–3305. [Google Scholar] [CrossRef] [Green Version]
- Romero, D.A.; Magill, D.; Millen, A.; Horvath, P.; Fremaux, C. Dairy lactococcal and streptococcal phage–host interactions: An industrial perspective in an evolving phage landscape. FEMS Microbiol. Rev. 2020, 44, 909–932. [Google Scholar] [CrossRef]
- McCabe, O.; Spinelli, S.; Farenc, C.; Labbé, M.; Tremblay, D.; Blangy, S.; Oscarson, S.; Moineau, S.; Cambillau, C. The targeted recognition of Lactococcus lactis phages to their polysaccharide receptors. Mol. Microbiol. 2015, 96, 875–886. [Google Scholar] [CrossRef]
- Spinelli, S.; Desmyter, A.; Verrips, C.T.; de Haard, H.J.W.; Moineau, S.; Cambillau, C. Lactococcal bacteriophage p2 receptor-binding protein structure suggests a common ancestor gene with bacterial and mammalian viruses. Nat. Struct. Mol. Biol. 2006, 13, 85–89. [Google Scholar] [CrossRef]
- Hanemaaijer, L.; Kelleher, P.; Neve, H.; Franz, C.M.A.P.; de Waal, P.P.; van Peij, N.N.M.E.; van Sinderen, D.; Mahony, J. Biodiversity of phages infecting the dairy bacterium Streptococcus thermophilus. Microorganisms 2021, 9, 1822. [Google Scholar] [CrossRef]
- Lavelle, K.; Martinez, I.; Neve, H.; Lugli, G.A.; Franz, C.M.A.P.; Ventura, M.; Bello, F.D.; Sinderen, D.V.; Mahony, J. Biodiversity of Streptococcus thermophilus phages in global dairy fermentations. Viruses 2018, 10, 577. [Google Scholar] [CrossRef]
- Hagens, S.; Loessner, M.J. Phages of Listeria offer novel tools for diagnostics and biocontrol. Front. Microbiol. 2014, 5, 159. [Google Scholar] [CrossRef]
- Xia, G.; Wolz, C. Phages of Staphylococcus aureus and their impact on host evolution. Inf. Genet. Evol. 2014, 21, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Łobocka, M.; Hejnowicz, M.S.; Dąbrowski, K.; Gozdek, A.; Kosakowski, J.; Witkowska, M.; Ulatowska, M.I.; Weber-Dąbrowska, B.; Kwiatek, M.; Parasion, S.; et al. Chapter 5—Genomics of Staphylococcal Twort-like Phages—Potential therapeutics of the post-antibiotic era. In Adv Virus Res; Łobocka, M., Szybalski, W., Eds.; Bacteriophages, Part B; Academic Press: Cambridge, MA, USA, 2012; Volume 83, pp. 143–216. [Google Scholar]
- Xia, G.; Maier, L.; Sanchez-Carballo, P.; Li, M.; Otto, M.; Holst, O.; Peschel, A. Glycosylation of Wall Teichoic Acid in Staphylococcus aureus by TarM. J. Bio. Chem. 2010, 285, 13405–13415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endl, J.; Seidl, P.H.; Fiedler, F.; Schleifer, K.H. Determination of cell wall teichoic acid structure of staphylococci by rapid chemical and serological screening methods. Arch. Microbiol. 1984, 137, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.; Xia, G.; Luhachack, L.G.; Campbell, J.; Meredith, T.C.; Chen, C.; Winstel, V.; Gekeler, C.; Irazoqui, J.E.; Peschel, A.; et al. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc. Natl. Acad. Sci. USA 2012, 109, 18909–18914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchiyama, J.; Taniguchi, M.; Kurokawa, K.; Takemura-Uchiyama, I.; Ujihara, T.; Shimakura, H.; Sakaguchi, Y.; Murakami, H.; Sakaguchi, M.; Matsuzaki, S. Adsorption of Staphylococcus viruses S13′ and S24-1 on Staphylococcus aureus strains with different glycosidic linkage patterns of wall teichoic acids. J. Gen. Virol. 2017, 98, 2171–2180. [Google Scholar] [CrossRef]
- Hrebík, D.; Štveráková, D.; Škubník, K.; Füzik, T.; Pantůček, R.; Plevka, P. Structure and genome ejection mechanism of Staphylococcus aureus phage P68. Sci. Adv. 2019, 5, eaaw7414. [Google Scholar] [CrossRef] [Green Version]
- Xia, G.; Corrigan, R.M.; Winstel, V.; Goerke, C.; Gründling, A.; Peschel, A. Wall teichoic acid-dependent adsorption of staphylococcal siphovirus and myovirus. J. Bacteriol. 2011, 193, 4006–4009. [Google Scholar] [CrossRef] [Green Version]
- Seul, A.; Brasilès, S.; Petitpas, I.; Lurz, R.; Campanacci, V.; Cambillau, C.; Weise, F.; Zairi, M.; Tavares, P.; Auzat, I. Biogenesis of a bacteriophage long non-contractile tail. J. Mol. Biol. 2021, 433, 167112. [Google Scholar] [CrossRef]
- São-José, C.; Lhuillier, S.; Lurz, R.; Melki, R.; Lepault, J.; Santos, M.A.; Tavares, P. The ectodomain of the viral receptor YueB forms a fiber that triggers ejection of bacteriophage SPP1 DNA. J. Bio. Chem. 2006, 281, 11464–11470. [Google Scholar] [CrossRef]
- Vinga, I.; Baptista, C.; Auzat, I.; Petipas, I.; Lurz, R.; Tavares, P.; Santos, M.A.; São-José, C. Role of bacteriophage SPP1 tail spike protein gp21 on host cell receptor binding and trigger of phage DNA ejection. Mol. Microbiol. 2012, 83, 289–303. [Google Scholar] [CrossRef]
- Carroll, L.M.; Cheng, R.A.; Wiedmann, M.; Kovac, J. Keeping up with the Bacillus cereus group: Taxonomy through the genomics era and beyond. Crit. Rev. Food Sci. Nutr. 2022, 62, 7677–7702. [Google Scholar] [CrossRef]
- Abshire, T.G.; Brown, J.E.; Ezzell, J.W. Production and validation of the use of Gamma phage for identification of Bacillus anthracis. J. Clin. Microbiol. 2005, 43, 4780–4788. [Google Scholar] [CrossRef] [Green Version]
- Schuch, R.; Fischetti, V.A. Detailed genomic analysis of the Wβ and γ phages infecting Bacillus anthracis: Implications for evolution of environmental fitness and antibiotic resistance. J. Bacteriol. 2006, 188, 3037–3051. [Google Scholar] [CrossRef] [Green Version]
- Kong, M.; Ryu, S. Bacteriophage PBC1 and its endolysin as an antimicrobial agent against Bacillus cereus. Appl. Env. Microbiol. 2015, 81, 2274–2283. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Yuan, X.; Li, N.; Wang, J.; Yu, S.; Zeng, H.; Zhang, J.; Wu, Q.; Ding, Y. Isolation and characterization of Bacillus cereus phage VB_BceP-DLc1 reveals the largest member of the φ29-like phages. Microorganisms 2020, 8, 1750. [Google Scholar] [CrossRef]
- Hussain, W.; Ullah, M.W.; Farooq, U.; Aziz, A.; Wang, S. Bacteriophage-based advanced bacterial detection: Concept, mechanisms, and applications. Biosens. Bioelect. 2021, 177, 112973. [Google Scholar] [CrossRef]
- Vikram, A.; Woolston, J.; Sulakvelidze, A. Phage biocontrol applications in food production and processing. Curr. Issues Mol. Biol. 2021, 40, 267–302. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, W.; Zhong, Q.; Chen, Q.; He, X.; Baker, J.L.; Xiong, K.; Jin, X.; Wang, J.; Hu, F.; et al. Development of a bacteriophage cocktail to constrain the emergence of phage-resistant Pseudomonas aeruginosa. Front. Microbiol. 2020, 11, 327. [Google Scholar] [CrossRef] [Green Version]
- Gurney, J.; Brown, S.P.; Kaltz, O.; Hochberg, M.E. Steering phages to combat bacterial pathogens. Trends Microbiol. 2020, 28, 85–94. [Google Scholar] [CrossRef]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef] [Green Version]
- Lenneman, B.R.; Fernbach, J.; Loessner, M.J.; Lu, T.K.; Kilcher, S. Enhancing phage therapy through synthetic biology and genome engineering. Curr. Op. Biotechnol. 2021, 68, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Ando, H.; Lemire, S.; Pires, D.P.; Lu, T.K. Engineering modular viral scaffolds for targeted bacterial population editing. Cell Syst. 2015, 1, 187–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goulet, A.; Cambillau, C. Structure and topology prediction of phage adhesion devices using AlphaFold2: The case of two Oenococcus oeni phages. Microorganisms 2021, 9, 2151. [Google Scholar] [CrossRef] [PubMed]
- Goulet, A.; Joos, R.; Lavelle, K.; Van Sinderen, D.; Mahony, J.; Cambillau, C. A structural discovery journey of streptococcal phages adhesion devices by AlphaFold2. Front. Mol. Biosci. 2022, 9, 1–9. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leprince, A.; Mahillon, J. Phage Adsorption to Gram-Positive Bacteria. Viruses 2023, 15, 196. https://doi.org/10.3390/v15010196
Leprince A, Mahillon J. Phage Adsorption to Gram-Positive Bacteria. Viruses. 2023; 15(1):196. https://doi.org/10.3390/v15010196
Chicago/Turabian StyleLeprince, Audrey, and Jacques Mahillon. 2023. "Phage Adsorption to Gram-Positive Bacteria" Viruses 15, no. 1: 196. https://doi.org/10.3390/v15010196
APA StyleLeprince, A., & Mahillon, J. (2023). Phage Adsorption to Gram-Positive Bacteria. Viruses, 15(1), 196. https://doi.org/10.3390/v15010196






