Deciphering the Factors for Nodulation and Symbiosis of Mesorhizobium Associated with Cicer arietinum in Northwest India
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Sites, Isolation, and Culture Condition
2.2. Validation, Nodulation, and Host-Specificity Test of Chickpea Nodule Rhizobia
2.3. PGP Traits
2.4. Biofilm Assay
2.5. Biolog Carbon Source Utilization and Numerical Analysis
2.6. Molecular Identification, Sequence Typing, Genetic Differentiation, and Gene Flow of (gap, edD, gnD, glnD, rpoB, and nodC)
2.6.1. Nucleotide Sequence Accession Number
2.6.2. Statistical Analysis
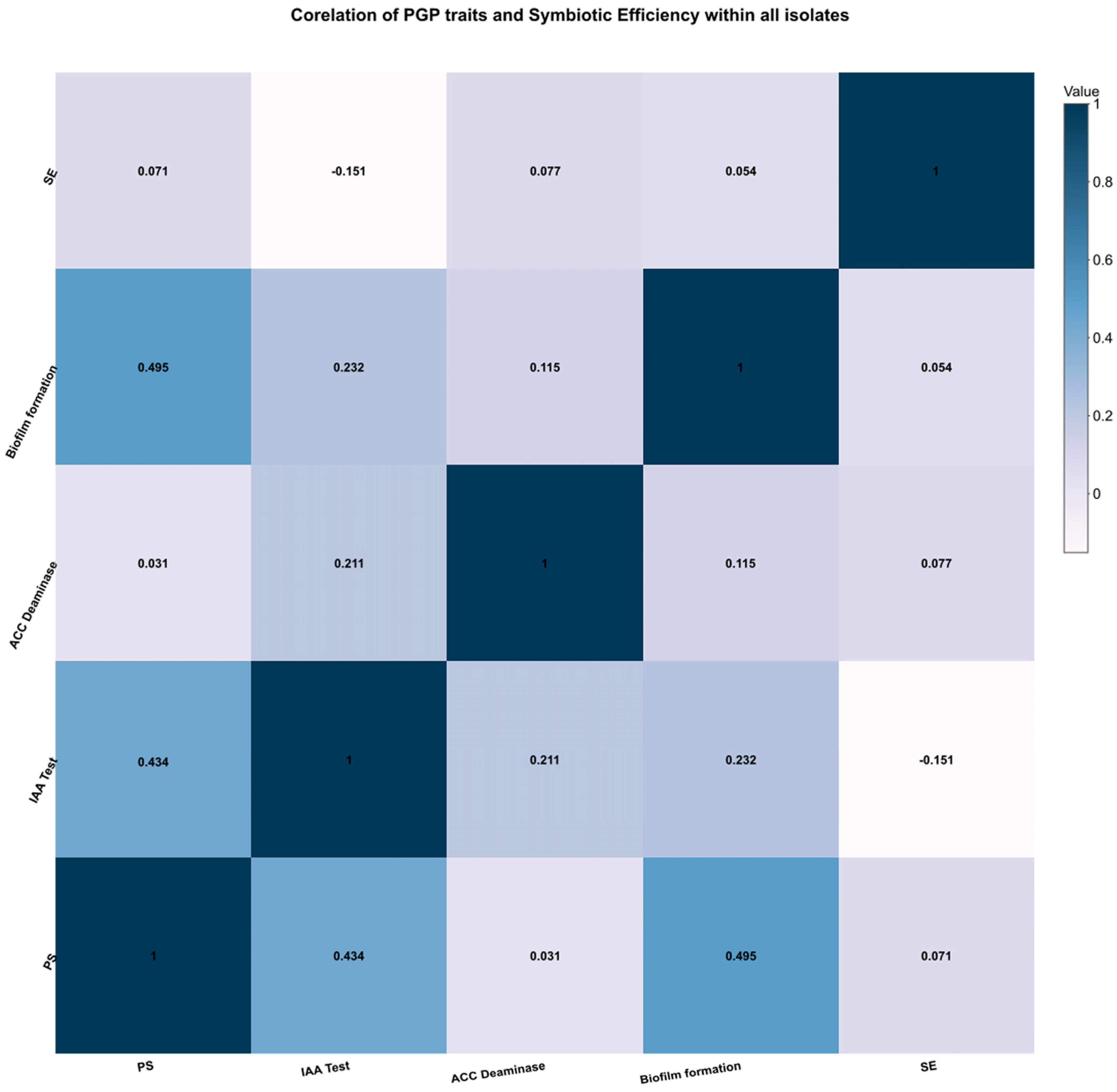
3. Results
3.1. Morph-Phenotypic Characters
3.2. Biofilm Efficiency
3.3. Nodulation and Symbiotic Efficiency Test
3.4. Carbon Utilization
3.5. Molecular Identification and Sequence Typing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Singh, R.P.; Manchanda, G.; Maurya, I.K.; Tiwari, P.K.; Maheshwari, N.K.; Rai, A.R. Streptomyces from rotten wheat straw endowed the high Plant growth potential traits and agro-active compounds. Biocatal. Agri. Biotechnol. 2019, 17, 507–513. [Google Scholar] [CrossRef]
- Pinski, A.; Betekhtin, A.; Hupert-Kocurek, K.; Luis, A.J.M.; Hasterok, R. Defining the Genetic Basis of Plant-Endophytic Bacteria Interactions. Int. J. Mol. Sci. 2019, 20, 1947. [Google Scholar] [CrossRef] [PubMed]
- Gage, D.J. Infection and Invasion of Roots by Symbiotic, Nitrogen-Fixing Rhizobia during Nodulation of Temperate Legumes. Microbiol. Mol. Biol. Rev. 2004, 68, 280–300. [Google Scholar] [CrossRef] [PubMed]
- Santoyo, G.; Moreno-Hagelsieb, G.; del Carmen Orozco-Mosqueda, M.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Rovira, A.D. Rhizosphere research, 85 years of progress and frustration. In The Rhizosphere and Plant Growth; Keister, D.L., Cregan, P.B., Eds.; Kluwer Academic Publisher: Dordrecht, The Netherlands, 1991; pp. 3–13. [Google Scholar]
- Andrews, J.H.; Harris, R.F. The ecology and biogeography of microorganisms of plant surfaces. Annual. Rev. Phytopathol. 2000, 38, 145–180. [Google Scholar] [CrossRef]
- Karatan, E.; Watnick, P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 2009, 73, 310–347. [Google Scholar] [CrossRef]
- Davey, M.E.; O’Toole, G.A. Microbial biofilms: From ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867. [Google Scholar] [CrossRef]
- Nocelli, N.; Bogino, P.C.; Banchio, E.; Giordano, W. Roles of Extracellular Polysaccharides and Biofilm Formation in Heavy Metal Resistance of Rhizobia. Materials 2016, 9, 418. [Google Scholar] [CrossRef]
- Rinaudi, L.V.; Giordano, W. An integrated view of biofilm formation in rhizobia. FEMS Microbiol. Lett. 2010, 304, 1–11. [Google Scholar] [CrossRef]
- Yuan, H.L.; Wang, E.T.; Jiao, Y.S.; Tian, C.F.; Wang, L.; Wang, Z.J.; Guan, J.J.; Singh, R.P.; Chen, W.X.; Che, W.F. Symbiotic characteristics of Bradyrhizobium diazoefficiens USDA 110 mutants associated with shrubby sophora (Sophora flavescens) and soybean (Glycine max). Microbiol. Res. 2018, 214, 19–27. [Google Scholar]
- Sturz, A.; Christie, B.; Nowak, J. Bacterial endophytes, potential role in developing sustainable systems of crop production. CRC Crit. Rev. Plant Sci. 2000, 19, 1–30. [Google Scholar] [CrossRef]
- Weyens, N.; Van-der, L.D.; Taghavi, S.; Vangronsveld, J. Phytoremediation, plant endophyte partnerships take the challenge. Curr. Opin. Biotechnol. 2009, 20, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Brígido, C.; Singh, S.; Menéndez, E.; Tavares, M.J.; Glick, B.R.; Félix, M.R.; Oliveira, S.; Carvalho, M. Diversity and Functionality of Culturable Endophytic Bacterial Communities in Chickpea Plants. Plants 2019, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Matos, I.; Schröder, E.C. Strain selection for pigeon pea Rhizobium under greenhouse conditions. Plant Soil. 1989, 116, 19–22. [Google Scholar] [CrossRef]
- FAOSTAT. FAO (Food and Agricultural Organization of the United Nations). Available online: http://faostat.fao.org (accessed on 11 December 2014).
- Upadhyaya, H.D.; Kashiwagi, J.; Varshney, R.K.; Gaur, P.M.; Saxena, K.B.; Krishnamurthy, L.; Gowda, C.L.L.; Pundir, R.P.S.; Chaturvedi, S.K.; Basu, P.S.; et al. Phenotyping chickpeas and pigeonpeas for adaptation to drought. Front. Physiol. 2012, 3, 179. [Google Scholar] [CrossRef]
- Dudeja, S.S.; Nidhi. Molecular diversity of rhizobial and non rhizobial bacteria from nodules of cool season legumes. In Biotechnology: Prospects and Applications; Salar, R.K., Gahlawat, S.K., Siwach, P., Duhan, J.S., Eds.; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar] [CrossRef]
- Pérez-Yépez, J.; Armas-Capote, N.; Velázquez, E.; Pérez-Galdona, R.; Rivas, R.; León-Barrios, M. Evaluation of seven housekeeping genes for multilocus sequence analysis of the genus Mesorhizobium: Resolving the taxonomic affiliation of the Cicer canariense rhizobia. Syst. Appl. Microbiol. 2014, 37, 553–559. [Google Scholar] [CrossRef]
- Nour, S.H.; Fernandez, M.P.; Normand, P.; Cleyet-Marel, J.C. Rhizobium ciceri sp. nov., consisting of strains that nodulate chickpeas (Cicer arietinum L.). Inter. J. Syst. Bacteriol. 1994, 44, 511–522. [Google Scholar] [CrossRef]
- Nour, S.M.; Cleyet-Marel, J.; Normand, C.P.; Fernandez, M.P. Genomic heterogeneity of strains nodulating chickpeas (Cicer arietinum L.) and description of Rhizobium mediterraneum sp. nov. Inter. J. Syst. Bacteriol. 1995, 45, 640–648. [Google Scholar] [CrossRef]
- Zgadzaj, R.; James, E.K.; Kelly, S.; Kawaharada, Y.; de Jonge, N.; Jensen, D.B.; Madsen, L.H.; Radutoiu, S. A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic Bacteria. PLoS Genet. 2015, 11, 1–21. [Google Scholar] [CrossRef]
- Andrews, M.; Andrews, M.E. Specificity in Legume-Rhizobia Symbioses. Int. J. Mol. Sci. 2017, 18, 705. [Google Scholar] [CrossRef]
- Geurts, R.; Franssen, H. Signal transduction in Rhizobium induced nodule formation. Plant Physiol. 1996, 112, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Cabanes, D.; Boistard, P.; Batut, J. Identification of Sinorhizobium meliloti genes regulated during symbiosis. J. Bacteriol. 2000, 182, 3632–3637. [Google Scholar] [CrossRef] [PubMed]
- Flores-Félix, J.D.; Carro, L.; Velázquez, E.; Valverde, A.; Cerda-Castillo, E.; García-Fraile, P.; Rivas, R. Phyllobacterium endophyticum sp. nov., isolated from nodules of Phaseolus vulgaris. Int. J. Syst. Evol. Microbiol. 2012, 63, 821–826. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.M. A Manual for the Practical Study of Root-Nodule Bacteria; IBP Handbook 15; Blackwell: Oxford, UK, 1970. [Google Scholar]
- Zhang, J.J.; Yang, X.; Chen, G.; de Lajudie, P.; Singh, R.P.; Wang, E.T.; Chen, W.F. Mesorhizobium muleiense and Mesorhizobium gsp. nov. are symbionts of Cicer arietinum L. in alkaline soils of Gansu, Northwest China. Plant Soil 2016, 410, 103–112. [Google Scholar] [CrossRef]
- Zhang, J.J.; Jing, X.Y.; de Lajudie, P.; Ma, C.; He, P.X.; Singh, R.P.; Chen, W.F.; Wang, E.T. Association of white clover (Trifolium repens L.) with rhizobia of sv. trifolii belonging to three genomic species in alkaline soils in North and East China. Plant Soil 2016, 407, 417–427. [Google Scholar]
- Gibson, A.H. Evaluation of nitrogen fixation by legumes in the greenhouse and growth chamber. In Symbiotic Nitrogen Fixation Technology; Gibson, A.H., Ed.; Marcel Dekker: New York, NY, USA, 1987; pp. 321–363. [Google Scholar]
- Nandasena, K.G.; O’Hara, G.W.; Tiwari, R.P.; Yates, R.J.; Kishinevsky, B.D.; Howieson, J.G. Symbiotic relationships and root nodule ultrastructure of the pasture legume Biserrula pelecinus L.; a new legume in agriculture. Soil Biol. Biochem. 2004, 36, 1309–1317. [Google Scholar] [CrossRef]
- Pikovskya, R.I. Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. Microbiology 1948, 17, 362–370. [Google Scholar]
- Vazquez, P.; Holguin, G.; Puente, M.E.; Lopez-Cortes, A.; Bashan, Y. Phosphate solubilizing microorganisms associated with the rhizosphere of Mangroves growing in a semiarid coastal lagoon. Biol. Fertil. Soils 2000, 30, 460–468. [Google Scholar] [CrossRef]
- Khanmna, S.; Yokota, A.; Lumyong, S. Actinomycetes isolated from medicinal plant rhizosphere soils: Diversity and screening of antifungal compounds, indole-3-acetic acid and siderophore production. World J. Microbiol. Biotechnol. 2009, 25, 649–655. [Google Scholar] [CrossRef]
- Smibert, R.M.; Krieg, N.R. Phenotypic characterization. In Methods for General and Molecular Bacteriology; Gerhardt, P., Murray, R.G.E., Wood, W.A., Krieg, N.R., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 607–654. [Google Scholar]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef]
- Mirani, Z.A.; Jamil, N. Effect of sub-lethal doses of vancomycin and oxacillin on biofilm formation by vancomycin intermediate resistant Staphylococcus aureus. J. Basic Microbiol. 2011, 51, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Garland, J.L.; Mills, A.L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 1991, 57, 2351–2359. [Google Scholar] [PubMed]
- Garland, J.L. Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol. Biochem. 1996, 28, 213–221. [Google Scholar] [CrossRef]
- Van Berkum, P.; Badri, Y.; Elia, P.; Aouani, M.E.; Eardly, B.D. Chromosomal and symbiotic relationships of rhizobia nodulating Medicago truncatula and M. laciniata. Appl. Environ. Microbiol. 2007, 73, 7597–7604. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6, molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.W.; Heenan, P.B.; De, M.; Sofie, E.; Willems, A.; Andrews, M. Diverse novel mesorhizobia nodulate New Zealand native Sophora species. Syst. Appl. Microbiol. 2015, 38, 91–98. [Google Scholar] [CrossRef]
- Degefu, T.; Wolde-meskel, E.; Woliy, K.; Frostegård, A. Phylogenetically diverse groups of Bradyrhizobium isolated from nodules of tree and annual legume species growing in Ethiopia. Syst. Appl. Microbiol. 2017, 40, 205–214. [Google Scholar] [CrossRef]
- Villesen, P. FaBox, an online toolbox for Fasta sequences. Mol. Ecol. Res. 2007, 7, 965–968. [Google Scholar] [CrossRef]
- Duncan, D.B. Multiple range and multiple F-tests. Biometrics 1955, 11, 1–42. [Google Scholar] [CrossRef]
- Koskey, G.; Mburu, S.W.; Kimiti, J.M.; Ombori, O.; Maingi, J.M.; Njeru, E.M. Genetic Characterization and Diversity of Rhizobium Isolated from Root Nodules of Mid-Altitude Climbing Bean (Phaseolus vulgaris L.) Varieties. Front. Microbiol. 2018, 9, 968. [Google Scholar] [CrossRef] [PubMed]
- Saeki, Y.; Aimi, N.; Tsukamoto, S.; Yamakawa, T.; Nagatomo, Y.; Akao, S. Diversity and geographical distribution of indigenous soybean-nodulating bradyrhizobia in Japan. Soil Sci. Plant Nutri. 2006, 52, 418–426. [Google Scholar] [CrossRef]
- Somasegaran, P.; Hoben, H.J. Hand Book for Rhizobia; Springer: New York, NY, USA, 1994. [Google Scholar]
- Rodrigues, C.; Laranjo, M.; Oliveira, S. Effect of heat and pH stress in the growth of chickpea mesorhizobia. Curr. Microbiol. 2006, 53, 1–7. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nour, S.M.; Cleyet-Marel, J.B.; Effosse, C.D.A.; Fernandez, M.P. Genotypic and phenotypic diversity of Rhizobium isolated from chickpea (Cicer arietinum L.). Can. J. Microbiol. 1994, 40, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Peix, A.; Rivas-Boyero, A.A.; Mateos, P.F.; Rodriguez-Barrueco, C.; Martınez-Molina, E.; Velazquez, E. Growth promotion of chickpea and barley by a phosphate solubilizing strain of Mesorhizobium mediterraneum under growth. Soil Biol. Biochem. 2001, 33, 103–110. [Google Scholar] [CrossRef]
- Chen, L.; Luo, S.; Xiao, X.; Guo, H.; Chen, J.; Wan, Y.; Li, B.; Xu, T.; Xi, Q.; Rao, C.; et al. Application of plant growth-promoting endophytes (PGPE) isolated from Solanum nigrum L. for phytoextraction of Cd-polluted soils. Appl. Soil Ecol. 2010, 46, 383–389. [Google Scholar] [CrossRef]
- Pravin, V.; Rosazlin, A.; Tumirah, K.; Salmah, I.; Amru, N.B. Role of Plant Growth Promoting Rhizobacteria in Agricultural Sustainability—A Review. Molecules 2016, 21, 573. [Google Scholar]
- Chandra, S.; Choure, K.; Dubey, R.C.; Maheshwari, D.K. Rhizosphere competent Mesorhizobium loti MP6 induces root hair curling, inhibits Sclerotinia sclerotiorum and enhances growth of Indian mustard (Brassica campestris). Braz. J. Microbiol. 2007, 38, 124–130. [Google Scholar] [CrossRef]
- Adnan, M.; Shah, Z.; Fahad, S.; Arif, M.; Alam, M.; Khan, I.A.; Mian, I.A.; Basir, A.; Ullah, H.; Arshad, M.; et al. Phosphate-Solubilizing Bacteria Nullify the Antagonistic Effect of Soil Calcifcation on Bioavailability of Phosphorus in Alkaline Soils. Sci. Rep. 2017, 7, 16131. [Google Scholar] [CrossRef]
- Gupta, C.P.; Dubey, R.C.; Maheshwari, D.K. Plant growth enhancement and suppression of Macrophomina phaseolina causing charcoal rot of peanut by fluorescent Pseudomonas. Biol. Fertil. Soil. 2002, 35, 399–405. [Google Scholar]
- María, P.F.I.; Valentine, A. Enhanced Plant Performance in Cicer arietinum L. Due to the Addition of a Combination of Plant Growth-Promoting Bacteria. Agriculture 2017, 7, 40. [Google Scholar]
- Nakayama, M.; Nakajima-Kambe, T.; Katayama, H.; Higuchi, K.; Kawasaki, Y.; Fuji, R. High catalase production by Rhizobium radiobacter strain 2-1. J. Biosci. Bioeng. 2008, 106, 554–558. [Google Scholar] [CrossRef] [PubMed]
- Downie, A.J. Legume nodulation. Curr. Biol. 2014, 24, R184–R190. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R.; Todorovic, B.; Czarny, J.; Cheng, Z.; Duan, J.; Mc-Conkey, B. Promotion of plant growth by bacterial ACC deaminase. Crit. Rev. Plant Sci. 2007, 26, 227–242. [Google Scholar] [CrossRef]
- Anwar, S.; Ali, B.; Sajid, I. Screening of Rhizospheric Actinomycetes for Various In-vitro and In-vivo Plant Growth Promoting (PGP) Traits and for Agroactive Compounds. Front. Microbiol. 2016, 7, 1334. [Google Scholar] [CrossRef]
- Mahmood, A.; Athar, M. Cross inoculation studies: Response of Vigna mungo to inoculation with rhizobia from tree legumes growing under arid Environment. Int. J. Environ. Sci. Technol. 2008, 5, 135–139. [Google Scholar] [CrossRef]
- Jiao, Y.S.; Liu, Y.H.; Yan, H.; Wang, E.T.; Tian, C.F.; Chen, W.X.; Guo, B.L.; Chen, W.F. Rhizobial diversity and nodulation characteristics of the extremely promiscuous legume Sophora flavescens. Mol. Plant-Microbe Interact. 2015, 28, 1338–1352. [Google Scholar] [CrossRef]
- Singh, R.P.; Manchanda, G.; Singh, R.N.; Srivastava, A.K.; Dubey, R.C. Selection of alkalotolerant and symbiotically efficient chickpea nodulating rhizobia from North-West Indo Gangetic Plains. J. Basic Microbiol. 2016, 56, 14–25. [Google Scholar] [CrossRef]
- Fujishige, N.A.; Kapadia, N.N.; De Hoff, P.L.; Hirsch, A.M. Investigations of Rhizobium biofilm formation. FEMS Microbiol. Ecol. 2006, 56, 195–206. [Google Scholar] [CrossRef]
- Wang, H.; Zhong, Z.; Cai, T.; Li, S.; Zhu, J. Heterologous overexpression of quorum-sensing regulators to study cell density-dependent phenotypes in a symbiotic plant bacterium Mesorhizobium huakui. Arch. Microbiol. 2004, 182, 520–525. [Google Scholar] [CrossRef]
- Wang, D.; Xu, A.; Elmerich, C.; Ma, L.Z. Biofilm formation enables free-living nitrogen-fixing rhizobacteria to fix nitrogen under aerobic conditions. ISME J. 2017, 11, 1602–1613. [Google Scholar] [CrossRef] [PubMed]
- Govaerts, B.; Mezzalama, M.; Unno, Y.; Sayre, K.D.; Luna-Guido, M.; Vanherck, K.; Dendooven, L.; Deckers, J. Influence of tillage, residue management, and crop rotation on soil microbial biomass and catabolic diversity. Appl. Soil Ecol. 2007, 37, 18–30. [Google Scholar] [CrossRef]
- Grayston, S.J.; Wang, S.Q.; Campbell, C.D.; Edwards, A.C. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 1998, 30, 369–378. [Google Scholar] [CrossRef]
- Dighe, A.S.; Jangid, K.; González, J.M.; Pidiyar, V.J.; Patole, M.S.; Ranade, D.R.; Shouche, Y.S. Comparison of 16S rRNA gene sequences of genus Methanobrevibacter. BMC Microbiol. 2004, 4, 20. [Google Scholar] [CrossRef]
- Sato, M.; Miyazaki, K. Phylogenetic Network Analysis Revealed the Occurrence of Horizontal Gene Transfer of 16S rRNA in the Genus Enterobacter. Front. Microbiol. 2017, 8, 2225. [Google Scholar] [CrossRef]
- Elbannaa, K.; Elbadryb, M.; Gamal-Eldina, H. Genotypic and phenotypic characterization of rhizobia that nodulate snap bean (Phaseolus vulgaris L.) in Egyptian soils. Syst. Appl. Microbiol. 2009, 32, 522–530. [Google Scholar] [CrossRef]



| Isolates | Growth Condition | PS (S.I. Value) | IAA Test | Oxidase Test | Catalase Test | Ammonia Production | ACC Deaminase (mmol−l) | Biofilm Formation (OD) |
|---|---|---|---|---|---|---|---|---|
| IC-2058 | MS | 3.92 ± 0.04 | 99.54 ± 0.07 | + | + | + | 0.91 ± 0.08 | 0.487 ± 0.04 |
| IC-2018 | MS | 3.86 ± 0.07 | 91.71 ± 0.06 | + | + | + | 1.03 ± 0.11 | 0.565 ± 0.02 |
| CPN1 | MS | 3.63 ± 0.01 | 100.24 ± 0.09 | + | + | + | 1.57 ± 0.04 | 0.678 ± 0.05 |
| CPN2 | MS | 3.12 ± 0.03 | 79.31 ± 0.08 | + | + | + | 0.84 ± 0.06 | 0.405 ± 0.05 |
| CPN3 | MF | 3.33 ± 0.04 | 80.22 ± 0.07 | + | + | + | 1.04 ± 0.02 | 0.733 ± 0.04 |
| CPN4 | MS | 3.09 ± 0.04 | 94.12 ± 0.05 | + | + | + | 0.87 ± 0.12 | 0.532 ± 0.06 |
| CPN5 | MS | 2.92 ± 0.00 | 98.61 ± 0.07 | - | + | + | 1.02 ± 0.04 | 0.219 ± 0.04 |
| CPN6 | MS | 3.41 ± 0.01 | 103.90 ± 0.06 | + | + | + | 1.12 ± 0.06 | 0.374 ± 0.07 |
| CPN7 | MS | 3.29 ± 0.01 | 86.31 ± 0.04 | + | + | + | 1.05 ± 0.02 | 0.752 ± 0.04 |
| CPN8 | MS | 4.17 ± 0.07 | 104.52 ± 0.09 | + | + | + | 1.24 ± 0.12 | 0.502 ± 0.02 |
| CPN9 | MS | 3.99 ± 0.05 | 97.24 ± 0.11 | + | + | + | 1.32 ± 0.05 | 0.785 ± 0.05 |
| CPN10 | MS | 3.56 ± 0.05 | 93.61 ± 0.09 | + | + | + | 1.16 ± 0.05 | 0.692 ± 0.02 |
| CPN11 | MS | 3.68 ± 0.01 | 91.76 ± 0.07 | - | + | + | 1.27 ± 0.02 | 0.775 ± 0.03 |
| CPN12 | MS | 3.14 ± 0.03 | 86.35 ± 0.11 | + | + | + | 1.33 ± 0.08 | 0.523 ± 0.04 |
| CPN13 | MS | 3.51 ± 0.07 | 97.58 ± 0.06 | - | + | + | 1.21 ± 0.06 | 0.293 ± 0.01 |
| CPN14 | MS | 2.97 ± 0.01 | 93.41 ± 0.12 | - | + | + | 1.31 ± 0.05 | 0.465 ± 0.04 |
| CPN15 | MS | 2.88 ± 0.07 | 95.24 ± 0.07 | + | + | + | 1.42 ± 0.14 | 0.504 ± 0.08 |
| CPN16 | MF | 3.69 ± 0.08 | 99.47 ± 0.05 | + | + | + | 0.88 ± 0.04 | 0.802 ± 0.06 |
| CPN17 | MS | 3.05 ± 0.04 | 98.66 ± 0.06 | + | + | + | 0.67 ± 0.02 | 0.514 ± 0.06 |
| CPN18 | MS | 3.19 ± 0.03 | 90.05 ± 0.06 | + | + | + | 1.09 ± 0.02 | 0.308 ± 0.02 |
| CPN19 | MS | 3.25 ± 0.08 | 97.11 ± 0.09 | + | + | + | 1.18 ± 0.06 | 0.347 ± 0.03 |
| CPN20 | MS | 3.07 ± 0.05 | 82.41 ± 0.08 | + | + | + | 1.19 ± 0.07 | 0.289 ± 0.03 |
| CPN21 | MS | 3.61 ± 0.00 | 102.57 ± 0.08 | + | + | + | 0.98 ± 0.05 | 0.872 ± 0.05 |
| CPN22 | MS | 3.77 ± 0.02 | 105.38 ± 0.07 | + | + | + | 1.13 ± 0.03 | 0.759 ± 0.05 |
| CPN23 | MS | 3.47 ± 0.07 | 99.84 ± 0.05 | + | + | + | 1.28 ± 0.05 | 0.384 ± 0.04 |
| CPN24 | MS | 3.33 ± 0.02 | 84.61 ± 0.05 | + | + | + | 1.09 ± 0.04 | 0.405 ± 0.02 |
| CPN25 | MS | 3.43 ± 0.02 | 89.73 ± 0.07 | + | + | + | 1.09 ± 0.05 | 0.372 ± 0.02 |
| CPN26 | MS | 3.28 ± 0.04 | 90.51 ± 0.07 | + | + | + | 1.22 ± 0.04 | 0.325 ± 0.04 |
| CPN27 | MS | 3.16 ± 0.07 | 100.82 ± 0.05 | + | + | + | 1.28 ± 0.06 | 0.217 ± 0.05 |
| CPN28 | MS | 3.84 ± 0.03 | 100.25 ± 0.11 | + | + | + | 0.86 ± 0.02 | 0.374 ± 0.05 |
| CPN29 | MS | 4.01 ± 0.06 | 98.67 ± 0.12 | + | + | + | 1.34 ± 0.05 | 0.805 ± 0.08 |
| CPN30 | MS | 3.87 ± 0.04 | 99.58 ± 0.11 | + | + | + | 1.09 ± 0.02 | 0.541 ± 0.04 |
| CPN31 | MS | 3.61 ± 0.06 | 81.64 ± 0.07 | + | + | + | 0.96 ± 0.04 | 0.581 ± 0.03 |
| CPN32 | MS | 4.08 ± 0.06 | 107.41 ± 0.09 | + | + | + | 1.08 ± 0.0.6 | 0.791 ± 0.05 |
| CPN33 | MS | 3.63 ± 0.05 | 89.33 ± 0.05 | + | + | + | 0.84 ± 0.06 | 0.537 ± 0.04 |
| CPN34 | MS | 4.13 ± 0.06 | 92.56 ± 0.06 | + | + | + | 1.06 ± 0.02 | 0.861 ± 0.06 |
| CPN35 | MS | 3.5 ± 0.06 | 91.63 ± 0.06 | + | + | + | 1.18 ± 0.08 | 0.837 ± 0.02 |
| CPN36 | MS | 3.25 ± 0.01 | 87.64 ± 0.05 | + | + | + | 1.12 ± 0.06 | 0.465 ± 0.03 |
| CPN37 | MS | 3.61 ± 0.06 | 90.35 ± 0.08 | + | + | + | 1.04 ± 0.06 | 0.438 ± 0.06 |
| CPN38 | MS | 3.17 ± 0.03 | 88.62 ± 0.06 | + | + | + | 0.87 ± 0.04 | 0.483 ± 0.05 |
| CPN39 | MS | 3.34 ± 0.03 | 100.71 ± 0.09 | + | + | + | 1.34 ± 0.07 | 0.806 ± 0.04 |
| CPN40 | MS | 3.01 ± 0.01 | 85.61 ± 0.06 | + | + | + | 0.78 ± 0.02 | 0.308 ± 0.09 |
| CPN41 | MS | 2.95 ± 0.03 | 87.24 ± 0.05 | + | + | + | 1.03 ± 0.06 | 0.355 ± 0.07 |
| CPN42 | MS | 3.62 ± 0.05 | 101.13 ± 0.06 | + | + | + | 1.05 ± 0.08 | 0.428 ± 0.09 |
| CPN43 | MS | 3.19 ± 0.02 | 83.61 ± 0.07 | + | + | + | 1.05 ± 0.12 | 0.611 ± 0.06 |
| CPN44 | MS | 3.55 ± 0.01 | 89.34 ± 0.05 | + | + | + | 0.93 ± 0.06 | 0.489 ± 0.05 |
| CPN45 | MS | 3.29 ± 0.00 | 99.65 ± 0.06 | + | + | + | 1.02 ± 0.08 | 0.785 ± 0.07 |
| CPN46 | MS | 2.87 ± 0.03 | 92.04 ± 0.08 | + | + | + | 1.14 ± 0.06 | 0.523 ± 0.06 |
| CPN47 | MS | 2.96 ± 0.03 | 93.84 ± 0.12 | + | + | + | 1.07 ± 0.05 | 0.429 ± 0.05 |
| CPN48 | MS | 3.41 ± 0.02 | 88.06 ± 0.09 | + | + | + | 0.86 ± 0.03 | 0.367 ± 0.03 |
| CPN49 | MS | 3.64 ± 0.01 | 90.71 ± 0.05 | + | + | + | 1.22 ± 0.09 | 0.356 ± 0.04 |
| CPN50 | MS | 3.06 ± 0.04 | 87.42 ± 0.06 | + | + | + | 1.14 ± 0.12 | 0.503 ± 0.04 |
| CPN51 | MS | 3.88 ± 0.05 | 90.21 ± 0.08 | + | + | + | 0.96 ± 0.05 | 0.486 ± 0.06 |
| CPN52 | MS | 4.33 ± 0.03 | 101.08 ± 0.09 | + | + | + | 1.01 ± 0.05 | 0.806 ± 0.05 |
| CPN53 | MS | 3.44 ± 0.00 | 97.33 ± 0.05 | + | + | + | 0.97 ± 0.02 | 0.539 ± 0.07 |
| CPN54 | MS | 3.21 ± 0.01 | 91.82 ± 0.11 | + | + | + | 1.04 ± 0.05 | 0.211 ± 0.04 |
| S.No. | Isolates | Chlorophyll Content (mg g−1 F.W. of Leaves) | NN | SDW (g Plant-1) | SE (%) |
|---|---|---|---|---|---|
| 1. | T1 (Control) | 0.806 ± 0.444 | − | 0.419 ± 1.528 | − |
| 2. | T2 (Control) N-Supplemented | 1.111 ± 0.121 | − | 4.217 ± 0.639 | 100.00 |
| 3. | IC-2058 | 1.047 ± 0.459 | 15.31 ± 1.224 | 1.127 ± 0.558 | 26.72 |
| 4. | IC-2018 | 0.986 ± 0.194 | 12.21 ± 1.321 | 0.927 ± 0.161 | 21.98 |
| 5. | CPN1 | 1.240 ± 0.152 | 22.81 ± 0.423 | 1.203 ± 0.183 | 28.52 |
| 6. | CPN2 | 0.958 ± 0.408 | 7.37 ± 1.729 | 0.603 ± 0.199 | 14.29 |
| 7. | CPN3 | 1.121 ± 0.057 | 11.66 ± 2.854 | 1.186 ± 0.241 | 28.12 |
| 8. | CPN4 | 0.919 ± 0.158 | 6.24 ± 1.852 | 0.884 ± 0.114 | 20.96 |
| 9. | CPN5 | 0.911 ± 0.008 | 6.51 ± 2.010 | 0.653 ± 0.0838 | 15.48 |
| 10. | CPN6 | 1.126 ± 0.058 | 17.38 ± 0.874 | 1.119 ± 0.292 | 26.53 |
| 11. | CPN7 | 1.199 ± 0.122 | 13.57 ± 0.973 | 1.288 ± 0.541 | 30.54 |
| 12. | CPN8 | 1.201 ± 0.008 | 14.91 ± 1.233 | 1.303 ± 0.346 | 30.89 |
| 13. | CPN9 | 0.906 ± 0.002 | 7.61 ± 0.935 | 0.971 ± 0.154 | 23.02 |
| 14. | CPN10 | 1.128 ± 0.116 | 18.51 ± 1.900 | 1.266 ± 0.573 | 30.02 |
| 15. | CPN11 | 0.891 ± 0.048 | 9.22 ± 1.312 | 0.639 ± 0.113 | 15.15 |
| 16. | CPN12 | 0.931 ± 0.043 | 12.42 ± 0941 | 0.997 ± 0.157 | 23.64 |
| 17. | CPN13 | 0.933 ± 0.032 | 11.61 ± 2.078 | 0.999 ± 0.202 | 23.68 |
| 18. | CPN14 | 0.975 ± 0.038 | 8.33 ± 2.333 | 1.107 ± 0.244 | 26.25 |
| 19. | CPN15 | 0.897 ± 0.055 | 9.33 ± 0.334 | 0.769 ± 0.118 | 18.23 |
| 20. | CPN16 | 1.217 ± 0.330 | 16.00 ± 1.154 | 1.176 ± 0.421 | 27.88 |
| 21. | CPN17 | 0.968 ± 0.393 | 11.66 ± 0.881 | 0.894 ± 0.050 | 21.19 |
| 22. | CPN18 | 0.922 ± 0.051 | 13.24 ± 1.622 | 0.931 ± 0.157 | 22.07 |
| 23. | CPN19 | 0.938 ± 0.196 | 15.29 ± 2.299 | 1.091 ± 0.472 | 25.87 |
| 24. | CPN20 | 0.871 ± 0.055 | 6.33 ± 1.763 | 0.646 ± 0.563 | 15.31 |
| 25. | CPN21 | 1.211 ± 0.202 | 19.88 ± 3.489 | 1.207 ± 0.200 | 28.62 |
| 26. | CPN22 | 1.113 ± 0.289 | 17.79 ± 1.399 | 1.198 ± 0.435 | 28.40 |
| 27. | CPN23 | 0.839 ± 0.085 | 6.33 ± 1.763 | 0.693 ± 0.201 | 16.43 |
| 28. | CPN24 | 0.854 ± 0.096 | 10.41 ± 1.691 | 0.707 ± 0.064 | 16.76 |
| 29. | CPN25 | 0.819 ± 0.091 | 7.33 ± 0.881 | 0.697 ± 0.112 | 16.52 |
| 30. | CPN26 | 0.824 ± 0.069 | 6.33 ± 1.334 | 0.655 ± 0.227 | 15.53 |
| 31. | CPN27 | 0.971 ± 0.028 | 14.66 ± 1.333 | 1.009 ± 0.056 | 23.92 |
| 32. | CPN28 | 0.873 ± 0.043 | 7.66 ± 1.855 | 0.687 ± 0.047 | 16.29 |
| 33. | CPN29 | 1.218 ± 0.391 | 18.33 ± 1.763 | 1.200 ± 0.224 | 28.45 |
| 34. | CPN30 | 0.872 ± 0.143 | 7.66 ± 1.334 | 0.683 ± 0.098 | 16.19 |
| 35. | CPN31 | 0.962 ± 0.324 | 10.67 ± 1.452 | 0.916 ± 0.140 | 21.72 |
| 36. | CPN32 | 1.298 ± 0.166 | 27.65 ± 1.332 | 1.301 ± 0.165 | 31.01 |
| 37. | CPN33 | 0.888 ± 0.072 | 9.34 ± 1.201 | 0.982 ± 0.370 | 23.28 |
| 38. | CPN34 | 1.271 ± 0.173 | 17.66 ± 1.856 | 1.281 ± 0.214 | 30.37 |
| 39. | CPN35 | 1.259 ± 0.062 | 16.00 ± 1.154 | 1.266 ± 0.299 | 30.02 |
| 40. | CPN36 | 0.963 ± 0.361 | 11.34 ± 2.905 | 0.916 ± 0.185 | 21.72 |
| 41. | CPN37 | 0.847 ± 0.083 | 8.00 ± 1.527 | 0.729 ± 0.100 | 17.28 |
| 42. | CPN38 | 0.855 ± 0.023 | 10.67 ± 0.334 | 0.773 ± 0.093 | 18.33 |
| 43. | CPN39 | 1.115 ± 0.105 | 13.50 ± 1.756 | 1.181 ± 0.371 | 28.00 |
| 44. | CPN40 | 0.908 ± 0.215 | 12.67 ± 1.201 | 0.975 ± 0.196 | 23.12 |
| 45. | CPN41 | 0.914 ± 0.029 | 10.34 ± 0.667 | 0.862 ± 0.223 | 20.44 |
| 46. | CPN42 | 0.897 ± 0.051 | 10.34 ± 2.603 | 0.904 ± 0.182 | 21.43 |
| 47. | CPN43 | 0.895 ± 0.053 | 10.34 ± 2.334 | 0.899 ± 0.123 | 21.43 |
| 48. | CPN44 | 0.866 ± 0.060 | 9.67 ± 0.881 | 0.788 ± 0.205 | 18.68 |
| 49. | CPN45 | 1.171 ± 0.050 | 13.65 ± 1.452 | 1.152 ± 0.413 | 27.31 |
| 50. | CPN46 | 0.937 ± 0.026 | 7.67 ± 1.452 | 0.996 ± 0.351 | 23.61 |
| 51. | CPN47 | 0.841 ± 0.081 | 9.32 ± 1.453 | 0.711 ± 0.145 | 16.86 |
| 52. | CPN48 | 0.819 ± 0.034 | 6.65 ± 1.856 | 0.591 ± 0.099 | 14.01 |
| 53. | CPN49 | 0.842 ± 0.211 | 6.40 ± 1.763 | 0.603 ± 0.227 | 14.29 |
| 54. | CPN50 | 0.837 ± 0.022 | 7.34 ± 0.667 | 0.597 ± 0.246 | 14.15 |
| 55. | CPN51 | 0.869 ± 0.022 | 9.66 ± 1.334 | 0.759 ± 0.141 | 17.99 |
| 56. | CPN52 | 1.118 ± 0.058 | 13.34 ± 2.403 | 1.163 ± 0.354 | 27.57 |
| 57. | CPN53 | 0.803 ± 0.092 | 5.34 ± 1.452 | 0.587 ± 0.202 | 13.91 |
| 58. | CPN54 | 0.972 ± 0.053 | 13.67 ± 3.179 | 0.913 ± 0.303 | 21.65 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, R.P.; Manchanda, G.; Yang, Y.; Singh, D.; Srivastava, A.K.; Dubey, R.C.; Zhang, C. Deciphering the Factors for Nodulation and Symbiosis of Mesorhizobium Associated with Cicer arietinum in Northwest India. Sustainability 2019, 11, 7216. https://doi.org/10.3390/su11247216
Singh RP, Manchanda G, Yang Y, Singh D, Srivastava AK, Dubey RC, Zhang C. Deciphering the Factors for Nodulation and Symbiosis of Mesorhizobium Associated with Cicer arietinum in Northwest India. Sustainability. 2019; 11(24):7216. https://doi.org/10.3390/su11247216
Chicago/Turabian StyleSingh, Raghvendra Pratap, Geetanjali Manchanda, Yingjie Yang, Dipti Singh, Alok Kumar Srivastava, Ramesh Chandra Dubey, and Chengsheng Zhang. 2019. "Deciphering the Factors for Nodulation and Symbiosis of Mesorhizobium Associated with Cicer arietinum in Northwest India" Sustainability 11, no. 24: 7216. https://doi.org/10.3390/su11247216
APA StyleSingh, R. P., Manchanda, G., Yang, Y., Singh, D., Srivastava, A. K., Dubey, R. C., & Zhang, C. (2019). Deciphering the Factors for Nodulation and Symbiosis of Mesorhizobium Associated with Cicer arietinum in Northwest India. Sustainability, 11(24), 7216. https://doi.org/10.3390/su11247216





