Predicting Microbial Species in a River Based on Physicochemical Properties by Bio-Inspired Metaheuristic Optimized Machine Learning
Abstract
1. Introduction
2. Literature Review
2.1. Microbial Community in a River
2.2. Physicochemical and Deoxyribose Nucleic acid Sequencing Factors
2.3. Current Use of Artificial Intelligence to Predict Microbial Community
3. Methodology
3.1. Dimensionality Reduction and Handling of Missing Data
3.2. Hybrid of Multi-Output Model and Bio-Inspired Metaheuristic Optimization Algorithm
3.2.1. Multi-Output Least Squares Support Vector Regression
3.2.2. Accelerated Particle Swarm Optimization
3.3. Performance Evaluation
4. Model Development
4.1. Data Collection
4.2. Determining Critical Factors Related to Microbial Community in a River
4.3. Hybrid Model Development
5. Experimental Results
5.1. Metaheuristic Optimized Multiple-Input Multiple-Output Machine Learning
5.2. General Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bier, R.L.; Bernhardt, E.S.; Boot, C.M.; Graham, E.B.; Hall, E.K.; Lennon, J.T.; Nemergut, D.R.; Osborne, B.B.; Ruiz-González, C.; Schimel, J.P. Linking Microbial Community Structure and Microbial Processes: An Empirical and Conceptual Overview. FEMS Microbiol. Ecol. 2015, 91, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.E.; Field, D.; Gilbert, J.A. Predicting Bacterial Community Assemblages using an Artificial Neural Network Approach. Nat. Methods 2012, 9, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Freguia, S.; Logrieco, E.M.; Monetti, J.; Ledezma, P.; Virdis, B.; Tsujimura, S. Self-Powered Bioelectrochemical Nutrient Recovery for Fertilizer Generation from Human Urine. Sustainability 2019, 11, 5940. [Google Scholar] [CrossRef]
- Konopka, A. What is Microbial Community Ecology? ISME J. 2009, 3, 1223–1230. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Yang, T.; Zhang, W.; Lei, Y.; Zhang, C.; Ma, J.; Zhang, C. Preparation of NH2-Functionalized Fe2O3 and Its Chitosan Composites for the Removal of Heavy Metal Ions. Sustainability 2019, 11, 5186. [Google Scholar] [CrossRef]
- Baek, S.; Kim, S. Optimum Design and Energy Performance of Hybrid Triple Glazing System with Vacuum and Carbon Dioxide Filled Gap. Sustainability 2019, 11, 5543. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, Z.; Yin, X.; Wang, N.; Chen, D. Influences of Nitrogen Application Levels on Properties of Humic Acids in Chernozem Amended with Different Types of Organic Materials. Sustainability 2019, 11, 5405. [Google Scholar] [CrossRef]
- Heintz-Buschart, A.; Wilmes, P. Human Gut Microbiome: Function Matters. Trends Microbiol. 2017. [Google Scholar] [CrossRef]
- Sboner, A.; Mu, X.J.; Greenbaum, D.; Auerbach, R.K.; Gerstein, M.B. The Real Cost of Sequencing: Higher Than You Think! Genome Biol. 2011, 12, 1–10. [Google Scholar] [CrossRef]
- Heather, J.M.; Chain, B. The Sequence of Sequencers: The History of Sequencing DNA. Genomics 2016, 107, 1–8. [Google Scholar] [CrossRef]
- Janizadeh, S.; Avand, M.; Jaafari, A.; Phong, V.T.; Bayat, M.; Ahmadisharaf, E.; Prakash, I.; Pham, T.B.; Lee, S. Prediction Success of Machine Learning Methods for Flash Flood Susceptibility Mapping in the Tafresh Watershed, Iran. Sustainability 2019, 11, 5426. [Google Scholar] [CrossRef]
- Sivapriya, T.; Kamal, A.N.B.; Thavavel, V. Imputation and Classification of Missing Data using Least Square Support Vector Machines–A New Approach in Dementia Diagnosis. Int. J. Adv. Res. Artif. Intell. 2012, 1, 29–33. [Google Scholar] [CrossRef]
- Zhang, D.; Zhu, Q.; Zhang, D. Multi-Modal Dimensionality Reduction using Effective Distance. Neurocomputing 2017, 259, 130–139. [Google Scholar] [CrossRef]
- Shiklomanov, I.A. World Water Resources: A New Appraisal and Assessment for the 21st Century: A Summary of the Monograph World Water Resources; UNESCO International Hydrological Programme, UNESCO-IHP: Paris, France, 1998. [Google Scholar]
- Stanley, E.H.; Fisher, S.G.; Grimm, N.B. Ecosystem Expansion and Contraction in Streams. BioScience 1997, 47, 427–435. [Google Scholar] [CrossRef]
- Ghai, R.; Rodŕíguez-Valera, F.; McMahon, K.D.; Toyama, D.; Rinke, R.; de Oliveira, T.C.S.; Garcia, J.W.; de Miranda, F.P.; Henrique-Silva, F. Metagenomics of the Water Column in the Pristine Upper Course of the Amazon River. PLoS ONE 2011, 6, e23785. [Google Scholar] [CrossRef] [PubMed]
- Newton, R.J.; Bootsma, M.J.; Morrison, H.G.; Sogin, M.L.; McLellan, S.L. A Microbial Signature Approach to Identify Fecal Pollution in the Waters Off an Urbanized Coast of Lake Michigan. Microb. Ecol. 2013, 65, 1011–1023. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, Y.; Zhou, C.; Meng, J.; Sun, J.; Zhou, T.; Tao, J. Impact of climate factors on future distributions of Paeonia ostii across China estimated by MaxEnt. Ecol. Inform. 2019, 50, 62–67. [Google Scholar] [CrossRef]
- Ager, D.; Evans, S.; Li, H.; Lilley, A.K.; Van Der Gast, C.J. Anthropogenic Disturbance Affects the Structure of Bacterial Communities. Environ. Microbiol. 2010, 12, 670–678. [Google Scholar] [CrossRef]
- Kneip, C.; Lockhart, P.; Voß, C.; Maier, U.-G. Nitrogen Fixation in Eukaryotes–New Models for Symbiosis. BMC Evol. Biol. 2007, 7, 55. [Google Scholar] [CrossRef]
- Bernhard, A. The Nitrogen Cycle: Processes. Available online: https://www.nature.com/scitable/knowledge/library/the-nitrogen-cycle-processes-players-and-human-15644632 (accessed on 18 December 2018).
- Gougoulias, C.; Clark, J.M.; Shaw, L.J. The Role of Soil Microbes in the Global Carbon Cycle: Tracking the Below-Ground Microbial Processing of Plant-Derived Carbon for Manipulating Carbon Dynamics in Agricultural Systems. J. Sci. Food Agric. 2014, 94, 2362–2371. [Google Scholar] [CrossRef]
- Schlegel, H. Microorganisms Involved in the Nitrogen and Sulfur Cycles. In Biology of Inorganic Nitrogen and Sulfur; Springer: Berlin/Heidelberg, Germany, 1981; pp. 3–12. [Google Scholar] [CrossRef]
- Hu, A.; Yao, T.; Jiao, N.; Liu, Y.; Yang, Z.; Liu, X. Community Structures of Ammonia-Oxidising Archaea and Bacteria in High-Altitude Lakes on the Tibetan Plateau. Freshw. Biol. 2010, 55, 2375–2390. [Google Scholar] [CrossRef]
- Fenwick, A. Waterborne Infectious Diseases—Could They be Consigned to History? Science 2006, 313, 1077–1081. [Google Scholar] [CrossRef] [PubMed]
- Cabral, J.P. Water Microbiology. Bacterial Pathogens and Water. Int. J. Environ. Res. Public Health 2010, 7, 3657–3703. [Google Scholar] [CrossRef] [PubMed]
- Pirofski, L.-A.; Casadevall, A. Q and A What is a Pathogen? BMC Biol. 2012, 10, 6. [Google Scholar] [CrossRef]
- Breznak, J.A.; Costilow, R.N. Physicochemical Factors in Growth. In Methods for General and Molecular Microbiology, 3rd ed.; American Society of Microbiology: Washington DC, USA, 2007; pp. 309–329. [Google Scholar] [CrossRef]
- Alesheikh, S.; Shahtahmassebi, N.; Roknabadi, M.R.; Pilevar Shahri, R. Silicene Nanoribbon as a New DNA Sequencing Device. Phys. Lett. A 2018, 382, 595–600. [Google Scholar] [CrossRef]
- Yang, N.; Jiang, X. Nanocarbons for DNA Sequencing: A Review. Carbon 2017, 115, 293–311. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, Y.; Ying, C.; Wang, D.; Du, C. Nanopore-Based Fourth-Generation DNA Sequencing Technology. Genom. Proteom. Bioinform. 2015, 13, 4–16. [Google Scholar] [CrossRef]
- Ansorge, W.J. Next-Generation DNA Sequencing Techniques. New Biotechnol. 2009, 25, 195–203. [Google Scholar] [CrossRef]
- Kircher, M.; Heyn, P.; Kelso, J. Addressing Challenges in the Production and Analysis of Illumina Sequencing Data. BMC Genom. 2011, 12, 382. [Google Scholar] [CrossRef]
- Buermans, H.P.J.; den Dunnen, J.T. Next Generation Sequencing Technology: Advances and Applications. Biochim. Et Biophys. Acta (Bba) Mol. Basis Dis. 2014, 1842, 1932–1941. [Google Scholar] [CrossRef]
- Wu, S.G.; Wang, Y.; Jiang, W.; Oyetunde, T.; Yao, R.; Zhang, X.; Shimizu, K.; Tang, Y.J.; Bao, F.S. Rapid Prediction of Bacterial Heterotrophic Fluxomics using Machine Learning and Constraint Programming. PLoS Comput. Biol. 2016, 12, e1004838. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Kamber, M.; Pei, J. Data Preprocessing. In Data Mining, 3rd ed.; Morgan Kaufmann: Boston, MA, USA, 2012. [Google Scholar]
- Famili, A.; Shen, W.-M.; Weber, R.; Simoudis, E. Data Preprocessing and Intelligent Data Analysis. Intell. Data Anal. 1997, 1, 3–23. [Google Scholar] [CrossRef]
- Ngoc Thach, N.; Bao-Toan Ngo, D.; Xuan-Canh, P.; Hong-Thi, N.; Hang Thi, B.; Nhat-Duc, H.; Dieu, T.B. Spatial pattern assessment of tropical forest fire danger at Thuan Chau area (Vietnam) using GIS-based advanced machine learning algorithms: A comparative study. Ecol. Inform. 2018, 46, 74–85. [Google Scholar] [CrossRef]
- Rajeswari, K.; Vaithiyanathan, V.; Pede, S.V. Feature Selection for Classification in Medical Data Mining. Int. J. Emerg. Trends Technol. Comput. Sci. (Ijettcs) 2013, 2, 492–497. [Google Scholar] [CrossRef]
- Kwak, S.K.; Kim, J.H. Statistical Data Preparation: Management of Missing Values and Outliers. Korean J. Anesthesiol. 2017, 70, 407–411. [Google Scholar] [CrossRef]
- Qi, Z.; Wang, H.; Li, J.; Gao, H. FROG: Inference from Knowledge Base for Missing Value Imputation. Knowl. Based Syst. 2018, 145, 77–90. [Google Scholar] [CrossRef]
- Tsai, C.-F.; Li, M.-L.; Lin, W.-C. A Class Center based Approach for Missing Value Imputation. Knowl. -Based Syst. 2018, 151, 124–135. [Google Scholar] [CrossRef]
- Li, E.Y. Artificial Neural Networks and Their Business Applications. Inf. Manag. 1994, 27, 303–313. [Google Scholar] [CrossRef]
- Chou, J.-S.; Yang, K.-H.; Lin, J.-Y. Peak Shear Strength of Discrete Fiber-Reinforced Soils Computed by Machine Learning and Metaensemble Methods. J. Comput. Civ. Eng. 2016, 30, 04016036. [Google Scholar] [CrossRef]
- Zou, K.H.; Tuncali, K.; Silverman, S.G. Correlation and Simple Linear Regression. Radiology 2003, 227, 617–628. [Google Scholar] [CrossRef]
- Kumar, R. Decision Tree for the Weather Forecasting. Int. J. Comput. Appl. 2013, 76, 31–34. [Google Scholar] [CrossRef]
- Chou, J.-S.; Ho, C.-C.; Hoang, H.-S. Determining quality of water in reservoir using machine learning. Ecol. Inform. 2018, 44, 57–75. [Google Scholar] [CrossRef]
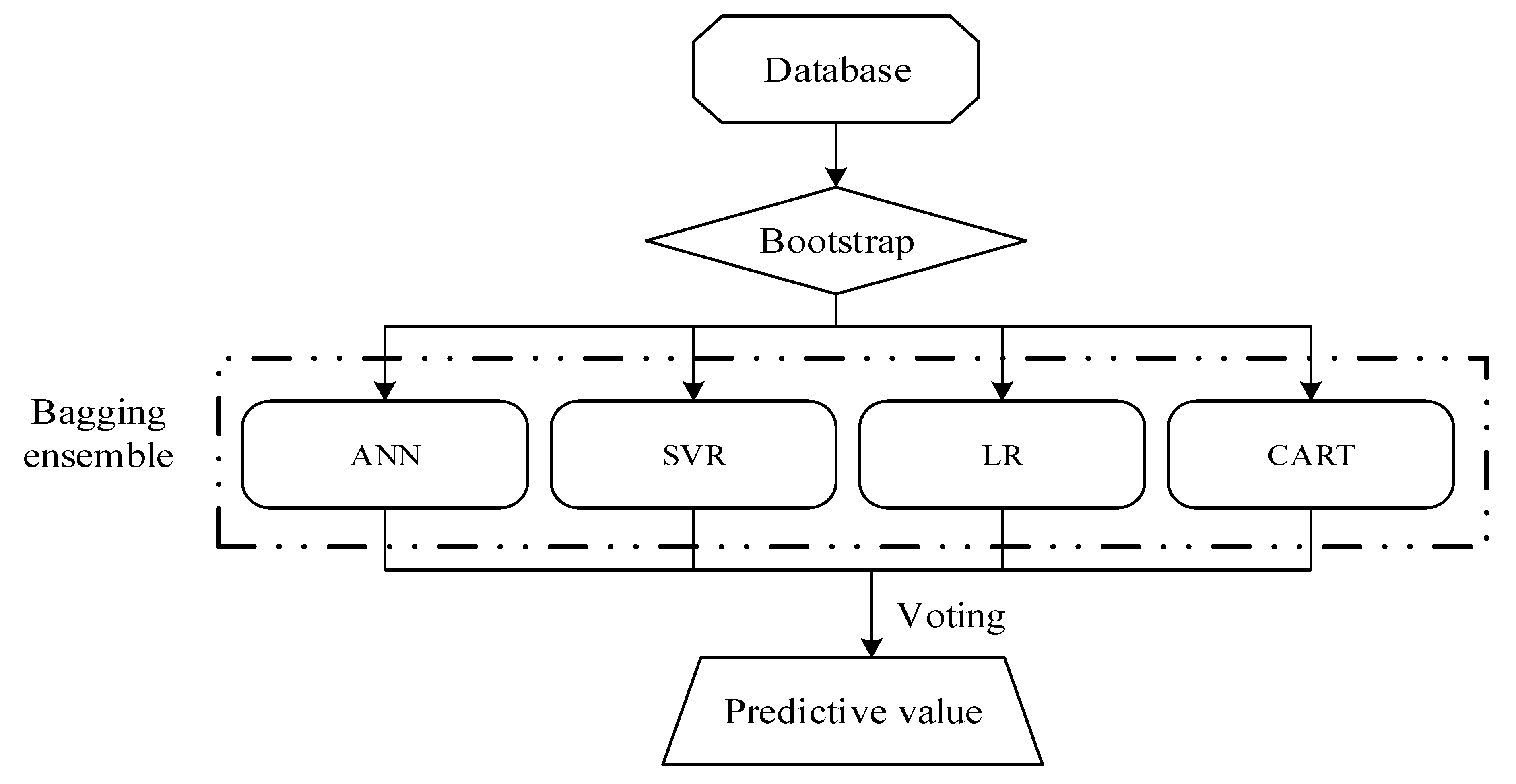
- Breiman, L. Bagging Predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Jiang, T.; Zhou, X. Gradient/Hessian-Enhanced Least Square Support Vector Regression. Inf. Process. Lett. 2018, 134, 1–8. [Google Scholar] [CrossRef]
- Xu, S.; An, X.; Qiao, X.; Zhu, L.; Li, L. Multi-Output Least-Squares Support Vector Regression Machines. Pattern Recognit. Lett. 2013, 34, 1078–1084. [Google Scholar] [CrossRef]
- Khennak, I.; Drias, H. An Accelerated PSO for Query Expansion in Web Information Retrieval: Application to Medical Dataset. Appl. Intell. 2017, 47, 793–808. [Google Scholar] [CrossRef]
- Yang, X.-S.; Deb, S.; Fong, S. Accelerated Particle Swarm Optimization and Support Vector Machine for Business Optimization and Applications. In Proceedings of the International Conference on Networked Digital Technologies, Macau, 11–13 July 2011; pp. 53–66. [Google Scholar]
- Colin Cameron, A.; Windmeijer, F.A.G. An R-Squared Measure of Goodness of Fit for Some Common Nonlinear Regression Models. J. Econom. 1997, 77, 329–342. [Google Scholar] [CrossRef]
- Huang, L.-S.; Chen, J. Analysis of Variance, Coefficient of Determination and F-Test for Local Polynomial Regression. Ann. Stat. 2008, 36, 2085–2109. [Google Scholar] [CrossRef]
- Park, Y.-S.; Lek, S. Artificial Neural Networks: Multilayer Perceptron for Ecological Modeling. In Developments in Environmental Modelling; Elsevier: Amsterdam, The Netherlands, 2016; Volume 28, pp. 123–140. [Google Scholar]
- Jerves-Cobo, R.; Córdova-Vela, G.; Iñiguez-Vela, X.; Díaz-Granda, C.; Van Echelpoel, W.; Cisneros, F.; Nopens, I.; Goethals, P. Model-based analysis of the potential of macroinvertebrates as indicators for microbial pathogens in rivers. Water 2018, 10, 375. [Google Scholar] [CrossRef]
- Jerves-Cobo, R.; Forio, M.A.E.; Lock, K.; Van Butsel, J.; Pauta, G.; Cisneros, F.; Nopens, I.; Goethals, P.L.M. Biological water quality in tropical rivers during dry and rainy seasons: A model-based analysis. Ecol. Indic. 2020, 108, 105769. [Google Scholar] [CrossRef]
- Damanik-Ambarita, M.N.; Everaert, G.; Forio, M.A.E.; Nguyen, T.H.T.; Lock, K.; Musonge, P.L.S.; Suhareva, N.; Dominguez-Granda, L.; Bennetsen, E.; Boets, P.; et al. Generalized linear models to identify key hydromorphological and chemical variables determining the occurrence of macroinvertebrates in the guayas river basin (ecuador). Water 2016, 8, 297. [Google Scholar] [CrossRef]
- Aazami, J.; Esmaili Sari, A.; Abdoli, A.; Sohrabi, H.; Van den Brink, P.J. Assessment of ecological quality of the tajan river in iran using a multimetric macroinvertebrate index and species traits. Environ. Manag. 2015, 56, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Forio, M.A.E.; Goethals, P.L.M.; Lock, K.; Asio, V.; Bande, M.; Thas, O. Model-based analysis of the relationship between macroinvertebrate traits and environmental river conditions. Environ. Model. Softw. 2018, 106, 57–67. [Google Scholar] [CrossRef]




| Data Collecting Period | Season | Average Temperature (°C) | Average Precipitation (mm) |
|---|---|---|---|
| 5–6 September 2012 | Normal | 26 | 141 |
| 15–17 January 2012 | Dry | 13 | 34 |
| 7–9 June 2013 | Wet | 26 | 187 |
| No. | Attribute | No. | Attribute | No. | Attribute | No. | Attribute |
|---|---|---|---|---|---|---|---|
| 1 | Temperature | 15 | ATP | 29 | GBL | 43 | OMC |
| 2 | pH | 16 | BP3 | 30 | IDT | 44 | PPB |
| 3 | DO | 17 | BPA | 31 | IPF | 45 | PPL |
| 4 | DO% | 18 | BPB | 32 | KPF | 46 | PPZ |
| 5 | Spc | 19 | CAF | 33 | LOR | 47 | SDZ |
| 6 | NH4-N | 20 | CDI | 34 | LOS | 48 | SFZ |
| 7 | NO2-N | 21 | CPP | 35 | MBB | 49 | SIL |
| 8 | NO3-N | 22 | CRO | 36 | MEF | 50 | SMT |
| 9 | DIN | 23 | CZP | 37 | MNZ | 51 | SMZ |
| 10 | TP | 24 | DAN | 38 | MPB | 52 | TBD |
| 11 | PO4-P | 25 | DFA | 39 | MPL | 53 | TCC |
| 12 | DIN/SRP | 26 | DZP | 40 | NPF | 54 | TCS |
| 13 | APR | 27 | ENX | 41 | OCT | - | |
| 14 | ASP | 28 | FPF | 42 | OF | - |
| No. | Attribute | No. | Attribute | No. | Attribute |
|---|---|---|---|---|---|
| 1 | Unclassified | 28 | Firmicutes | 55 | Betaproteobacteria |
| 2 | Other Archaea | 29 | Fusobacteria | 56 | Deltaproteobacteria |
| 3 | Crenarchaeota | 30 | GAL15 | 57 | Epsilonproteobacteria |
| 4 | Euryarchaeota | 31 | GN02 | 58 | Gammaproteobacteria |
| 5 | Parvarchaeota | 32 | GN04 | 59 | TA18 |
| 6 | Other Bacteria | 33 | GOUTA4 | 60 | Other Proteobacteria |
| 7 | Unclassified Bacteria | 34 | Gemmatimonadetes | 61 | SBR1093 |
| 8 | AC1 | 35 | H-178 | 62 | SC4 |
| 9 | AD3 | 36 | Hyd24-12 | 63 | SR1 |
| 10 | Acidobacteria | 37 | KSB3 | 64 | Spirochaetes |
| 11 | Actinobacteria | 38 | Kazan 3B-28 | 65 | Synergistetes |
| 12 | Aquificae | 39 | LCP-89 | 66 | TM6 |
| 13 | Armatimonadetes | 40 | LD1 | 67 | TM7 |
| 14 | BHI80-139 | 41 | Lentisphaerae | 68 | TPD-58 |
| 15 | BRC1 | 42 | MVP-21 | 69 | Tenericutes |
| 16 | Bacteroidetes | 43 | MVS-104 | 70 | Thermotogae |
| 17 | Caldiserica | 44 | NC10 | 71 | Verrucomicrobia |
| 18 | Caldithrix | 45 | NKB19 | 72 | WPS-2 |
| 19 | Chlamydiae | 46 | Nitrospirae | 73 | WS1 |
| 20 | Chlorobi | 47 | OC31 | 74 | WS2 |
| 21 | Chloroflexi | 48 | OD1 | 75 | WS3 |
| 22 | Cyanobacteria | 49 | OP11 | 76 | WS4 |
| 23 | Deferribacteres | 50 | OP3 | 77 | WS5 |
| 24 | Elusimicrobia | 51 | OP8 | 78 | WWE1 |
| 25 | FBP | 52 | OP9 | 79 | ZB3 |
| 26 | FCPU426 | 53 | Planctomycetes | 80 | Caldithrix |
| 27 | Fibrobacteres | 54 | Alphaproteobacteria | 81 | Thermi |
| Variable | Unit | Parameter (Abbreviation) | Direction | Min. | Max. | Mean |
|---|---|---|---|---|---|---|
| X1 | °C | Temperature (Temp) | Input | 14.00 | 30.67 | 22.75 |
| X2 | - | pH | Input | 6.58 | 8.40 | 7.39 |
| X3 | mg/L | Dissolved Oxygen (DO) | Input | 4.37 | 11.11 | 7.42 |
| X4 | mg/L | Ammonium Nitrogen (NH4-N) | Input | 0.00 | 9.24 | 1.38 |
| X5 | mg/L | Nitrate Nitrogen (NO3-N) | Input | 0.27 | 18.65 | 6.03 |
| X6 | mg/L | Total Phosphorus (TP) | Input | 0.08 | 1.82 | 0.41 |
| X7 | mg/L | Orthophosphate (PO4-P) | Input | 0.02 | 0.68 | 0.20 |
| X8 | mg/L | Inorganic Nitrogen (DIN) | Input | 1.99 | 24.92 | 7.74 |
| Y1 | - | Actinobacteria | Output | 0.00 | 0.54 | 0.11 |
| Y2 | - | Bacteroidetes | Output | 0.01 | 0.30 | 0.10 |
| Y3 | - | Cyanobacteria | Output | 0.00 | 0.24 | 0.02 |
| Y4 | - | Firmicutes | Output | 0.00 | 0.75 | 0.12 |
| Y5 | - | Planctomycetes | Output | 0.00 | 0.20 | 0.03 |
| Y6 | - | Proteobacteria | Output | 0.19 | 0.90 | 0.57 |
| Y7 | - | Others | Output | 0.01 | 0.13 | 0.05 |
| Stage | Amount of Inputs and Outputs | Dimensionality Reduction Method |
|---|---|---|
| I | 54 and 81 | Expert engineering judgement |
| 38 and 17 | ||
| II | 38 and 17 | Expert engineering judgement, feature creation, feature subset selection |
| 10 and 17 | ||
| III | 10 and 17 | Expert engineering judgement, feature creation |
| 10 and 7 | ||
| IV | 10 and 7 | Feature subset selection |
| 8 and 7 |
| Stage | Parameter (Abbreviation) | Feature Creation |
|---|---|---|
| II | Antipyrine (APR) | Pharmaceuticals and Personal Care Products (PPCP) |
| Paracetamol (ATP) | ||
| BP-3 (BP3) | ||
| BPA | ||
| Caffeine (CAF) | ||
| Carbamazepine (CZP) | ||
| Diclofenac (DFA) | ||
| Diazepam (DZP) | ||
| Fenoprofen (FPF) | ||
| Gemfibrozil (GBL) | ||
| Indomethacine (IDT) | ||
| Ibuprofen (IPF) | ||
| Ketoprofen (KPF) | ||
| Mefenamic (MEF) | ||
| Methylparaben (MPB) | ||
| Metoprolol (MPL) | ||
| Naprofen (NPF) | ||
| Octocrylene (OCT) | ||
| OMC | ||
| Propyl Paraben (PPB) | ||
| Sulfadiazine (SDZ) | ||
| Sulfamethoxazole (SFZ) | ||
| Sulfamethazine (SMT) | ||
| Thiabendazole (TBD) | ||
| TCC | ||
| TCS | ||
| IV | Alphaproteobacteria | Proteobacteria |
| Betaproteobacteria | ||
| Deltaproteobacteria | ||
| Gammaproteobacteria |
| R2 | Sig. | Temp | pH | DIN | DO | NH4-N | NO3-N | TP | PO4-P | NO2-N | PPCP | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteroidetes | 0.309 | 0.000 | √ | √ | ||||||||
| Firmicutes | 0.474 | 0.000 | √ | √ | √ | |||||||
| Proteobacteria | 0.315 | 0.000 | √ | √ | √ | |||||||
| Actinobacteria | 0.140 | 0.003 | √ | |||||||||
| Cyanobacteria | 0.335 | 0.000 | √ | √ | ||||||||
| Planctomycetes | 0.237 | 0.000 | √ | |||||||||
| Others | 0.266 | 0.000 | √ | |||||||||
| Score | 2 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| Field | Value |
| Dataset characteristics | Multivariate |
| Attributes characteristics | Real |
| Number of instances | 40 |
| Input attributes (26) | Temperature (Temp) pH Ammonium Nitrogen (NH4-N) Nitrate Nitrogen (NO3-N) Total Phosphorus (TP) Orthophosphate (PO4-P) Inorganic Nitrogen (DIN) Pharmaceuticals and Personal Care Products (PPCP) Crenarchaeota Euryarchaeota Acidobacteria Actinobacteria Bacteroidetes Cholorobi Choloroflexi Cyanobacteria Firmicutes Nitrospirae Planctomycetes Alphaproteobacteria Betaproteobacteria Gammaproteobacteria Verrucomicrobia Others |
| Output attribute (1) | Dissolved Oxygen (DO) |
| Parameters | Value |
|---|---|
| Number of Iteration | 50 |
| Number of Swarm Population | 20 |
| 0.2 | |
| 0.5 | |
| Number of Folds | 10 |
| Upper Bound | [2 2 2] |
| Lower Bound | [−2 −2 −2] |
| Field | Value |
|---|---|
| Number of data points | 60 |
| Training/testing data proportions | 80/20 (%) |
| Number of input attributes | 8 |
| Number of output attributes | 7 |
| Hyperparameters: | |
| 0.1553 | |
| 0.7752 | |
| 1.3620 |
| Number of Inputs | Performance Measure | ||||||
|---|---|---|---|---|---|---|---|
| MAAPE (%) | MAE | R2 | Overall SI | ||||
| Training | Testing | Training | Testing | Training | Testing | ||
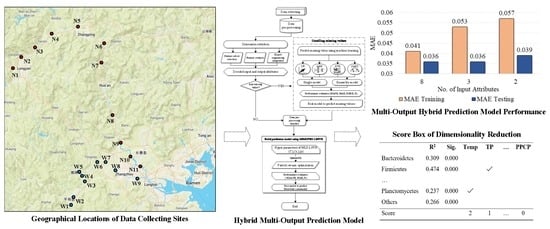
| 8 | 53 | 35 | 0.041 | 0.036 | 0.660 | 0.357 | 1 (1) |
| 3 | 58 | 37 | 0.053 | 0.036 | 0.442 | 0.223 | 0.415 (2) |
| 2 | 62 | 39 | 0.057 | 0.039 | 0.351 | 0.239 | 0.012 (3) |
| Literature | Machine Learning Technique | Parameter Setting | Model Type | Multiple Outputs |
|---|---|---|---|---|
| Larsen et al. [2] | Artificial neural network (ANN) | Manual experiment | Single | No |
| Jerves-Cobo et al. [56] | Decision tree models (DTMs) | Manual experiment | Single | No |
| Jerves-Cobo et al. [57] | Generalized linear model (GLM) | Manual experiment | Single | No |
| M.N. Damanik-Ambarita et al. [58] | Generalized linear model (GLM) | Manual experiment | Single | No |
| Aazami et al. [59] | Multimetric Macroinvertebrate Index (MMI) | Manual experiment | Single | No |
| Forio et al. [60] | Negative binomial regression (NBM) | Manual experiment | Single | No |
| This study | Multiple-input multiple-output machine learning | Automatically identified by bio-inspired metaheuristic optimization algorithms | Hybrid | Yes |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chou, J.-S.; Yu, C.-P.; Truong, D.-N.; Susilo, B.; Hu, A.; Sun, Q. Predicting Microbial Species in a River Based on Physicochemical Properties by Bio-Inspired Metaheuristic Optimized Machine Learning. Sustainability 2019, 11, 6889. https://doi.org/10.3390/su11246889
Chou J-S, Yu C-P, Truong D-N, Susilo B, Hu A, Sun Q. Predicting Microbial Species in a River Based on Physicochemical Properties by Bio-Inspired Metaheuristic Optimized Machine Learning. Sustainability. 2019; 11(24):6889. https://doi.org/10.3390/su11246889
Chicago/Turabian StyleChou, Jui-Sheng, Chang-Ping Yu, Dinh-Nhat Truong, Billy Susilo, Anyi Hu, and Qian Sun. 2019. "Predicting Microbial Species in a River Based on Physicochemical Properties by Bio-Inspired Metaheuristic Optimized Machine Learning" Sustainability 11, no. 24: 6889. https://doi.org/10.3390/su11246889
APA StyleChou, J.-S., Yu, C.-P., Truong, D.-N., Susilo, B., Hu, A., & Sun, Q. (2019). Predicting Microbial Species in a River Based on Physicochemical Properties by Bio-Inspired Metaheuristic Optimized Machine Learning. Sustainability, 11(24), 6889. https://doi.org/10.3390/su11246889