Immobilization of Metanil Yellow Decolorizing Mixed Culture FN3 Using Gelling Gum as Matrix for Bioremediation Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultivation of Mixed Culture FN3
2.2. Immobilization of the Mixed Culture Using Gellan Gum
2.3. Optimization of Immobilized Beads Using RSM
2.4. Metabolites Analysis Using High-Performance Liquid Chromatography (HPLC)
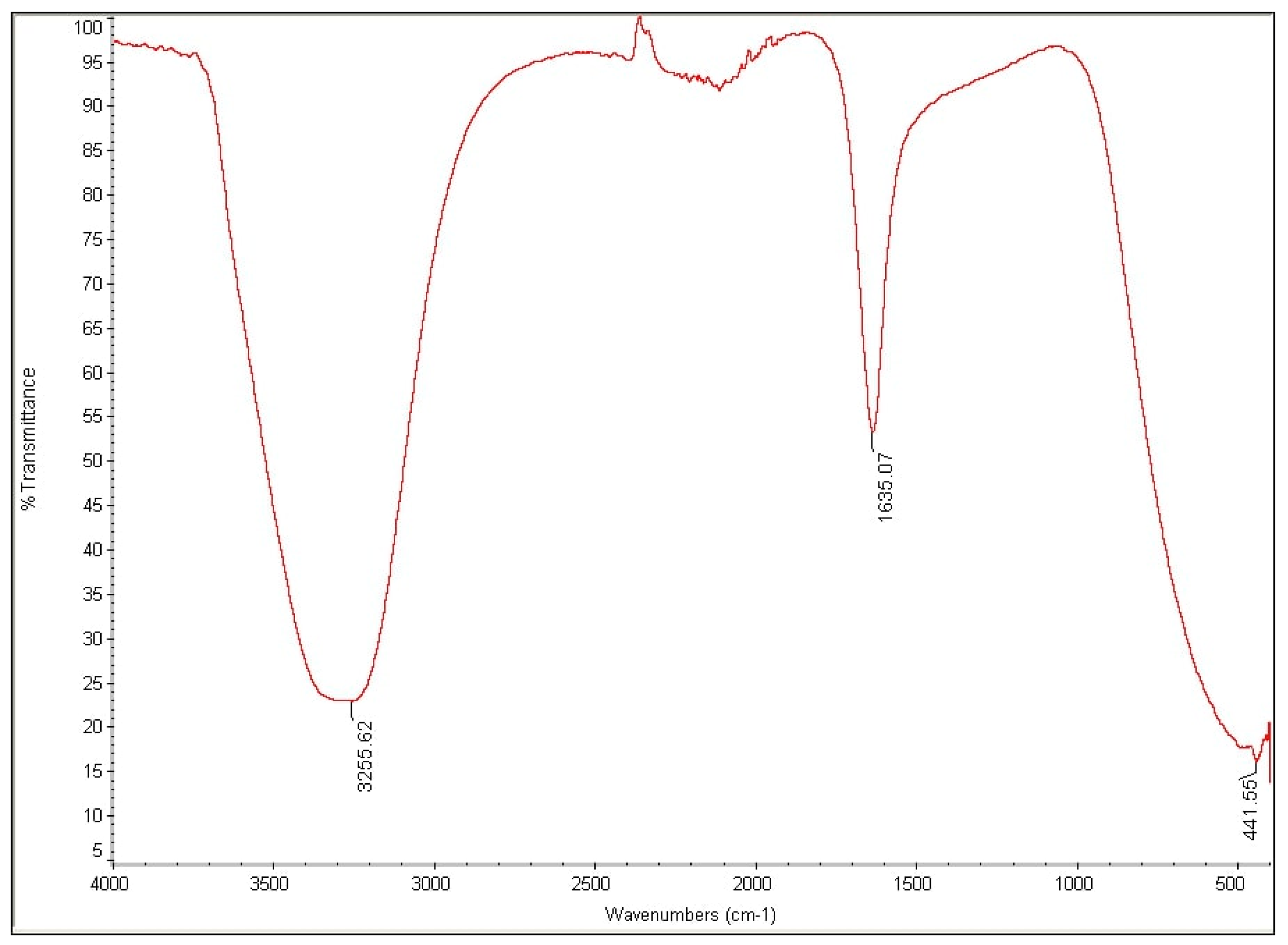
2.5. Metabolites Analysis Using Fourier-Transform Infrared Spectroscopy (FTIR)
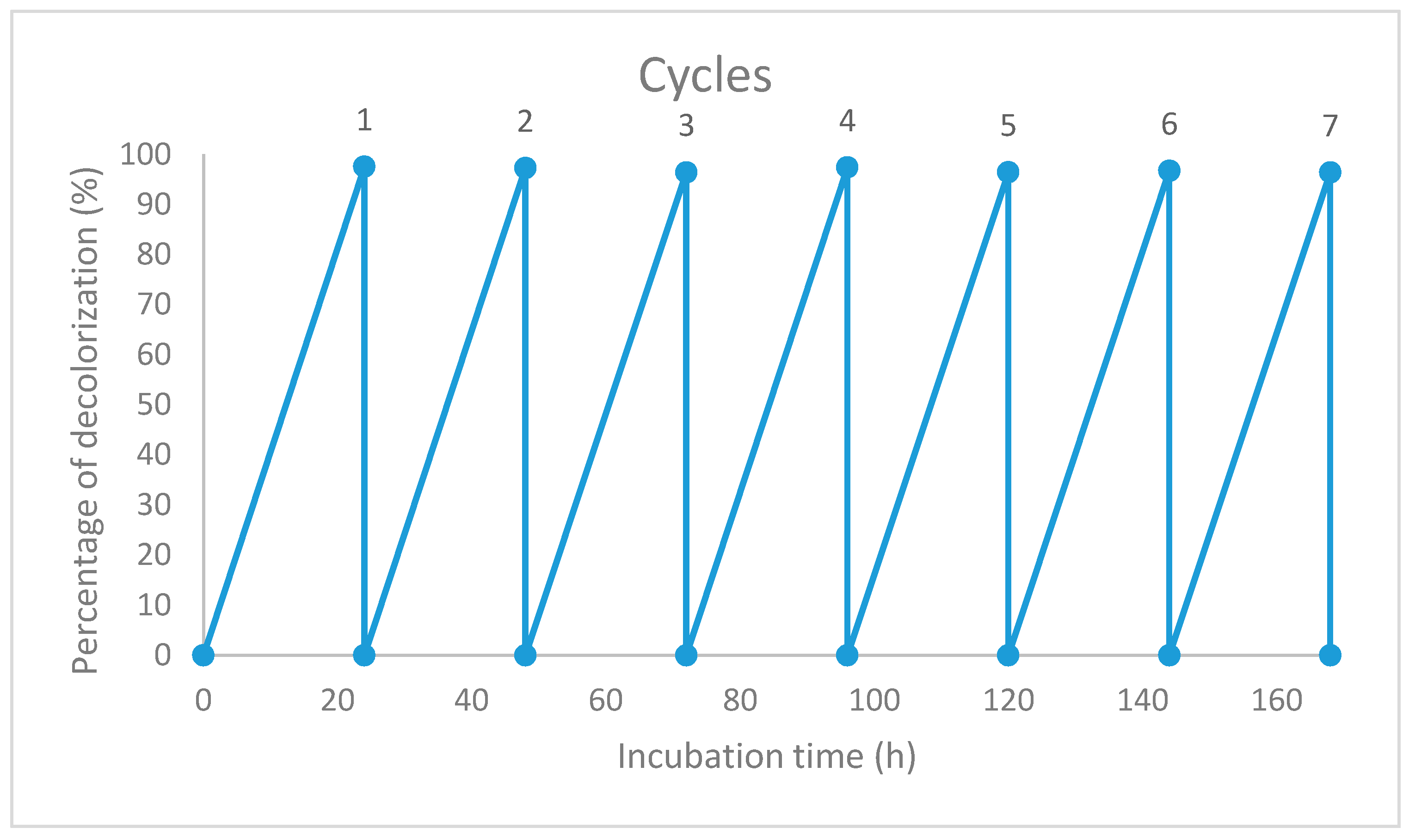
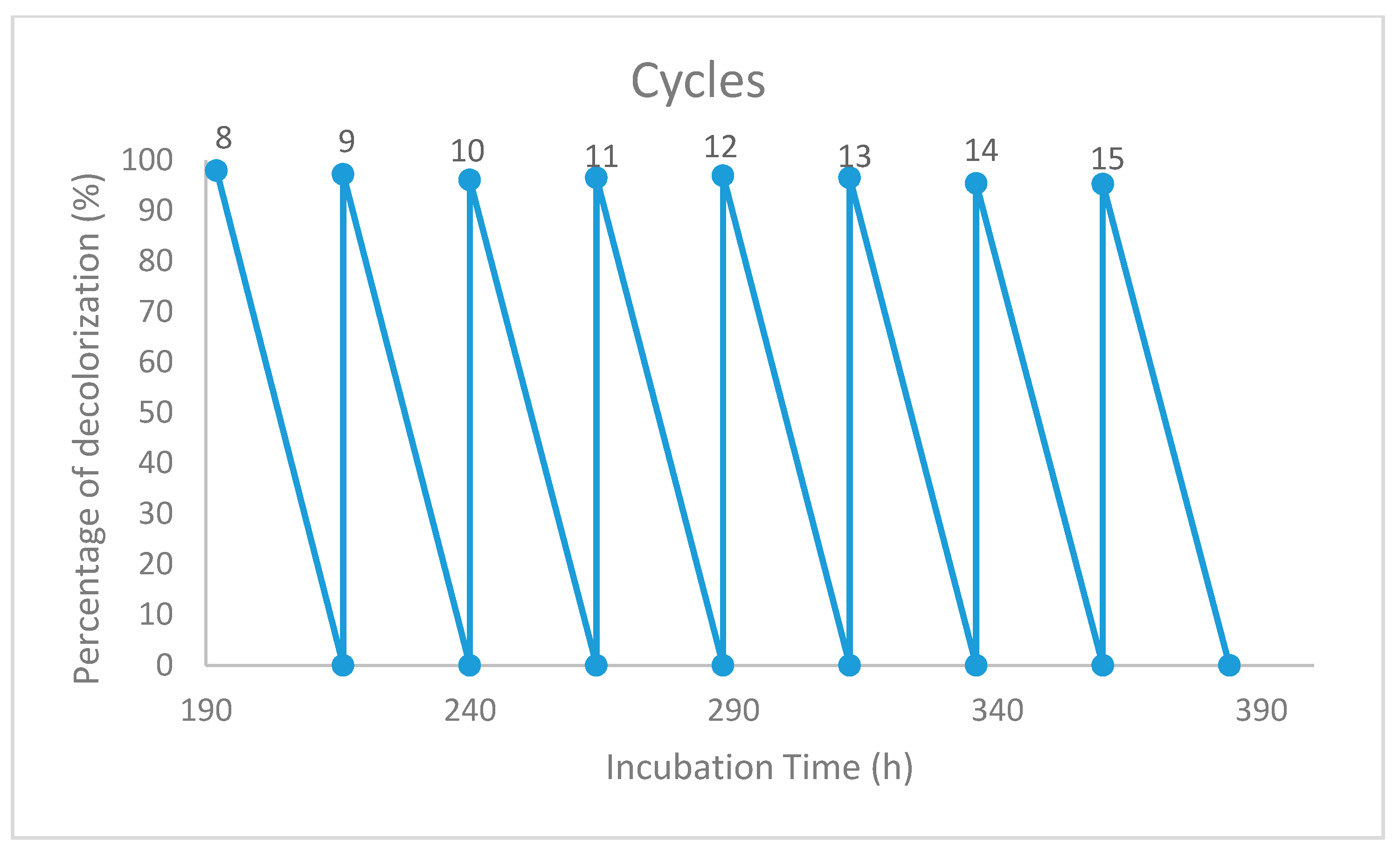
2.6. Reusability of the Immobilized Beads
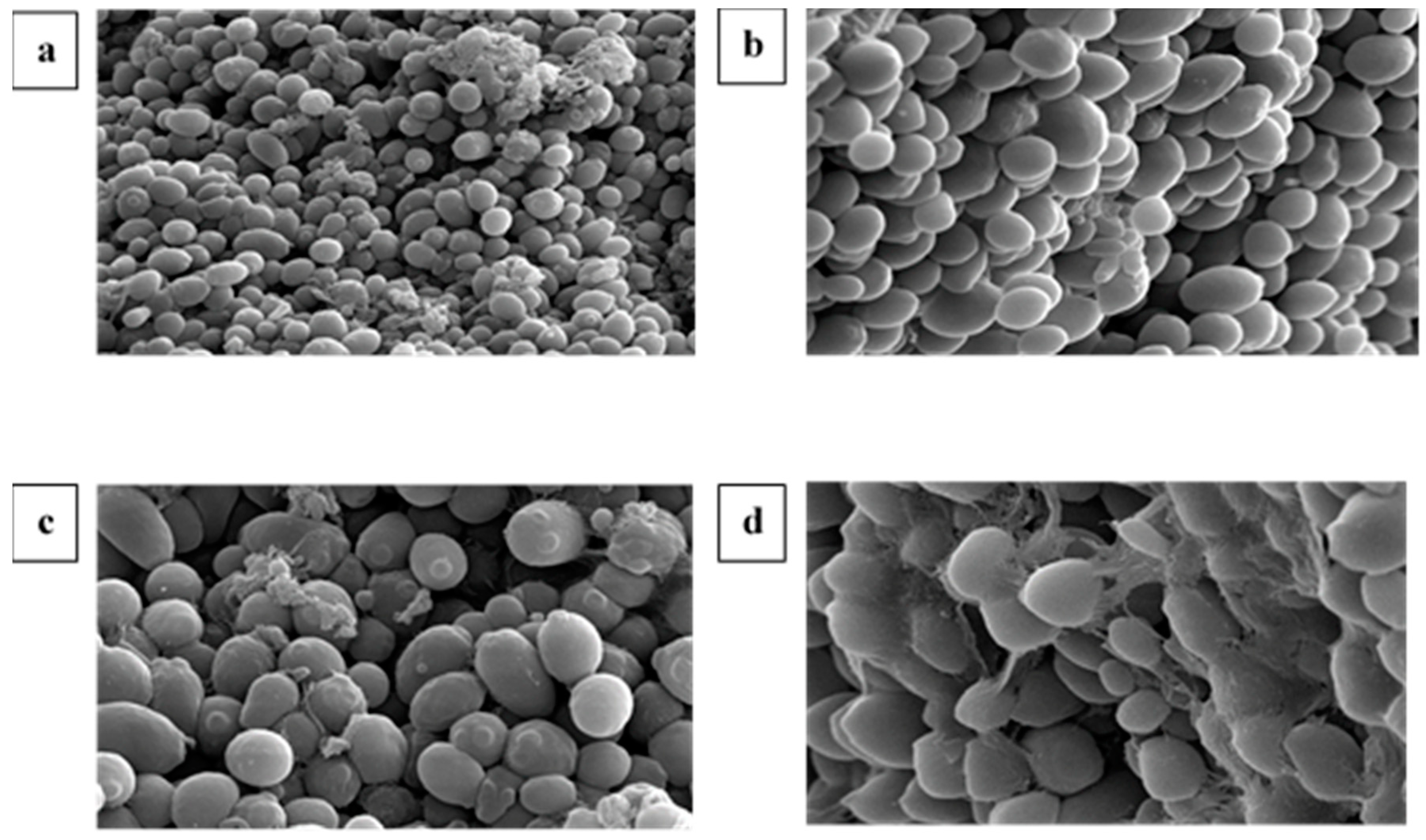
2.7. Scanning Electron Microscopy (SEM) of Immobilized Mixed Culture FN3
2.8. Effects of Heavy Metal Ions on Dye Decolorization of Immobilised Cells
3. Results and Discussions
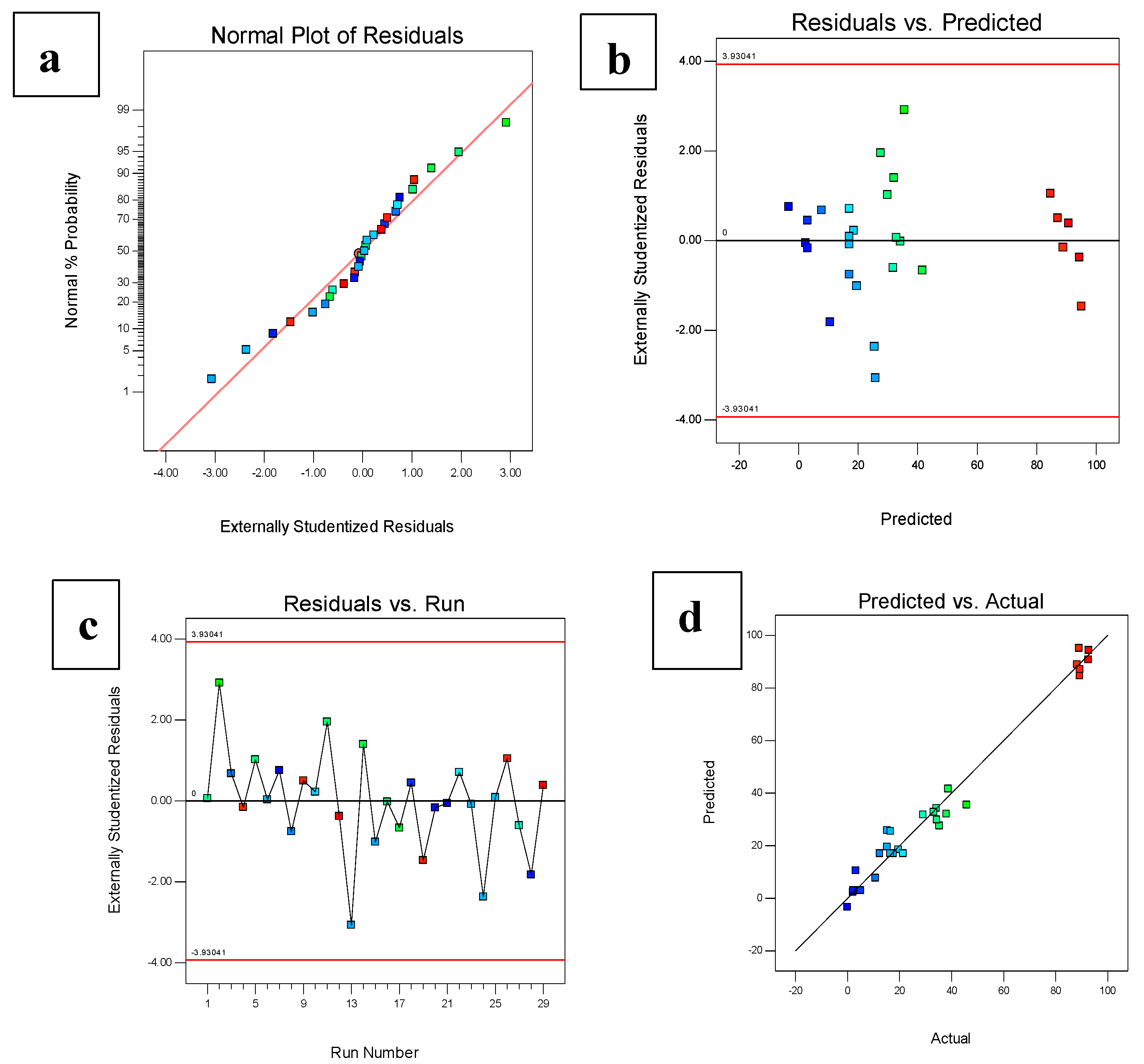
3.1. Optimization of Immobilized Beads Using RSM
3.2. Determination and Validation of Optimal Conditions
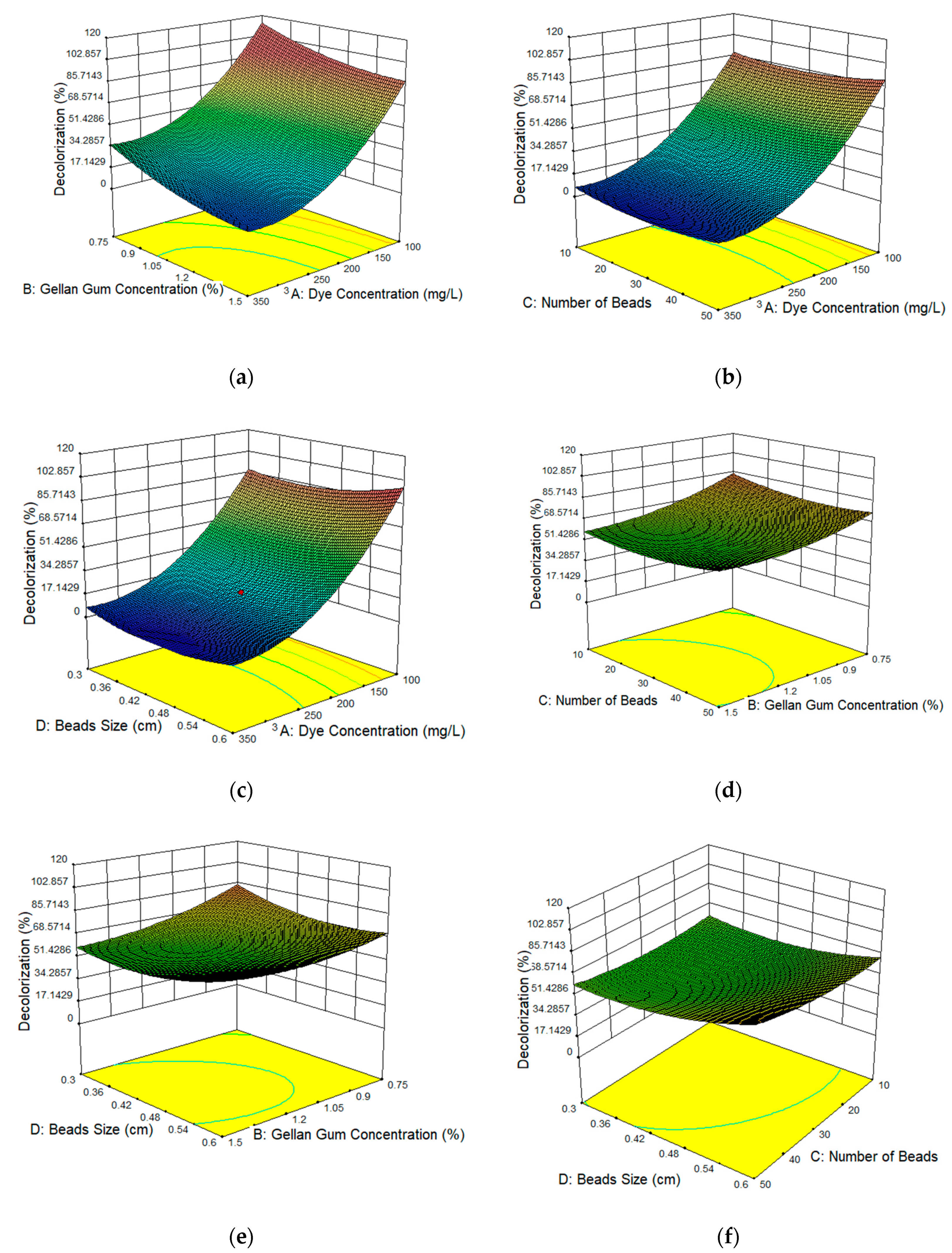
3.3. Response Surface Plots of the Affecting Parameters
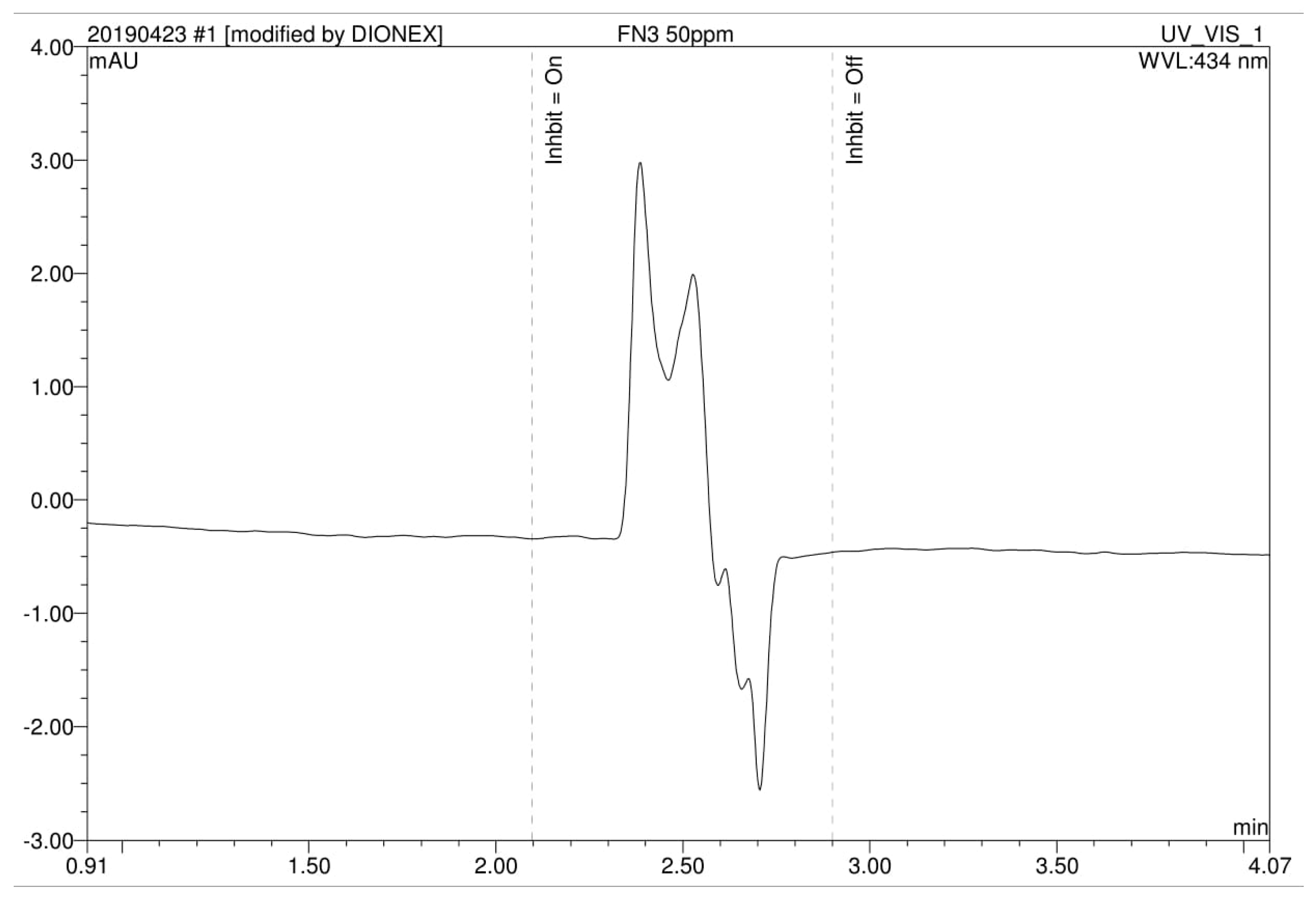
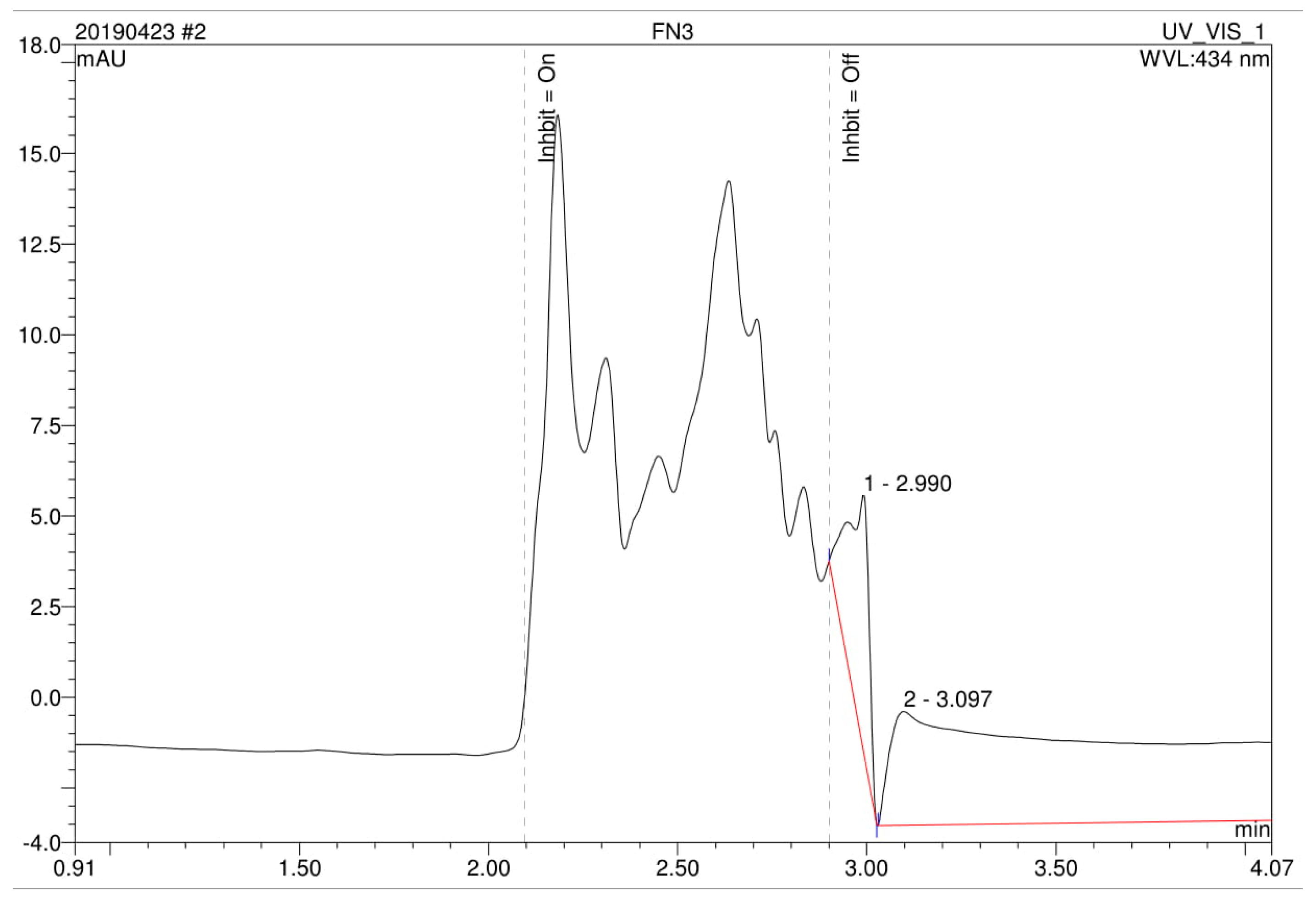
3.4. Analysis of High Performance Liquid Chromatography (HPLC) and Fourier-Transform Infrared Spectroscopy (FTIR)
3.5. Reusability of the Immobilized Beads
3.6. Scanning Electron Microscopy (SEM) of Immobilized Mixed Culture FN3
3.7. Effects of Metal Ions on Dye Decolorization of Immobilised FN3
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chang, J.-S.; Chou, C.; Lin, Y.-C.; Lin, P.-J.; Ho, J.-Y.; Hu, T.-L. Kinetic characteristics of bacterial azo-dye decolorization by Pseudomonas luteola. Water Res. 2001, 35, 2841–2850. [Google Scholar] [CrossRef]
- Singh, R.P.; Singh, P.K.; Singh, R.L. Bacterial Decolorization of Textile Azo Dye Acid Orange by Staphylococcus hominis RMLRT03. Toxicol. Int. 2014, 21, 160–166. [Google Scholar] [CrossRef]
- Puvaneswari, N.; Jayarama, M.; Gunasekaran, P. Toxicity assessment and microbial degradation of azo dyes. Indian J. Exp. Boil. 2006, 44, 618–626. [Google Scholar]
- Sarkar, S.; Banerjee, A.; Halder, U.; Biswas, R.; Bandopadhyay, R. Degradation of Synthetic Azo Dyes of Textile Industry: A Sustainable Approach Using Microbial Enzymes. Water Conserv. Sci. Eng. 2017, 2, 121–131. [Google Scholar] [CrossRef]
- Sudha, M.; Saranya, A.; Gopal, S.; Natesan, S. Microbial degradation of Azo Dyes: A review. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 670–690. [Google Scholar]
- Sivashankar, R.; Velmurugan, S.; Sathya, A.B.; Pallipad, S. Biosorption of Hazardous Azo Dye Metanil Yellow using Immobilized Aquatic weed. In Proceedings of the International Conference on Future Trends in Structural, Civil, Environmental and Mechanical Engineering–FTCEM, Bangkok, Thailand, 13–14 July 2013. [Google Scholar]
- Anjaneya, O.; Souche, S.Y.; Santoshkumar, M.; Karegoudar, T.B. Decolorization of sulfonated azo dye Metanil Yellow by newly isolated bacterial strains: Bacillus sp. strain AK1 and Lysinibacillus sp. strain AK2. J. Hazard. Mater. 2011, 190, 351–358. [Google Scholar] [CrossRef]
- Ghosh, D.; Singha, P.; Firdaus, S.; Ghosh, S. Metanil yellow: The toxic food colorant. Asian Pac. J. Health Sci. 2017, 4, 65–66. [Google Scholar] [CrossRef]
- Aksu, Z.; Kılıç, N.K.; Ertuğrul, S.; Dönmez, G. Inhibitory effects of chromium(VI) and Remazol Black B on chromium(VI) and dyestuff removals by Trametes versicolor. Enzym. Microb. Technol. 2007, 40, 1167–1174. [Google Scholar] [CrossRef]
- Hao, O.; Kim, H.; Chiang, P.-C. Decolorization of Wastewater. Crit. Rev. Environ. Sci. Technol. 1999, 30, 449–505. [Google Scholar] [CrossRef]
- Moreira, R.; Peruch, M.G.; Kuhnen, N.C. Adsorption of textile dyes on alumina. Equilibrium studies and contact time effects. Braz. J. Chem. Eng. 1998, 15, 21–28. [Google Scholar] [CrossRef]
- Sarioglu, M.; Bali, U.; Bisgin, T. The removal of C.I. Basic Red 46 in a mixed methanogenic anaerobic culture. Dye. Pigments 2007, 74, 223–229. [Google Scholar] [CrossRef]
- Wang, H.; Su, J.; Zheng, X.; Tian, Y.; Xiong, X.; Zheng, T. Bacterial decolorization and degradation of the reactive dye Reactive Red 180 by Citrobacter sp. CK3. Int. Biodeter. Biodegr. 2009, 63, 395–399. [Google Scholar] [CrossRef]
- Pandey, A.; Singh, P.; Iyengar, L. Bacterial decolorization and degradation of azo dyes. Int. Biodeter. Biodegr. 2007, 59, 73–84. [Google Scholar] [CrossRef]
- Alabdraba, W.; Bayati, M. Biodegradation of Azo Dyes a Review. Int. J. Environ. Eng. Nat. Resour. 2014, 1, 179–189. [Google Scholar]
- Jain, K.; Shah, V.; Chapla, D.; Madamwar, D. Decolorization and degradation of azo dye–Reactive Violet 5R by an acclimatized indigenous bacterial mixed cultures-SB4 isolated from anthropogenic dye contaminated soil. J. Hazard. Mater. 2012, 213-214, 378–386. [Google Scholar] [CrossRef]
- Partovinia, A.; Rasekh, B. Review of the immobilized microbial cell systems for bioremediation of petroleum hydrocarbons polluted environments. Crit. Rev. Environ. Sci. Technol. 2018, 48, 1–38. [Google Scholar] [CrossRef]
- Unnikrishnan, S.; Ramamoorthi, P.; Sridhar, S. Studies on decolorization of malachite green using immobilized Pseudomonas putida. J. Chem. Pharm. Res. 2015, 7, 589–596. [Google Scholar]
- Ge, X.; Yang, L.; Xu, J. Cell Immobilization: Fundamentals, Technologies, and Applications. Ind. Biotechnol. 2017, 205–235. [Google Scholar] [CrossRef]
- Moslemy, P.; Guiot, S.R.; Neufeld, R.J. Encapsulation of bacteria for biodegradation of gasoline hydrocarbons. In Immobilization of Enzymes and Cells; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2006; pp. 415–426. [Google Scholar]
- Moslemy, P.; Neufeld, R.J.; Millette, D.; Guiot, S.R. Transport of gellan gum microbeads through sand: An experimental evaluation for encapsulated cell bioaugmentation. J. Environ. Manag. 2003, 69, 249–259. [Google Scholar] [CrossRef]
- Ashtaputre, A.A.; Shah, A.K. Studies on a Viscous, Gel-Forming Exopolysaccharide from Sphingomonas paucimobilis GS1. Appl. Environ. Microbiol. 1995, 61, 1159–1162. [Google Scholar] [CrossRef]
- Sharma, D.C.; Satyanarayana, T. A marked enhancement in the production of a highly alkaline and thermostable pectinase by Bacillus pumilus dcsr1 in submerged fermentation by using statistical methods. Bioresour. Technol. 2006, 97, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Muliadi, F.N.A.; Halmi, M.I.E.; Abdul Wahid, S.; Abd Gani, S.S.; Uswatun, H.; Mahmod, K.; Abd Shukor, M.Y. Biostimulation of Microbial Communities from Malaysia Agriculture Soil for Detoxification of Metanil Yellow Dye; Response Surface Methodological Approach. Sustainability 2020. (Accepted). [Google Scholar]
- Survase, S.; Annapure, U.; Singhal, R.S. Gellan Gum as Immobilization Matrix for Production of Cyclosporin A. J. Microbiol. Biotechnol. 2010, 20, 1086–1091. [Google Scholar] [CrossRef]
- Murugesan, K.; Dhamija, A.; Nam, I.-H.; Kim, Y.-M.; Chang, Y.-S. Decolourization of reactive black 5 by laccase: Optimization by response surface methodology. Dyes Pigm. 2007, 75, 176–184. [Google Scholar] [CrossRef]
- Bezerra, M.A.; Santelli, R.E.; Oliveira, E.P.; Villar, L.S.; Escaleira, L.A. Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta 2008, 76, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Lade, H.; Govindwar, S.; Paul, D. Low-Cost Biodegradation and Detoxification of Textile Azo Dye C.I. Reactive Blue 172 by Providencia rettgeri Strain HSL1. J. Chem. 2015, 2015, 894109. [Google Scholar] [CrossRef]
- Saratale, R.G.; Saratale, G.D.; Kalyani, D.C.; Chang, J.S.; Govindwar, S.P. Enhanced decolorization and biodegradation of textile azo dye Scarlet R by using developed microbial consortium-GR. Bioresour. Technol. 2009, 100, 2493–2500. [Google Scholar] [CrossRef]
- Ajaz, M.; Rehman, A.; Khan, Z.; Nisar, M.A.; Hussain, S. Degradation of azo dyes by Alcaligenes aquatilis 3c and its potential use in the wastewater treatment. AMB Express 2019, 9, 64. [Google Scholar] [CrossRef]
- Kurade, M.B.; Waghmode, T.R.; Jadhav, M.U.; Jeon, B.-H.; Govindwar, S.P. Bacterial–yeast consortium as an effective biocatalyst for biodegradation of sulphonated azo dye Reactive Red 198. RSC Adv. 2015, 5, 23046–23056. [Google Scholar] [CrossRef]
- He, F.; Hu, W.; Li, Y. Biodegradation Mechanisms and Kinetics of Azo Dye 4BS by a Microbial Consortium. Chemosphere 2004, 57, 293–301. [Google Scholar] [CrossRef]
- Ozdemir, G.; Pazarbasi, B.; Kocyigit, A.; Ersoy Omeroglu, E.; Yaşa, İ.; Karaboz, I. Decolorization of Acid Black 210 by Vibrio harveyi TEMS1 a Newly Isolated Bioluminescent Bacterium Enhanced Decolorization of Cibacron Red FN-2BL by Schizophyllum Commune from Izmir Bay. World J. Microbiol. Biotechnol. 2007, 24, 1375–1381. [Google Scholar] [CrossRef]
- Johari, W.L.W.; Isa, R.I.M.; Ghazali, N.; Arif, N.M.; Shukor, M.Y.A. Decolorization of Azo Dyes by Local Microorganisms. In Proceedings of the From Sources to Solution; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2013; pp. 357–361. [Google Scholar]
- El Ahwany, A.M.D. Decolorization of Fast red by metabolizing cells of Oenococcus oeni ML34. World J. Microbiol. Biotechnol. 2008, 24, 1521–1527. [Google Scholar] [CrossRef]
- Mansur, R.; Gusmanizar, N.; Roslan, M.A.H.; Ahmad, S.A.; Shukor, M.Y. Isolation and Characterisation of a Molybdenum-reducing and Metanil Yellow Dye-decolourising Bacillus sp. strain Neni-10 in Soils from West Sumatera, Indonesia. Trop. Life Sci. Res. 2017, 28, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Sanusi, S.N.A.; Halmi, M.I.E.; Abdullah, S.R.S.; Hassan, H.A.; Hamzah, F.M.; Idris, M. Comparative process optimization of pilot-scale total petroleum hydrocarbon (TPH) degradation by Paspalum scrobiculatum L. Hack using response surface methodology (RSM) and artificial neural networks (ANNs). Ecol. Eng. 2016, 97, 524–534. [Google Scholar] [CrossRef]
- Halmi, M.I.E.b.; Abdullah, S.R.S.; Wasoh, H.; Johari, W.L.W.; Ali, M.S.b.M.; Shaharuddin, N.A.; Shukor, M.Y. Optimization and maximization of hexavalent molybdenum reduction to Mo-blue by Serratia sp. strain MIE2 using response surface methodology. Rendiconti Lincei 2016, 27, 697–709. [Google Scholar] [CrossRef]
- Toolabi, A.; Malakootian, M.; Taghi Ghaneian, M.; Esrafili, A.; Ehrampoush, M.; Askarishahi, M.; Tabatabaei, M.; Khatami, M. Optimizing the photocatalytic process of removing diazinon pesticide from aqueous solutions and effluent toxicity assessment via a response surface methodology approach. Rendiconti Lince- 2019, 30, 155–165. [Google Scholar] [CrossRef]
- Gunawan, E.; Basri, M.; Rahman, M.; Bakar Salleh, A.; Rahman, R. Study on response surface methodology (RSM) of lipase-catalyzed synthesis of palm-based wax ester. Enzym. Microb. Technol 2005, 37, 739–744. [Google Scholar] [CrossRef]
- Beshay, U. Production of alkaline protease by Teredinobacter turnirae cells immobilized in Ca-alginate beads. Afr. J. Biotechnol. 2003, 2, 60–65. [Google Scholar] [CrossRef][Green Version]
- Pokharia, A.; Singh, S. Biodecolorization and degradation of xenobiotic azo dye -Basic Red 46 by Staphylococcus epidermidis MTCC 10623. Int. J. Res. Biosci. 2016, 5, 10–23. [Google Scholar]
- Chang, J.-S.; Chou, C.; Chen, S.-Y. Decolorization of azo dyes with immobilized Pseudomonas luteola. Process. Biochem. 2001, 36, 757–763. [Google Scholar] [CrossRef]
- Nallapan maniyam, M.; Latif Ibrahim, A.; Cass, T. Enhanced cyanide biodegradation by immobilized crude extract of Rhodococcus UKMP-5M. Environ. Technol. 2017, 40, 1–41. [Google Scholar] [CrossRef]
- Jafari, N.; Kasra-Kermanshahi, R.; Soudi, M.R. Screening, identification and optimization of a yeast strain, Candida palmioleophila JKS4, capable of azo dye decolorization. Iran J. Microbiol. 2013, 5, 434. [Google Scholar] [PubMed]
- Saratale, R.G.; Saratale, G.D.; Chang, J.S.; Govindwar, S.P. Bacterial decolorization and degradation of azo dyes: A review. J. Taiwan Inst. Chem. Eng. 2011, 42, 138–157. [Google Scholar] [CrossRef]
- Gauthier, P.T.; Norwood, W.P.; Prepas, E.E.; Pyle, G.G. Metal–PAH mixtures in the aquatic environment: A review of co-toxic mechanisms leading to more-than-additive outcomes. Aquat. Toxicol. 2014, 154, 253–269. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.P.; Patel, K.A.; Nair, S.S.; Darji, A. Microbial decolorization of methyl orange dye by Pseudomonas spp. ETL-M. Int. J. Environ. 2013, 1, 54–59. [Google Scholar]


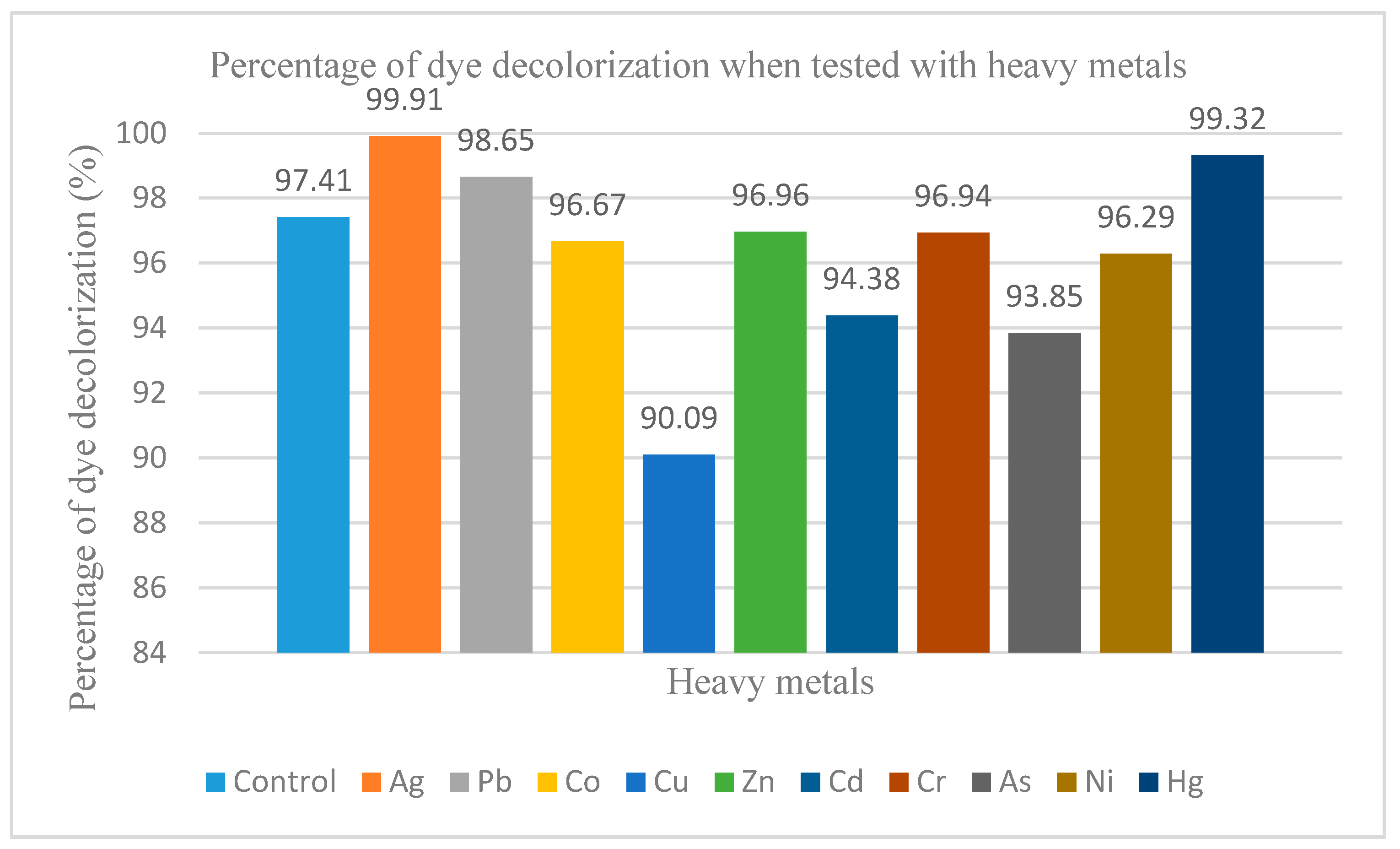
| Run | A: Dye Concentration(mg/L) | B: Gellan Gum Concentration (%) | C: Number of Beads | D: Beads Size (cm) | Decolorization (%) | Predicted RSM Decolorization (%) |
|---|---|---|---|---|---|---|
| 1 | 225 | 0.750 | 50 | 0.45 | 33.12 | 32.82 |
| 2 | 225 | 0.750 | 10 | 0.45 | 45.80 | 35.55 |
| 3 | 350 | 1.125 | 30 | 0.30 | 10.75 | 7.74 |
| 4 | 100 | 1.125 | 30 | 0.30 | 88.23 | 88.93 |
| 5 | 225 | 1.500 | 30 | 0.60 | 34.31 | 29.87 |
| 6 | 225 | 1.125 | 30 | 0.45 | 17.27 | 17.04 |
| 7 | 350 | 1.500 | 30 | 0.45 | 0.00 | -3.35 |
| 8 | 225 | 1.125 | 30 | 0.45 | 12.40 | 17.04 |
| 9 | 100 | 1.125 | 10 | 0.45 | 89.36 | 87.11 |
| 10 | 225 | 1.500 | 10 | 0.45 | 19.53 | 18.52 |
| 11 | 225 | 1.125 | 50 | 0.3 | 35.38 | 27.61 |
| 12 | 100 | 1.125 | 30 | 0.6 | 92.67 | 94.37 |
| 13 | 225 | 1.125 | 10 | 0.6 | 15.26 | 25.82 |
| 14 | 225 | 0.750 | 30 | 0.6 | 38.00 | 32.10 |
| 15 | 225 | 1.500 | 30 | 0.3 | 15.18 | 19.59 |
| 16 | 225 | 1.125 | 50 | 0.6 | 34.16 | 34.25 |
| 17 | 225 | 0.750 | 30 | 0.3 | 38.73 | 41.69 |
| 18 | 350 | 1.125 | 30 | 0.6 | 5.00 | 2.99 |
| 19 | 100 | 0.750 | 30 | 0.45 | 88.96 | 95.10 |
| 20 | 350 | 1.125 | 50 | 0.45 | 2.20 | 2.96 |
| 21 | 350 | 1.125 | 10 | 0.45 | 2.10 | 2.35 |
| 22 | 225 | 1.125 | 30 | 0.45 | 21.40 | 17.04 |
| 23 | 225 | 1.125 | 30 | 0.45 | 16.53 | 17.04 |
| 24 | 225 | 1.500 | 50 | 0.45 | 16.57 | 25.52 |
| 25 | 225 | 1.125 | 30 | 0.45 | 17.59 | 17.04 |
| 26 | 100 | 1.500 | 30 | 0.45 | 89.23 | 84.67 |
| 27 | 225 | 1.125 | 10 | 0.3 | 29.07 | 31.77 |
| 28 | 350 | 0.75 | 30 | 0.45 | 3.20 | 10.55 |
| 29 | 100 | 1.125 | 50 | 0.45 | 92.50 | 90.77 |
| Microorganisms | Optimal pH and Temperature for Reduction | Optimal C Source | Dye Concentration Tested (mg/L) | References |
|---|---|---|---|---|
| Vibrio harveyi TEMS1 | 20 °C | Glucose | 100 | [33] |
| Bacillus sp AK1 Lysinibacillus sp AK2 | 7.2 37 °C | Metanil Yellow | 200 | [7] |
| Unknown local isolates (NII and RHG) | - | Glucose | 50 | [34] |
| Oenococcus oeni ML34 | 7.0 30 °C | Glucose | 1000 | [35] |
| Bacillus sp Neni-10 | 6.3 34 °C | Glucose | 150 | [36] |
| ANOVA for Response Surface Quadratic Model | ||||||
|---|---|---|---|---|---|---|
| Analysis of Variance Table [Partial Sum of Squares—Type III] | ||||||
| Source | Sum of Squares | df | Mean Square | F Value | p-Value Prob > F | |
| Model | 26,709.35 | 14 | 1907.81 | 41.84 | <0.0001 | significant |
| A: Dye concentration | 22,334.7 | 1 | 22,334.7 | 489.87 | <0.0001 | |
| B: Gellan Gum Concentration | 444.07 | 1 | 444.07 | 9.74 | 0.0075 | |
| C: Number of beads | 13.67 | 1 | 13.67 | 0.3 | 0.5927 | |
| D: Beads size | 0.35 | 1 | 0.35 | 7.75−3 | 0.9311 | |
| AB | 3 | 1 | 3 | 0.066 | 0.8014 | |
| AC | 2.31 | 1 | 2.31 | 0.051 | 0.8252 | |
| AD | 25.94 | 1 | 25.94 | 0.57 | 0.4632 | |
| BC | 23.64 | 1 | 23.64 | 0.52 | 0.4833 | |
| BD | 98.62 | 1 | 98.62 | 2.16 | 0.1635 | |
| CD | 39.62 | 1 | 39.62 | 0.87 | 0.367 | |
| A2 | 3643.94 | 1 | 3643.94 | 79.92 | <0.0001 | |
| B2 | 233.92 | 1 | 233.92 | 5.13 | 0.0399 | |
| C2 | 165.93 | 1 | 165.93 | 3.64 | 0.0772 | |
| D2 | 391.46 | 1 | 391.46 | 8.59 | 0.011 | |
| Residual | 638.3 | 14 | 45.59 | |||
| Lack of fit | 597.15 | 10 | 59.71 | 5.8 | 0.0524 | Not significant |
| Pure Error | 41.15 | 4 | 10.29 | |||
| Cor Total | 27,347.66 | 28 | ||||
| Dye Concentration (mg/L) | Gellan Gum Concentration (%) | Number of Beads | Beads Size (cm) | Decolorization (%) | Desirability |
|---|---|---|---|---|---|
| 130 | 1.478 | 50 | 0.600 | 90.378 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muliadi, F.N.A.; Halmi, M.I.E.; Wahid, S.B.A.; Gani, S.S.A.; Mahmud, K.; Shukor, M.Y.A. Immobilization of Metanil Yellow Decolorizing Mixed Culture FN3 Using Gelling Gum as Matrix for Bioremediation Application. Sustainability 2021, 13, 36. https://doi.org/10.3390/su13010036
Muliadi FNA, Halmi MIE, Wahid SBA, Gani SSA, Mahmud K, Shukor MYA. Immobilization of Metanil Yellow Decolorizing Mixed Culture FN3 Using Gelling Gum as Matrix for Bioremediation Application. Sustainability. 2021; 13(1):36. https://doi.org/10.3390/su13010036
Chicago/Turabian StyleMuliadi, Fatin Natasha Amira, Mohd Izuan Effendi Halmi, Samsuri Bin Abdul Wahid, Siti Salwa Abd Gani, Khairil Mahmud, and Mohd Yunus Abd Shukor. 2021. "Immobilization of Metanil Yellow Decolorizing Mixed Culture FN3 Using Gelling Gum as Matrix for Bioremediation Application" Sustainability 13, no. 1: 36. https://doi.org/10.3390/su13010036
APA StyleMuliadi, F. N. A., Halmi, M. I. E., Wahid, S. B. A., Gani, S. S. A., Mahmud, K., & Shukor, M. Y. A. (2021). Immobilization of Metanil Yellow Decolorizing Mixed Culture FN3 Using Gelling Gum as Matrix for Bioremediation Application. Sustainability, 13(1), 36. https://doi.org/10.3390/su13010036





