Remediation of Petroleum-Contaminated Soils with Microbial and Microbial Combined Methods: Advances, Mechanisms, and Challenges
Abstract
:1. Introduction
2. Petroleum-Contaminated Soil
2.1. Sources of Petroleum Pollutants
2.2. Composition of Petroleum Pollutants
2.3. Toxic Effects of Petroleum on the Environment
2.3.1. Toxic Effects of Petroleum on Soil
2.3.2. Toxic Effects of Petroleum on Plants
2.3.3. Toxic Effects of Petroleum on Human Health
3. Advances in the Utilization of Microorganisms in Petroleum Remediation
4. Microbial Remediation
5. Combined Microbial Methods Remediation
5.1. Microorganism–Biochar Interactions in Remediation of Hydrocarbons
5.2. Microorganism–Nutrients Interactions in Remediation of Hydrocarbons
5.3. Microorganism–Plant Interactions in Remediation of Hydrocarbons
6. Advantages and Challenges in Combined Microbial Methods Application for Hydrocarbon Removal
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abbaspour, A.; Zohrabi, F.; Dorostkar, V.; Faz, A.; Acosta, J.A. Remediation of an oil-contaminated soil by two native plants treated with biochar and mycorrhizae. J. Environ. Manag. 2020, 254, 109755. [Google Scholar] [CrossRef]
- Freedman, B. Oil pollution. In Environmental Ecology, 2nd ed.; Academic Press: San Diego, CA, USA, 1995; pp. 159–188. [Google Scholar]
- Suganthi, S.H.; Murshid, S.; Sriram, S. Enhanced biodegradation of hydrocarbons in petroleum tank bottom oil sludge and characterization of biocatalysts and biosurfactants. J. Environ. Manag. 2018, 220, 87–95. [Google Scholar] [CrossRef]
- Ansari, N.; Hassanshahian, M.; Ravan, H. Study the microbial communities’ changes in desert and farmland soil after crude oil pollution. Int. J. Environ. Res. 2018, 12, 391–398. [Google Scholar] [CrossRef]
- Ekundayo, E.O.; Emede, T.O.; Osayande, D.I. Effects of crude oil spillage on growth and yield of maize (Zea mays L.) in soils of midwestern Nigeria. Plant Foods Hum. Nutr. 2001, 56, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Regar, R.K.; Manickam, N. Improved polycyclic aromatic hydrocarbon degradation in a crude oil by individual and a consortium of bacteria. Bioresour. Technol. 2018, 254, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Ozigis, M.S.; Kaduk, J.D.; Jarvis, C.H. Detection of oil pollution impacts on vegetation using multifrequency SAR, multispectral images with fuzzy forest and random forest methods. Environ. Pollut. 2020, 256, 113360. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan-Gordo, C.; Orta-Martínez, M.; Kogevinas, M. Health effects of non-occupational exposure to oil extraction. Environ. Health 2016, 15, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Maletić, S.; Dalmacija, B.; Rončević, S. Petroleum hydrocarbon biodegradability in soil–implications for bioremediation. Intech 2013, 43, 43–64. [Google Scholar]
- Sankaran, S.; Pandey, S.; Sumathy, K. Experimental investigation on waste heat recovery by refinery oil sludge incineration using fluidised-bed technique. Environ. Lett. 1998, 33, 829–845. [Google Scholar] [CrossRef]
- Liao, X.; Wu, Z.; Li, Y.; Cao, H.; Su, C. Effect of various chemical oxidation reagents on soil indigenous microbial diversity in remediation of soil contaminated by PAHs. Chemosphere 2019, 226, 483–491. [Google Scholar] [CrossRef]
- Giannopoulos, D.; Kolaitis, D.I.; Togkalidou, A.; Skevis, G.; Founti, M.A. Quantification of emissions from the co-incineration of cutting oil emulsions in cement plants—Part II: Trace species. Fuel 2007, 86, 2491–2501. [Google Scholar] [CrossRef]
- Vidonish, J.E.; Zygourakis, K.; Masiello, C.A.; Sabadell, G.; Alvarez, P.J.J. Thermal treatment of hydrocarbon-impacted soils: A Review of technology innovation for sustainable remediation. Engineering 2016, 2, 426–437. [Google Scholar] [CrossRef]
- Kang, C.-U.; Kim, D.-H.; Khan, M.A.; Kumar, R.; Ji, S.-E.; Choi, K.-W.; Paeng, K.-J.; Park, S.; Jeon, B.-H. Pyrolytic remediation of crude oil-contaminated soil. Sci. Total Environ. 2020, 713, 136498. [Google Scholar] [CrossRef]
- Ku-Fan, C.; Yu-Chen, C.; Wan-Ting, C. Remediation of diesel-contaminated soil using in situ chemical oxidation (ISCO) and the effects of common oxidants on the indigenous microbial community: A comparison study. J. Chem. Technol. Biotechnol. 2016, 91, 1877–1888. [Google Scholar]
- Das, N.; Chandran, P. Microbial degradation of petroleum hydrocarbon contaminants: An overview. Biotechnol. Res. Int. 2011, 10, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senko, J.M.; Campbell, B.S.; Henriksen, J.R.; Elshahed, M.S.; Dewers, T.A.; Krumholz, L.R. Barite deposition resulting from phototrophic sulfide-oxidizing bacterial activity. Geochim. Cosmochim. Acta 2004, 68, 773–780. [Google Scholar] [CrossRef]
- Velacano, M.; Castellanohinojosa, A.; Vivas, A.F.; Toledo, M.V.M. Effect of Heavy Metals on the Growth of Bacteria Isolated from Sewage Sludge Compost Tea. Adv. Microbiol. 2014, 4, 644–655. [Google Scholar] [CrossRef] [Green Version]
- Chaîneau, C.H.; Rougeux, G.; Yéprémian, C.J. Oudot. Effects of nutrient concentration on the biodegradation of crude oil and associated microbial populations in the soil. Soil Biol. Biochem. 2005, 37, 1490–1497. [Google Scholar] [CrossRef]
- ChaIneau, C.-H.; Morel, J.-L.; Oudot, J. Microbial degradation in soil microcosms of fuel oil hydrocarbons from drilling cuttings. Environ. Sci. Technol. 1995, 29, 1615–1621. [Google Scholar] [CrossRef]
- Pino, N.J.; Muñera, L.M.; Peñuela, G.A. Bioaugmentation with immobilized microorganisms to enhance phytoremediation of PCB-contaminated soil. Soil Sediment Contam. Int. J. 2016, 25, 419–430. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Xu, Y.; Wang, H.-Y.; Zhao, J.; Gao, D.-M.; Li, F.-M.; Xing, B. Biodegradation of crude oil in contaminated soils by free and immobilized microorganisms. Pedosphere 2012, 22, 717–725. [Google Scholar] [CrossRef]
- Hamamura, N.; Fukui, M.; Ward, D.M.; Inskeep, W.P. Assessing soil microbial populations responding to crude-oil amendment at different temperatures using phylogenetic, functional gene (alkB) and physiological analyses. Environ. Sci. Technol. 2008, 42, 7580–7586. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Wang, M.; Xing, Y.; Lan, G.; Ge, T.; Yan, X.; Gu, T. Characterization and optimization of biosurfactants produced by Acinetobacter baylyi ZJ2 isolated from crude oil-contaminated soil sample toward microbial enhanced oil recovery applications. Biochem. Eng. J. 2014, 90, 49–58. [Google Scholar] [CrossRef]
- Das, M.; Adholeya, A. Role of Microorganisms in Remediation of Contaminated Soil. In Microorganisms in Environmental Management; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Wu, M.; Wu, J.; Zhang, X.; Ye, X. Effect of bioaugmentation and biostimulation on hydrocarbon degradation and microbial community composition in petroleum-contaminated loessal soil. Chemosphere 2019, 237, 124456. [Google Scholar] [CrossRef] [PubMed]
- Fanaei, F.; Moussavi, G.; Shekoohiyan, S. Enhanced treatment of the oil-contaminated soil using biosurfactant-assisted washing operation combined with H2O2-stimulated biotreatment of the effluent. J. Environ. Manag. 2020, 271, 110941. [Google Scholar] [CrossRef] [PubMed]
- Abioye, O.P. Biological remediation of hydrocarbon and heavy metals contaminated soil. Soil Contam. 2011, 7, 127–142. [Google Scholar]
- Adipah, S. Introduction of petroleum hydrocarbons contaminants and its human effects. J. Environ. Sci. Public Health 2018, 3, 001–009. [Google Scholar] [CrossRef]
- Wang, S.; Xu, Y.; Lin, Z.; Zhang, J.; Norbu, N.; Liu, W. The harm of petroleum-polluted soil and its remediation research. Aip Conf. Proc. 2017, 1864, 020222. [Google Scholar]
- Kachieng’a, L.; Momba, M.N.B. Kinetics of petroleum oil biodegradation by a consortium of three protozoan isolates (Aspidisca sp., Trachelophyllum sp. and Peranema sp.). Biotechnol. Rep. 2017, 15, 125–131. [Google Scholar] [CrossRef]
- Sarma, H.; Nava, A.R.; Prasad, M.N.V. Mechanistic understanding and future prospect of microbe-enhanced phytoremediation of polycyclic aromatic hydrocarbons in soil. Environ. Technol. Innov. 2019, 13, 318–330. [Google Scholar] [CrossRef]
- Brakstad, O.G.; Ribicic, D.; Winkler, A.; Netzer, R. Biodegradation of dispersed oil in seawater is not inhibited by a commercial oil spill dispersant. Mar. Pollut. Bull. 2018, 129, 555–561. [Google Scholar] [CrossRef]
- Włodarczyk-Makuła, M. Half-Life of carcinogenic polycyclic aromatic hydrocarbons in stored sewage sludge. Arch. Environ. Prot. 2012, 38, 33–44. [Google Scholar] [CrossRef] [Green Version]
- Elumalai, P.; Parthipan, P.; Karthikeyan, O.P.; Rajasekarcorresponding, A. Enzyme-mediated biodegradation of long-chain n-alkanes (C32 and C40) by thermophilic bacteria. 3 Biotech 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguelmous, A.; Zegzouti, Y.; Khadra, A.; Fels, L.E.; Souabi, S.; Hafidi, M. Landfilling and composting efficiency to reduce genotoxic effect of petroleum sludge. Environ. Technol. Innov. 2020, 20, 101047. [Google Scholar] [CrossRef]
- Schirmer, A.; Rude, M.A.; Li, X.; Popova, E.; Cardayre, S.B.D. Microbial Biosynthesis of Alkanes. Science 2010, 329, 559–562. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Bellemans, A. Molecular dynamics of liquid alkanes. Faraday Discuss. Chem. Soc. 1978, 66, 95–106. [Google Scholar] [CrossRef]
- Zakaria, M.P.; Bong, C.-W.; Vaezzadeh, V. Fingerprinting of petroleum hydrocarbons in malaysia using environmental forensic techniques: A 20-year field data review. In Oil Spill Environmental Forensics Case Studies; Butterworth-Heinemann: Oxford, UK, 2018; Chapter 16; pp. 345–372. [Google Scholar]
- Sadeghbeigi, R. Chapter 3—FCC Feed Characterization. In Fluid Catalytic Cracking Handbook, 3rd ed.; Butterworth-Heinemann: Oxford, UK, 2012. [Google Scholar]
- Tissot, B.P.; Welte, D.H. Composition of crude oils. In Petroleum Formation and Occurrence; Springer: Berlin/Heidelberg, Germany, 1984; Chapter 1; pp. 333–368. [Google Scholar]
- Vasconcelos, I.U.; FrançaI, F.P.D.; OliveiraII, F.J.S. Removal of high-molecular weight polycyclic aromatic hydrocarbons. Química Nova 2011, 34, 218–221. [Google Scholar] [CrossRef]
- Haines, J.R.; Alexander, M. Microbial degradation of high-molecular-weight alkanes. Appl. Microbiol. Biotechnol. 1974, 28, 1084–1085. [Google Scholar] [CrossRef] [Green Version]
- Imam, A.; Suman, S.K.; Ghosh, D.K. Kanaujia, P. Analytical approaches used in monitoring the bioremediation of hydrocarbons in petroleum-contaminated soil and sludge. TrAC Trends Anal. Chem. 2019, 118, 50–64. [Google Scholar] [CrossRef]
- Abdel-Shafy, H.I.; Mansour, M.S.M. A review on polycyclic aromatic hydrocarbons: Source, environmental impact, effect on human health and remediation. Egypt. J. Pet. 2016, 25, 107–123. [Google Scholar] [CrossRef] [Green Version]
- Fooladi, M.; Moogouei, R.; Jozi, S.A.; Golbabaei, F.; Tajadod, G. Phytoremediation of BTEX from indoor air by Hyrcanian plants. Environ. Health Eng. Manag. J. 2019, 6, 233–240. [Google Scholar] [CrossRef] [Green Version]
- Wei, Z.; Wang, J.J.; Meng, Y.; Li, J.; Gaston, L.A.; Fultz, L.M.; DeLaune, R.D. Potential use of biochar and rhamnolipid biosurfactant for remediation of crude oil-contaminated coastal wetland soil: Ecotoxicity assessment. Chemosphere 2020, 253, 126617. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Eyles, J.L.; Collins, C.D.; Hodson, M.E. Using deuterated PAH amendments to validate chemical extraction methods to predict PAH bioavailability in soils. Environ. Pollut. 2011, 159, 918–923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Achuba, F.I.; Peretiemo-Clarke, B.O. Effect of spent engine oil on soil catalase and dehydrogenase activities. Int. Agrophys. 2008, 22, 1–4. [Google Scholar]
- Polyak, Y.; Bakina, L.G.; Bakina, L.G.; Chugunova, M.V.; Bure, V. Effect of remediation strategies on biological activity of oil-contaminated soil—A field study. Int. Biodeterior. Biodegrad. 2018, 126, 57–68. [Google Scholar] [CrossRef]
- Wei, Y.; Li, G. Effect of oil pollution on water characteristics of loessial soil. IOP Conf. Ser. Earth Environ. Sci. 2018, 170, 032154. [Google Scholar] [CrossRef] [Green Version]
- Barua, D.; Buragohain, J.; Sarma, S.K. Certain physico-chemical changes in the soil brought about by contamination of crude oil in two oil fields of Assam. NE India Pelagia Res. Libr. 2011, 1, 154–161. [Google Scholar]
- Osuji, L.C.; Idung, I.D.; Ojinnaka, C.M. Preliminary investigation on Mgbede-20 oil-polluted site in Niger Delta, Nigeria. Chem. Biodivers. 2006, 3, 568–577. [Google Scholar] [CrossRef]
- Xin, L.; Hui-hui, Z.; Bing-bing, Y.; Nan, X.; Wen-xu, Z.; Ju-wei, H.; Guang-yu, S. Effects of Festuca arundinacea on the microbial community in crude oil-contaminated saline-alkaline soil. Chin. J. Appl. Ecol. 2012, 23, 3414–3420. [Google Scholar]
- Steliga, T.; Kluk, D. Application of Festuca arundinacea in phytoremediation of soils contaminated with Pb, Ni, Cd and petroleum hydrocarbons. Ecotoxicol. Environ. Saf. 2020, 194, 110409. [Google Scholar] [CrossRef] [PubMed]
- Mangse, G.; Werner, D.; Meynet, P.; Ogbaga, C.C. Microbial community responses to different volatile petroleum hydrocarbon class mixtures in an aerobic sandy soil. Environ. Pollut. 2020, 264, 114738. [Google Scholar] [CrossRef]
- Buzmakov, S.A.; Khotyanovskaya, Y.V. Degradation and pollution of lands under the influence of oil resources exploitation. Appl. Geochem. 2020, 113, 104443. [Google Scholar] [CrossRef]
- Uzoho, B.; Oti, N.; Onweremadu, E. Effect of crude oil pollution on maize growth and soil properties in Ihiagwa, Imo State, Nigeria. Int. J. Agric. Rural Dev. 2006, 5, 91–100. [Google Scholar] [CrossRef]
- Lorestani, B.; Kolahchi, N.; Ghasemi, M.; Cheraghi, M.; Yousefi, N. Survey the effect of oil pollution on morphological characteristics in Faba Vulgaris and Vicia Ervilia. J. Chem. Health Risks 2012, 2, 2251–6727. [Google Scholar]
- Zhen, M.; Chen, H.; Liu, Q.; Song, B.; Wang, Y.; Tang, J. Combination of rhamnolipid and biochar in assisting phytoremediation of petroleum hydrocarbon contaminated soil using Spartina anglica. J. Environ. Sci. 2019, 85, 107–118. [Google Scholar] [CrossRef]
- Kuppusamy, S.; Raju, M.N.; Mallavarapu, M.; Kadiyala, V. Impact of Total Petroleum Hydrocarbons on Human Health. In Total Petroleum Hydrocarbons; Springer: Cham, Germany, 2020; pp. 139–165. [Google Scholar]
- Haller, H.; Jonsson, A. Growing food in polluted soils: A review of risks and opportunities associated with combined phytoremediation and food production (CPFP). Chemosphere 2020, 254, 126826. [Google Scholar] [CrossRef] [PubMed]
- Hussain, I.; Puschenreiter, M.; Gerhard, S.; Sani, S.G.A.S.; Khan, W.-u.-d.; Reichenauer, T.G. Differentiation between physical and chemical effects of oil presence in freshly spiked soil during rhizoremediation trial. Environ. Sci. Pollut. Res. 2019, 26, 18451–18464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, K.; Liu, X.; Chen, X.; Hu, X.; Cao, L.; Yuan, X. Biodegradation of crude oil by a defined co-culture of indigenous bacterial consortium and exogenous Bacillus subtilis. Bioresour. Technol. 2016, 224, 327–332. [Google Scholar] [CrossRef]
- Abena, M.T.B.; Li, T.; Shah, M.N.; Zhong, W. Biodegradation of total petroleum hydrocarbons (TPH) in highly contaminated soils by natural attenuation and bioaugmentation. Chemosphere 2019, 234, 864–874. [Google Scholar] [CrossRef] [PubMed]
- Bacosa, H.P.; Suto, K.; Inoue, C. Bacterial community dynamics during the preferential degradation of aromatic hydrocarbons by a microbial consortium. Int. Biodeterior. Biodegrad. 2012, 74, 109–115. [Google Scholar] [CrossRef]
- Lee, D.W.; Lee, H.; Kwon, B.-O.; Khim, J.S.; Yim, U.H.; Kim, B.S.; Kim, J.-J. Biosurfactant-assisted bioremediation of crude oil by indigenous bacteria isolated from Taean beach sediment. Environ. Pollut. 2018, 241, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Teng, Y.; Yao, H.; Christie, P. Detection of functional microorganisms in benzene [a] pyrene-contaminated soils using DNA-SIP technology. J. Hazard. Mater. 2021, 407, 124788. [Google Scholar] [CrossRef]
- Pi, Y.; Chen, B.; Bao, M.; Fan, F.; Cai, Q.; Ze, L.; Zhang, B. Microbial degradation of four crude oil by biosurfactant producing strain Rhodococcus sp. Bioresour. Technol. 2018, 232, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Zhang, X.; Chen, X.; Kong, D.; Liu, X.; Shen, S. Synergistic degradation of crude oil by indigenous bacterial consortium and T exogenous fungus Scedosporium boydii. Bioresour. Technol. 2018, 264, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Varjani, S.J.; Upasani, V.N. Biodegradation of petroleum hydrocarbons by oleophilic strain of Pseudomonas aeruginosa NCIM 5514. Bioresour. Technol. 2016, 222, 195–201. [Google Scholar] [CrossRef]
- Wang, L.; Wang, W.; Lai, Q.; Shao, Z. Gene diversity of CYP153A and AlkB alkane hydroxylases in oil-degrading bacteria isolated from the Atlantic Ocean. Environ. Microbiol. 2010, 12, 1230–1242. [Google Scholar] [CrossRef]
- Cao, B.; Nagarajan, K.; Loh, K.C. Biodegradation of aromatic compounds: Current status and opportunities for biomolecular approaches. Appl. Microbiol. Biotechnol. 2009, 85, 207–228. [Google Scholar] [CrossRef]
- Margesin, R.; Zimmerbauer, A.; Schinner, F. Soil lipase activity–A useful indicator of oil biodegradation. Biotechnol. Tech. 1999, 13, 859–863. [Google Scholar] [CrossRef]
- Sirajuddin, S.; Rosenzweig, A.C. Enzymatic Oxidation of Methane. Biochemistry 2015, 54, 2283–2294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.-C.; Li, L.-Z.; Wu, Y.; Tian, W.; Zhang, L.-P.; Xu, L.; Shen, Q.-R.; Shen, B. Isolation of an alkane-degrading Alcanivorax sp. strain 2B5 and cloning of the alkB gene. Bioresour. Technol. 2010, 101, 310–316. [Google Scholar]
- Huan, L.; Jing, X.; Rubing, L.; Jianhua, L.; ONE, D.A.J.P. Characterization of the Medium- and Long-Chain n-Alkanes Degrading Pseudomonas aeruginosa Strain SJTD-1 and Its Alkane Hydroxylase Genes. PLoS ONE 2014, 9, e105506. [Google Scholar]
- Liu, J.; Zhao, B.; Lan, Y.; Ma, T. Enhanced degradation of different crude oils by defined engineered consortia of Acinetobacter venetianus RAG-1 mutants based on their alkane metabolism. Bioresour. Technol. 2021, 327, 124787. [Google Scholar] [CrossRef]
- Mimmi, T.-H.; Alexander, W.; Trond, E.E.; Hans-Kristian, K.; Sergey, B.Z. Identification of novel genes involved in long-chain n-alkane degradation by Acinetobacter sp. strain DSM 17874. Appl. Environ. Microbiol. 2007, 73, 3327–3332. [Google Scholar]
- Abouseoud, M.; Yataghene, A.; Amrane, A.; Maachi, R. Biosurfactant production by free and alginate entrapped cells of Pseudomonas fluorescens. J. Ind. Microbiol. Biotechnol. 2008, 35, 1303–1308. [Google Scholar] [CrossRef] [PubMed]
- Hesham, E.L.; Mawad, A.M.M.; Mostafa, Y.M.; Shoreit, A.J.M. Study of enhancement and inhibition phenomena and genes relating to degradation of petroleum polycyclic aromatic hydrocarbons in isolated bacteria. Microbiology 2014, 83, 599–607. [Google Scholar] [CrossRef]
- Varjani, S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2016, 223, 277–286. [Google Scholar] [CrossRef]
- Champion, P.M.; Gunsalus, I.C.; Wagner, G.C. Resonance Raman investigations of cytochrome P450CAM from Pseudomonas putida. J. Am. Chem. Soc. 1978, 100, 3743–3751. [Google Scholar] [CrossRef]
- Noble, M.E.M.; Cleasby, A.; Johnson, L.N.; Egmond, M.R.; Frenken, L.G.J. The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate. FEBS Lett. 1993, 331, 123–128. [Google Scholar] [CrossRef] [Green Version]
- Rojo, F. Degradation of alkanes by bacteria. Environ. Microbiol. 2009, 11, 2477–2490. [Google Scholar] [CrossRef]
- Xia, Y.; Min, H.; Rao, G.; Lv, Z.; Liu, J.; Ye, Y.; Duan, X. Isolation and character- ization of phenanthrene-degrading Sphingomonas paucimobilis strain ZX4. Biodegradation 2005, 16, 393–402. [Google Scholar] [CrossRef]
- Oyetibo, G.O.; Chien, M.F.; Ikeda-Ohtsubo, W.; Suzuki, H.; Obayori, O.S.; Adebusoye, S.A.; Ilori, M.O.; Amund, O.O.; Endo, G. Biodegradation of crude oil and phenanthrene by heavy metal resistant Bacillus subtilis isolated from a multi-polluted industrial wastewater creek. Int. Biodeterior. Biodegrad. 2017, 120, 143–151. [Google Scholar] [CrossRef]
- Deng, M.-C.; Li, J.; Liang, F.-R.; Wang, J.-H. Isolation and characterization of a novel hydrocarbon-degrading bacterium Achromobacter sp HZ01 from the crude oil-contaminated seawater at the Daya Bay, southern China. Mar. Pollut. Bull. 2014, 83, 79–86. [Google Scholar] [CrossRef]
- Shao, Y.; Wang, Y.; Wu, X.; Li, B. Biodegradation of PAHs by Acinetobacter isolated from karst groundwater in a coal-mining area. Environ. Earth Sci. 2015, 73, 7479–7488. [Google Scholar] [CrossRef]
- Lily, M.K.; Bahuguna, A.; Dangwal, K.; Garg, V. Degradation of Benzo [a] Pyrene by a novel strain Bacillus subtilis BMT4i (MTCC 9447). Braz. J. Microbiol. 2009, 40, 884–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Thukair, A.A.; Malik, K. Pyrene metabolism by the novel bacterial strains Burkholderia fungorum (T3A13001) and Caulobacter sp (T2A12002) isolated from an oil-polluted site in the Arabian Gulf. Int. Biodeterior. Biodegrad. 2016, 110, 32–37. [Google Scholar] [CrossRef]
- Darma, U.Z.; Aziz, N.A.A.; Zulkefli, S.Z.; Mustafa, M. Identification of Phenanthrene and Pyrene degrading bacteria from used engine oil contaminated soil. Int. J. Sci. Eng. Res. 2016, 7, 680–686. [Google Scholar]
- Rajkumari, J.; Singha, L.P.; Pandey, P. Genomic insights of aromatic hydrocarbon degrading Klebsiella pneumoniae AWD5 with plant growth promoting attributes: A paradigm of soil isolate with elements of biodegradation. 3 Biotech 2018, 8, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Heitkamp, M.A.; Freeman, J.P.; Miller, D.W.; Cerniglia, C.E. Pyrene degradation by a Mycobacterium sp.: Identification of ring oxidation and ring fission products. Appl. Environ. Microbiol. 1988, 54, 2556–2565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ping, L.; Guo, Q.; Chen, X.; Yuan, X.; Zhang, C.; Zhao, H. Biodegradation of pyrene and benzo[a]pyrene in the liquid matrix and soil by a newly identified Raoultella planticola strain. 3 Biotech 2017, 7, 56. [Google Scholar] [CrossRef] [Green Version]
- Song, X.; Xu, Y.; Li, G.; Zhang, Y.; Huang, T.; Hu, Z. Isolation, characterization of Rhodococcus sp. P14 capable of degrading high-molecular-weight polycyclic aromatic hydrocarbons and aliphatic hydrocarbons. Mar. Pollut. Bull. 2011, 62, 2122–2128. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, H.; Zanaroli, G.; Xu, P.; Tang, H. A Pseudomonas sp. strain uniquely degrades PAHs and heterocyclic derivatives via lateral dioxygenation pathways. J. Hazard. Mater. 2021, 403, 123956. [Google Scholar] [CrossRef]
- Patel, V.; Cheturvedula, S.; Madamwar, D. Phenanthrene degradation by Pseudoxanthomonas sp. DMVP2 isolated from hydrocarbon contaminated sediment of Amlakhadi canal, Gujarat, India. J. Hazard. Mater. 2012, 201, 43–51. [Google Scholar] [CrossRef]
- Chen, J.; Wong, M.H.; Wong, Y.S.; Tam, N.F.Y. Multi-factors on biodegradation kinetics of polycyclic aromatic hydrocarbons (PAHs) by Sphingomonas sp. a bacterial strain isolated from mangrove sediment. Mar. Pollut. Bull. 2008, 57, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, B.; Manickam, N.; Kumari, S.; Tiwari, A. Biodegradation and dissolution of polyaromatic hydrocarbons by Stenotrophomonas sp. Bioresour. Technol. 2016, 216, 1102–1105. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, C.; Duraipandiyan, V.; Balakrishna, K.; Ignacimuthu, S. Petroleum and polycyclic aromatic hydrocarbons (PAHs) degradation and naphthalene metabolism in Streptomyces sp. (ERI-CPDA-1) isolated from oil contaminated soil. Bioresour. Technol. 2012, 112, 83–90. [Google Scholar] [CrossRef]
- Al-Hawash, A.B.; Zhang, X.; Ma, F. Removal and biodegradation of different petroleum hydrocarbons using the filamentous fungus Aspergillus sp. RFC-1. Microbiologyopen 2019, 8, e00619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baek, K.-H.; Yoon, B.-D.; Oh, H.-M.; Kim, H.-S.; Lee, I.-S. Biodegradation of aliphatic and aromatic hydrocarbons by Nocardia sp. H17-1. Geomicrobiol. J. 2007, 23, 253–259. [Google Scholar] [CrossRef]
- Govarthanan, M.; Fuzisawa, S.; Hosogai, T.; Chang, Y.-C. Biodegradation of aliphatic and aromatichydrocarbons using the filamentous fungusPenicillium sp. CHY-2 and characterization of itsmanganese peroxidase activity. RSC Adv. 2017, 7, 20716–20723. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Gong, Z.; Li, P.; Zhang, L.; Hu, X. Degradation of pyrene and benzo(a)pyrene in contaminated soil by immobilized fungi. Environ. Eng. Sci. 2008, 25, 677–684. [Google Scholar] [CrossRef]
- Liu, Y.; Han, H.; Fang, F. Degradation of long-chain n-alkanes by Acinetobacter sp. Adv. Mater. Res. 2013, 726–731, 2151–2155. [Google Scholar] [CrossRef]
- Wang, D.; Lin, J.; Lin, J.; Wang, W.; Li, S. Biodegradation of petroleum hydrocarbons by bacillus subtilis BL-27, a strain with weak hydrophobicity. Molecules 2019, 24, 3021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, W.; Du, Z.; Cui, Q.; Dong, H.; Wang, F.; He, P.; Tang, Y. Biosurfactant produced by novel Pseudomonas sp. WJ6 with biodegradation of n-alkanes and polycyclic aromatic hydrocarbons. J. Hazard. Mater. 2014, 276, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Whyte, L.G.; Hawari, J.; Zhou, E.; Bourbonnière, L.; Inniss, W.E.; Greer, C.W. Biodegradation of Variable-Chain-Length Alkanes at Low Temperatures by a Psychrotrophic Rhodococcussp. Appl. Environ. Microbiol. 1998, 64, 2578–2584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teh, J.S.; Lee, K.H. Utilization of n-Alkanes by Cladosporium resinae. Appl. Microbiol. 1973, 25, 454–457. [Google Scholar] [CrossRef] [PubMed]
- Quatrini, P.; Scaglione, G.; Pasquale, C.D.; Riela, S.; Puglia, A.M. Isolation of Gram-positive n-alkane degraders from a hydrocarbon-contaminated Mediterranean shoreline. J. Appl. Microbiol. 2007, 104, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Chiciudean, I.; Nie, Y.; Tănase, A.-M.; Stoica, I.; Wu, X.-L. Complete genome sequence of Tsukamurella sp. MH1: A wide-chain length alkane-degrading actinomycete. J. Biotechnol. 2018, 268, 1–5. [Google Scholar] [CrossRef]
- Atlas, R.M. Microbial degradation of petroleum hydrocarbons: An environmental perspective. Microbiol. Rev. 1981, 45, 180–209. [Google Scholar] [CrossRef]
- Chen, Y.-A.; Liu, P.-W.G.; Whang, L.-M.; Wu, Y.-J.; Cheng, S.-S. Effect of soil organic matter on petroleum hydrocarbon degradation in diesel/fuel oil-contaminated soil. J. Biosci. Bioeng. 2020, 129, 603–612. [Google Scholar] [CrossRef]
- Zhang, H.; Hu, Z.; Hou, S.; Xu, T. Effects of microbial degradation on morphology, chemical compositions and microstructures of bitumen. Constr. Build. Mater. 2020, 248, 118569. [Google Scholar] [CrossRef]
- Hajieghrari, M.; Hejazi, P. Enhanced biodegradation of n-Hexadecane in solid-phase of soil by employing immobilized Pseudomonas Aeruginosa on size-optimized coconut fibers. J. Hazard. Mater. 2020, 389, 122134. [Google Scholar] [CrossRef]
- Premnath, N.; Mohanrasu, K.; Rao, R.G.R.; Dinesh, G.H.; Prakash, G.S.; Ananthi, V.; Ponnuchamy, K.; Muthusamy, G.; Arun, A. A crucial review on polycyclic aromatic Hydrocarbons–Environmental occurrence and strategies for microbial degradation. Chemosphere 2021, 280, 130608. [Google Scholar] [CrossRef]
- Heitkamp, M.A.; Cerniglia, C.E. Effects of chemical structure and exposure on the microbial degradation of polycyclic aromatic hydrocarbons in freshwater and estuarine ecosystems. Environ. Toxicol. Chem. 1987, 6, 535–546. [Google Scholar] [CrossRef]
- Li, Q.; Huang, Y.; Wen, D.; Fu, R.; Feng, L. Application of alkyl polyglycosides for enhanced bioremediation of petroleum hydrocarbon-contaminated soil using Sphingomonas changbaiensis and Pseudomonas stutzeri. Sci. Total Environ. 2020, 719, 137456. [Google Scholar] [CrossRef]
- Lladó, S.; Solanas, A.M.; Lapuente, J.D.; Borràs, M.; Viñas, M. A diversified approach to evaluate biostimulation and bioaugmentation strategies for heavy-oil-contaminated soil. Sci. Total Environ. 2012, 435, 262–269. [Google Scholar] [CrossRef] [Green Version]
- Mukome, F.N.D.; Buelowa, M.C.; Shang, u.; Peng, J.; Rodriguez, M.; Mackay, D.M.; Pignatello, J.J.; Sihota, N.; Hoelen, T.P.; Parikh, S.J. Biochar amendment as a remediation strategy for surface soils impacted by crude oil. Environ. Pollut. 2020, 265, 115006. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.-Y.; Huang, W.-H.; Xing, D.-F.; Zhang, H.-F. Remediation of soil co-contaminated with petroleum and heavy metals by the integration of electrokinetics and biostimulation. J. Hazard. Mater. 2013, 260, 399–408. [Google Scholar] [CrossRef]
- Tahseen, R.; Afzal, M.; Iqbal, S.; Shabir, G.; Khan, Q.M.; Khalid, Z.M.; Banat, I.M. Rhamnolipids and nutrients boost remediation of crude oil-contaminated soil by enhancing bacterial colonization and metabolic activities. Int. Biodeterior. Biodegrad. 2016, 115, 192–198. [Google Scholar] [CrossRef]
- Qin, G.; Gong, D.; Fan, M.-Y. Bioremediation of petroleum-contaminated soil by biostimulation amended with biochar. Int. Biodeterior. Biodegrad. 2013, 85, 150–155. [Google Scholar] [CrossRef]
- Zhao, L.; Deng, J.; Hou, H.; Li, J.; Yang, Y. Investigation of PAH and oil degradation along with electricity generation in soil using an enhanced plant-microbial fuel cell. J. Clean. Prod. 2019, 221, 678–683. [Google Scholar] [CrossRef]
- Rhykerd, R.L.; Crews, B.; Mclnnes, K.J.; Weaver, R.W. Impact of bulking agents, forced aeration, and tillage on remediation of oil-contaminated soil. Bioresour. Technol. 1999, 67, 279–285. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, X.; Dai, D.; Li, G. Porous biocarrier-enhanced biodegradation of crude oil contaminated soil. Int. Biodeterior. Biodegrad. 2009, 63, 80–87. [Google Scholar] [CrossRef]
- Xu, J.; Zheng, Y.; Fan, P.; Xu, L. Oil-addicted biodegradation of macro-alkanes in soils with activator. Biochem. Eng. J. 2020, 159, 107578. [Google Scholar] [CrossRef]
- Tang, J.; Wang, R.; Niu, X.; Zhou, Q. Enhancement of soil petroleum remediation by using a combination of ryegrass (Lolium perenne) and different microorganisms. Soil Tillage Res. 2010, 110, 87–93. [Google Scholar] [CrossRef]
- Shahzad, A.; Siddiqui, S.; Bano, A.; Sattar, S.; Hashmi, M.Z.; Qin, M.; Shakoor, A. Hydrocarbon degradation in oily sludge by bacterial consortium assisted with alfalfa (Medicago sativa L.) and maize (Zea mays L.). Arab. J. Geosci. 2020, 13, 1–12. [Google Scholar] [CrossRef]
- Yateem, A.; Balba, M.T.; El-Nawawy, A.S.; Al-Awadhi, N. Plants-associated microflora and the remediation of oil-contaminated soil. Int. J. Phytoremediat. 2008, 2, 183–191. [Google Scholar] [CrossRef]
- Jørgensen, K.S.; Puustinen, J.; Suortti, A.-M. Bioremediation of petroleum hydrocarbon-contaminated soil by composting in biopiles. Environ. Pollut. 2000, 107, 245–254. [Google Scholar] [CrossRef]
- Xu, Y.; Lu, M. Bioremediation of crude oil-contaminated soil: Comparison of different biostimulation and bioaugmentation treatments. J. Hazard. Mater. 2010, 183, 395–401. [Google Scholar] [CrossRef]
- Joo, H.-S.; Ndegwa, P.M.; Shoda, M.; Phae, C.-G. Bioremediation of oil-contaminated soil using Candida catenulata and food waste. Environ. Pollut. 2008, 156, 891–896. [Google Scholar] [CrossRef]
- Schaefer, M.; Petersen, S.O.; Filser, J. Effects of Lumbricus terrestris, Allolobophora chlorotica and Eisenia fetida on microbial community dynamics in oil-contaminated soil. Soil Biol. Biochem. 2005, 37, 2065–2076. [Google Scholar] [CrossRef]
- Luo, C.; Lü, F.; Shao, L.; He, P. Application of eco-compatible biochar in anaerobic digestion to relieve acid stress and promote the selective colonization of functional microbes. Water Res. 2014, 70, 710–718. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, L.; Zhang, X. Bioremediation of petroleum hydrocarbon-contaminated soil by petroleum-degrading bacteria immobilized on biochar. RSC Adv. 2019, 9, 35304–35311. [Google Scholar] [CrossRef] [Green Version]
- Margesin, R.; Schinner, F. Bioremediation (natural attenuation and biostimulation) of diesel-oil-contaminated soil in an alpine glacier skiing area. Appl. Environ. Microbiol. 2001, 67, 3127–3133. [Google Scholar] [CrossRef] [Green Version]
- Kanissery, R.G.; Sims, G.K. Biostimulation for the enhanced degradation of herbicides in soil. Appl. Environ. Soil Sci. 2011, 2011, 988–1027. [Google Scholar] [CrossRef] [Green Version]
- Hazen, T.C. Bioremediation. In Handbook of Hydrocarbon and Lipid Microbiology; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, W.; Wang, Y.; Guo, J.; Hu, X. Nutrient depletion is the main limiting factor in the crude oil bioaugmentation process. J. Environ. Sci. 2021, 100, 317–327. [Google Scholar] [CrossRef]
- Sarkar, J.; Kazy, S.K.; Gupta, A.; Dutta, A.; Mohapatra, B.; Roy, A.; Bera, P.; Mitra, A.; Sar, P. Biostimulation of Indigenous Microbial Community for Bioremediation of Petroleum Refinery Sludge. Front. Microbiol. 2016, 7, 1407. [Google Scholar] [CrossRef] [Green Version]
- Bento, F.M.; Camargo, F.A.O.; Okeke, B.C.; Frankenberger, W.T. Comparative bioremediation of soils contaminated with diesel oil by natural attenuation, biostimulation and bioaugmentation. Bioresour. Technol. 2005, 96, 1049–1055. [Google Scholar] [CrossRef]
- Varjani, S.; Upasani, V.N. Influence of abiotic factors, natural attenuation, bioaugmentation and nutrient supplementation on bioremediation of petroleum crude contaminated agricultural soil. J. Environ. Manag. 2019, 245, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Rehman, K.; Imran, A.; Amin, I.; Afzal, M. Inoculation with bacteria in floating treatment wetlands positively modulates the phytoremediation of oil field wastewater. J. Hazard. Mater. 2018, 349, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Weyens, N.; Lelie, D.V.D.; Taghavi, S.; Newman, L.; Vangronsveld, J. Exploiting plant–microbe partnerships to improve biomass production and remediation. Trends Biotechnol. 2009, 27, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, A.; Sohail, Y.; Thomas, G.R.; Angela, S. The inoculation method affects colonization and performance of bacterial inoculant strains in the phytoremediation of soil contaminated with diesel oil. Int. J. Phytoremediat. 2018, 14, 35–47. [Google Scholar]
- Shehzadi, M.; Afzal, M.; Khan, M.U.; Islam, E.; Mobin, A.; Anwar, S.; Khan, Q.M. Enhanced degradation of textile effluent in constructed wetland system using Typha domingensis and textile effluent-degrading endophytic bacteria. Water Res. 2014, 58, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Kadam, S.K.; Watharkar, A.D.; Chandanshive, V.V.; Khandare, R.V.; Jeon, B.-H.; Jadhav, J.P.; Govindwar, S.P. Co-planted floating phyto-bed along with microbial fuel cell for enhanced textile effluent treatment. J. Clean. Prod. 2018, 203, 788–798. [Google Scholar] [CrossRef]
- Wang, B.; Xie, H.-L.; Ren, H.-Y.; Li, X.; Chen, L.; Wu, B.-C. Application of AHP, TOPSIS, and TFNs to plant selection for phytoremediation of petroleum-contaminated soils in shale gas and oil fields. J. Clean. Prod. 2019, 233, 13–22. [Google Scholar] [CrossRef]
- Cai, B.; Ma, J.; Yan, G.; Dai, X.; Li, M.; Guo, S. Comparison of phytoremediation, bioaugmentation and natural attenuation for remediating saline soil contaminated by heavy crude oil. Biochem. Eng. J. 2016, 112, 170–177. [Google Scholar] [CrossRef]
- Mang, L.; Zhong-Zhi, Z.; Jing-Xiu, W.; Min, Z.; Yu-Xin, X.; Xue-Jiao, W. Interaction of heavy metals and pyrene on their fates in soil and tall fescue (Festuca arundinacea). Environ. Sci. Technol. 2014, 48, 1158–1165. [Google Scholar]
- Wilhantoro, Y.; Lowe, A. Phytoremediation of crude oil contaminated soil: The effect of growth of Glycine max on the physico-chemistry and crude oil contents of soil. Molecules 2015, 20, 22–30. [Google Scholar]
- Cherian, S.; Oliveira, M.M. Transgenic plants in phytoremediation: recent advances and new possibilities. Environ. Sci. Technol. 2005, 39, 9377–9390. [Google Scholar] [CrossRef]
- Abdullah, S.R.S.; Al-Baldawi, I.A.; Almansoory, A.F.; Purwanti, I.F.; Al-Sbani, N.H.; Sharuddin, S.S.N. Plant-assisted remediation of hydrocarbons in water and soil: Application, mechanisms, challenges and opportunities. Chemosphere 2020, 247, 125932. [Google Scholar] [CrossRef]
- Kiamarsi, Z.; Kafi, M.; Soleimani, M.; Nezami, A.; Lutts, S. Conjunction of Vetiveria zizanioides L. and oil-degrading bacteria as a promising technique for remediation of crude oil-contaminated soils. J. Clean. Prod. 2020, 253, 119719. [Google Scholar] [CrossRef]
- Rodríguez, M.D.F.; Gómez, M.C.G.; Blazquez, N.A.; Tarazona, J.V. Soil pollution remediation. Encycl. Toxicol. 2014, 4, 344–355. [Google Scholar]
- Li, X.; Li, J.; Qu, C.; Yu, T.; Du, M. Bioremediation of clay with high oil content and biological response after restoration. Sci. Rep. 2021, 11, 9725. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, P.; Deka, H. Enzymatic defense of Cyperus brevifolius in hydrocarbons stress environment and changes in soil properties. Sci. Rep. 2021, 11, 718. [Google Scholar] [CrossRef] [PubMed]
- Dekang, K.; Hongqi, W.; Zili, L.; Jie, X.; Ying, X. Remediation of Petroleum Hydrocarbon Contaminated Soil by Plant-Microbe and the Change of Rhizosphere Microenvironment. Asian J. Ecotoxicol. 2017, 12, 644–651. [Google Scholar]
- Iffis, B.; St-Arnaud, M.; Hijri, M. Petroleum Contamination and Plant Identity Influence Soil and Root Microbial Communities While AMF Spores Retrieved from the Same Plants Possess Markedly Different Communities. Front. Plant Sci. 2017, 8, 1381. [Google Scholar] [CrossRef]
- Yaashikaaa, P.R.; Senthil Kumar, P.; Varjanic, S.; Saravanan, A. A critical review on the biochar production techniques, characterization, stability and applications for circular bioeconomy. Biotechnol. Rep. 2020, 28, e00570. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, X.; Zhang, P.; Zhao, L.; Qiu, H.; Cao, X. Pyrolysis-temperature depended quinone and carbonyl groups as the electron accepting sites in barley grass derived biochar. Chemosphere 2019, 232, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Nan, H.; Yin, J.; Yang, F.; Luo, Y.; Zhao, L.; Cao, X. Pyrolysis temperature-dependent carbon retention and stability of biochar with participation of calcium: Implications to carbon sequestration. Environ. Pollut. 2021, 287, 117566. [Google Scholar] [CrossRef]
- Bianco, F.; Race, M.; Papirio, S.; Oleszczuk, P.; Esposito, G. The addition of biochar as a sustainable strategy for the remediation of PAH–contaminated sediments. Chemosphere 2021, 263, 128274. [Google Scholar] [CrossRef]
- Iqbal, A.; Mukherjee, M.; Rashid, J.; Khan, S.A.; Ali, M.A.; Arshad, M. Development of plant-microbe phytoremediation system for petroleum hydrocarbon degradation: An insight from alkb gene expression and phytotoxicity analysis. Sci. Total Environ. 2019, 671, 696–704. [Google Scholar] [CrossRef]
- Abedi-Koupai, J.; Vossoughi-Shavari, M.; Yaghmaei, S.; Borghei, M.; Ezzatian, A.R. The effects of microbial population on phytoremediation of petroleum contaminated soils using tall fescue. Int. J. Agric. Biol. 2007, 9, 242–246. [Google Scholar]
- Tang, J.; Wang, R.; Niu, X.; Wang, M.; Zhou, Q. Characterization on the rhizoremediation of petroleum contaminated soil as affected by different influencing factors. Biogeosci. Discuss. 2010, 7, 4665–4688. [Google Scholar]
- Zhang, M.; Guo, P.; Wu, B.; Guo, S. Change in soil ion content and soil water-holding capacity during electro-bioremediation of petroleum contaminated saline soil. J. Hazard. Mater. 2020, 387, 122003. [Google Scholar] [CrossRef] [PubMed]






| Substrates | Microorganisms | Source of Strain | Main Findings | Reference | |||
|---|---|---|---|---|---|---|---|
| Substrate Concentration | Incubation Conditions | Degradation Rate | |||||
| PAHs | bacteria | Achromobacter sp. HZ01 | Petroleum-contaminated seawater, China. | 100 mg/kg anthracene, phenanthrene and pyrene. | 109 cells mL−1/28 °C/150 rpm/30 days. | Strain remove anthracene, phenanthrene and pyrene about 29.8%, 50.6%, and 38.4%, respectively. | [88] |
| Acinetobacter sp. WSD | Petroleum-contaminated groundwater, Shanxi province of northern China. | 1 mg/kg phenanthrene, 2 mg/kg fluorine, and 0.14 mg/kg pyrene. | 5% cells suspension/33 °C/150 rpm/6 days. | Approximately 90% of fluorine, 90% of phenanthrene, and 50% of pyrene were degraded. | [89] | ||
| Bacillus subtilis BMT4i (MTCC 9447) | Automobile contaminated soil, Uttarakhand, India. | 50 g/mL Benzo[a]Pyrene. | 1 × 108 cells mL−1/37 °C/120 rpm/28 days. | Strain started degrading Benzo[a]Pyrene achieving maximum degradation of approximately 84.66%. | [90] | ||
| Caulobacter sp. (T2A12002) | From King Fahd University of Petroleum and Minerals Department of Life Sciences laboratory. | 100 ppm pyrene. | 2% cells suspension/37 °C and 25 °C/120 rpm/18 days/pH 5.0 and pH 9.0. | Strain degraded 35% and 36% of pyrene at 25 °C and 37 °C, respecitvely. | [91] | ||
| Enterobacter sp. (MM087) | Engine-oil-contaminated soil, Puchong and Seri Kembangan, Selangor Malaysia. | 500 mg/L phenanthrene and 250 mg/L pyrene. | 5% cells suspension and 1 × 10⁶cells mL−1/37 ± 0.5 °C/200 rpm/24 h. | Strain with 80.2% degradations for phenanthrene and 59.7% degradations for pyrene. | [92] | ||
| Klebsiella pneumoniae AWD5 | Automobile-contaminated soil, Silchar, Assam. | 0.005% PAH (Pyrene, Chrysene, Benzo(a)pyrene). | Cells(OD600 = 0.4)/30 °C/140 rpm/9 days. | Strain degraded pyrene (56.9%), chrysene (36.5%) and benzo(a)pyrene (50.5%), respectively. | [93] | ||
| Mycobacterium vanbaalenii PYR-1 | Petroleum-contaminated sediment and water, the watershed of Redfish Bay near Port Aransas, Tex. | 0.5 ug/mL pyrene. | 1.5 × 106 cells mL−1/24 °C /150 rpm/48 to 96 h. | After incubation, 47.3 to 52.4% of pyrene was mineralized to CO2. | [94] | ||
| Raoultella planticola | Near a car repair station, Hangzhou, China. | 20 mg L−1 pyrene and 10 mg L−1 benzo[a]pyrene. | 2.0 × 108 cells mL−1/30 °C /180 rpm/10 days. | Strain degraded 52.0% of pyrene and 50.8% of benzo[a]pyrene. | [95] | ||
| Rhodococcus sp. P14 | Petroleum-contaminated sediments, Xiamen, China. | 50 mg/L phenanthrene, pyrene and benzo[a]pyrene. | 1% cells suspension/30°C/150 rpm/30 days. | Strain degraded 34% of the pyrene, about 43% of the phenanthrene and 30% of the Benzo[a]pyrene. | [96] | ||
| Pseudomonas sp. MPDS | PAH- and petrochemical-contaminated soil and mud, Tianjin. | 1 mg/mL naphthalene, 0.1 mg/mL dibenzofuran, 0.1 mg/mL dibenzothiophene, 0.1mg/mL fluorene. | Cells(OD600 = 5.0)/25°C/200 rpm/84 h, 96 h, and 72 h. | Strain could completely degrade naphthalene in 84 h. A total of 65.7% dibenzofuran and 32.1% dibenzothiophene could be degraded in 96 h and 40.3% fluorene could be degraded in 72 h. | [97] | ||
| Pseudoxanthomonas sp. DMVP2 | Petroleum-contaminated sediment, Gujarat, India. | 300 ppm phenanthrene | 4% cells suspension/37 °C/150 rpm/72 h. | Strain was able to degrade 86% phenanthrene. | [98] | ||
| Sphinogmonas sp. | Typical mangrove swamp(surface sediment (0–2 cm)), Ho Chung, Hong Kong. | 5000 mg L−1 phenanthrene. | 180 rpm/7 days. | Strain was obtained to degrade 99.4% phenanthrene at the end of 7 days. | [99] | ||
| Stenotrophomonas sp. IITR87 | — 1 | Phenanthrene(10 ppm), pyrene(10 ppm), and benzo-α-pyrene(10 ppm). | 0.8% cells suspension/30 °C/175 rpm/15 days. | Strain showed >99, 98, and <50% degradation of phenanthrene, pyrene, and benzo-α-pyrene respectively. | [100] | ||
| Streptomyces sp. (ERI-CPDA-1) | Petroleum-contaminated soil, Chennai, India. | Naphthalene(0.1%), phenanthrene(0.1%). | 3% cells suspension/30 °C/200 rpm/7 days. | Strain could remove 99.14% naphthalene and 17.5% phenanthrene. | [101] | ||
| fungus | Aspergillus sp. RFC-1 | Rumaila oilfield(surface polluted sludge (1–10 cm)), Basra, Iraq. | 50 mg/L naphthalene, 20 mg/L phenanthrene, 20 mg/L pyrene. | 10% cells suspension/30 °C/120 rpm/7 days. | Biodegradation efficiencies of crude oil, naphthalene, phenanthrene, and pyrene were 84.6%, 50.3%, and 55.1%, respectively. | [102] | |
| Nocardia sp. H17-1 | Petroleum-contaminated soil | Aliphatic and aromatic (1%, w/v). | 30 °C/6 days. | The aliphatic and aromatic fractions were degraded 99.0 ± 0.1% and 23.8 ± 0.8%, respectively. | [103] | ||
| Penicillium sp. CHY-2 | Soil, Antarctic. | 100 mg L−1 butylbenzene, naphthalene, acenaphthene, ethylbenzene, and benzo[a]pyrene. | 20 °C/110 rpm/28 days. | Strain showed the level of degradation for butylbenzene (42.0%), naphthalene (15.0%), acenaphthene (10.0%), ethylbenzene (4.0%), and benzo[a]pyrene (2.0%). | [104] | ||
| Trichoderma sp. | — 1 | 100 mg kg−1 pyrene and benzo(a)pyrene. | 240 h | Strain degraded 63% of pyrene (100 mg kg−1) and 34% of benzo(a)pyrene (100 mg kg−1) after 240 h of incubation. | [105] | ||
| Fusarium sp. | — 1 | 100 mg kg−1 pyrene and benzo(a)pyrene. | 240 h | Strain degraded 69% of pyrene (100 mg kg−1) and 37% of benzo(a)pyrene (100 mg kg−1) after 240 h of incubation. | [105] | ||
| alkanes | bacteria | Achromobacter sp. HZ01 | Petroleum-contaminated seawater, China. | 2% (w/v) diesel oil | 28 °C/150 rpm/10 days. | Strain degraded the total n-alkanes reached up to 96.6%. | [88] |
| Acinetobacter sp. (KC211013) | Coal chemical industry wastewater treatment plant, northeast China. | 700 mg/L alkanes. | 35 °C | The degradation rate reached 58.7%. | [106] | ||
| Bacillus subtilis | Petroleum-polluted soil, Shengli Oilfield, China. | 0.3% (w/v) crude oil. | 6% cells suspension/30 °C/150 rpm /5 days. | The results indicated that 30–80% of the n-alkanes (C13–C30) were degraded by strain. | [107] | ||
| Pseudomonas sp. WJ6 | Xinjiang oilfield, China. | 0.5% (w/v) n-alkanes. | 1010 CFU mL−1/37 °C/180 rpm/20 days. | N-dodecane (C12) was degraded by 46.65%. 42.62%, 31.69%, and 23.62% of C22, C32, and C40 were degraded, respectively. | [108] | ||
| Rhodococcus sp. | Bay of Quinte, Ontario, Canada. | 0.1% (v/v) diesel fuel. | Cells(OD600 = 0.025)/0 °C/150 rpm/102 days. | After 102 days of incubation at 0 °C, strain mineralized C12 (8%), C16 (6.1%), C28 (1.6%), and C32 (4.3%). | [109] | ||
| fungus | Cladosporium Resinae | Soil, Australian. | 12.5%(v/v) n-alkanes. | 0.75–1.25% cells suspension/35 °C/35 days. | All higher n-alkanes from n-nonane to n-octadecane were assimilated by the fungus. | [110] | |
| Penicillium sp. CHY-2 | Soil, Antarctic. | 100 mg L−1 decane, dodecane and octane. | 20 °C/110 rpm/28 days. | Strain was degraded decane (49.0%), dodecane (33.0%), and octane (8.0%). | [104] | ||
| actinomycetes | Gordonia sp. | Hydrocarbon-contaminated Mediterranean shoreline, west coast of Sicily, Italy. | 1 g L−1 eicosane and octacosane. | 30 °C /28 days. | Eicosane and octacosane were degraded from 53% to 99% in 28 days. | [111] | |
| Tsukamurella sp. MH1 | Petroleum-contaminated soil, Pitești, Romania. | 0.5% (v/v) liquid alkanes. | 30 °C | Strain capable to use a wide range of n-alkanes as the only carbon source for growth. | [112] | ||
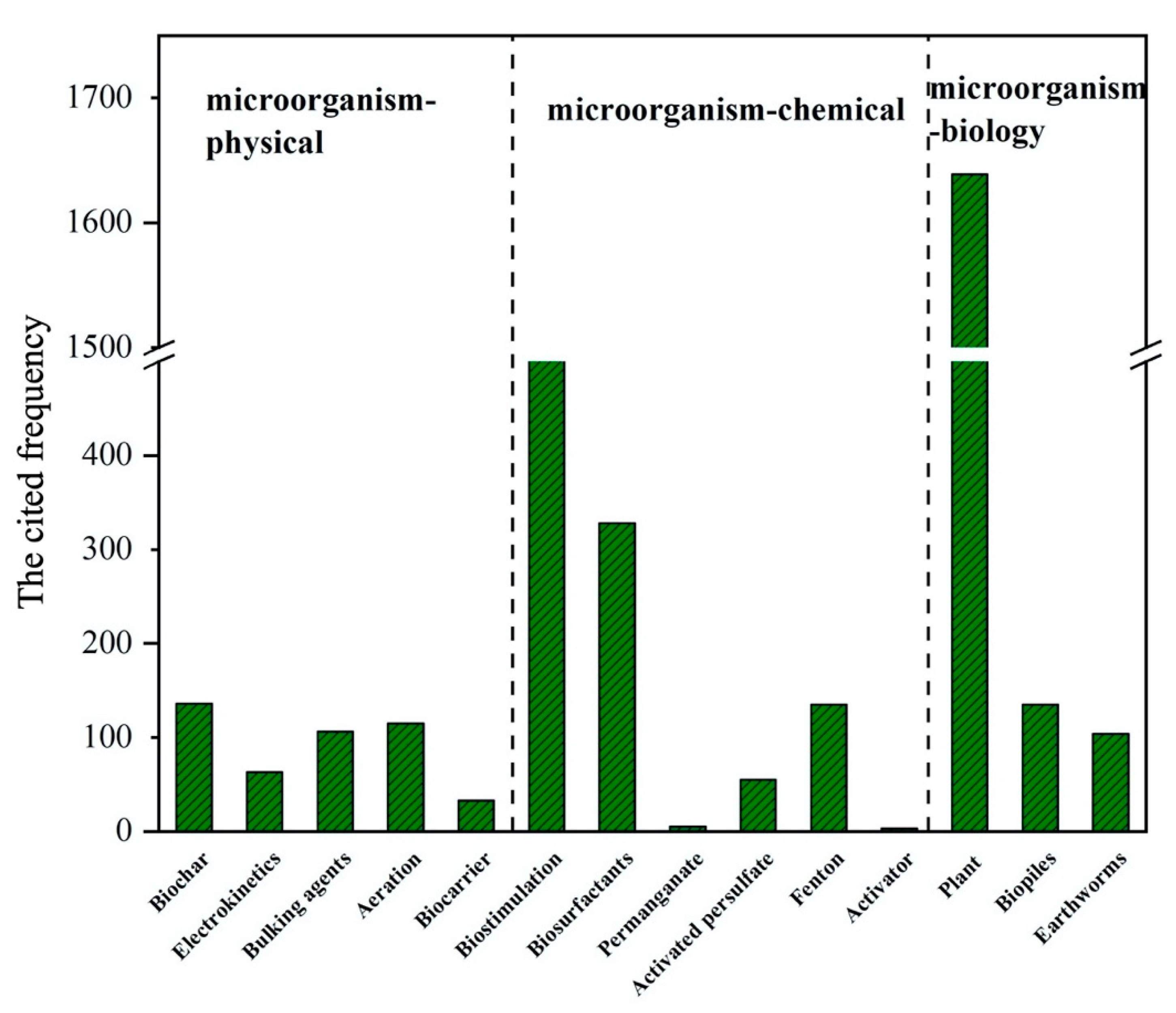
| Methods | Materials | Main Findings | Reference | ||
|---|---|---|---|---|---|
| Substrate Concentration | Incubation Conditions | Degradation Rate | |||
| Microorganism–physical | Biochar (walnut shell biochar (900 °C)/pinewood biochar (900 °C)) | 24,000, 16,000 and 21,000 mg/kg total petroleum hydrocarbons (TPH). | 50 g soil/5% pinewood biochar/C:N:P at 800:13.3:1/25 °C/60 days. | The combined remediation of biochar and fertilizer reduces the TPH in the soil to 10,000 mg/kg (the US EPA clean up standard). | [121] |
| Biochar (rice straw (500 °C)) | 16,300 mg kg−1 TPH (saturated hydrocarbons, 8260 mg kg−1; aromatic hydrocarbons, 5130 mg kg−1; polar components, 2910 mg kg−1). | 1000 g soil/2% (w/w) biochar/60% water holding capacity/C:N:P ratio 100:10:5/80 days. | TPH removal rate was 84.8%. | [124] | |
| electrokinetics | 12,500 mg/kg TPH. | 600 g soil/C:N:P 100:10:1/30 days. | The degradation rate of TPH was 88.3%. | [122] | |
| β-cyclodextrin | 1000 mg/kg PAHs | 1.5, 3.0, 5.0 mmol kg−1 β-cyclodextrin/25 °C. | Compared with the co-metabolism of glucose, the addition of β-cyclodextrin more strongly enhanced oil remediation in soil. | [125] | |
| bulking agents (chopped bermudagrass-hay/sawdust/vermiculite) | 10% TPH | C:N:P 1000:10:1/15–35 °C/12 weeks. | Tillage and adding bulking agents enhanced remediation of oil-contaminated soil. The most rapid rate of remediation occurred during the first 12 weeks, where the TPH decreased 82% and the initial concentration of TPH was 10%. | [126] | |
| aeration (tillage/forced aeration). | |||||
| Biocarrier (activated carbon/zeolite) | 49.81 mg g−1 TPH | 800 g soil/50 g biocarrier + 150 mL planktonic bacterial culture/C:N:P 100:10:1/30 °C/33 days. | Biocarrier enhanced the biodegradation of TPH, with 48.89% removal, compared to natural attenuation with 13.0% removal. | [127] | |
| biostimulation | 19.8 ± 0.38 g kg−1 TPH | 0.8 kg soil/108 cfu g−1 petroleum degrading flora/15% soil moisture/C:N:P 100:10:1/24 °C/12 weeks. | Biostimulation achieved 60% oil hydrocarbon degradation. | [26] | |
| biosurfactants(rhamnolipids) | 47.5 g kg−1 TPH | 500 g soil/7 g of rhamnolipids (dissolved in 1 L deionized water)/500 mL bacterial consortium (in sterile 0.9% NaCl solution)/20% (w/w) moisture content/C:N:P 100:10:1/30 days. | TPH degradation of 77.6% was observed in the soil inoculated with hydrocarbon-degrading bacteria supplemented with rhamnolipids and nutrients. | [123] | |
| permanganate/activated persulfate/modified-Fenton/Fenton | 263.6 ± 73.3 and 385.2 ± 39.6 mg·kg−1Σ16 PAHs. | 50 g soil/the final volume of the Milli-Q water and oxidant was 100 mL/150 rpm/15 days. | The removal efficiency of PAHs was ordered: permanganate (90.0–92.4%) > activated persulfate (81.5–86.54%) > modified Fenton (81.5–85.4%) > Fenton (54.1–60.0%). | [11] | |
| activator (low ammonia and acetic acid) | 29,500 mg kg−1 TPH. | 18–20% moisture content/12 weeks. | Macro-alkanes in soils were efficiently degraded. | [128] | |
| Microorganism–biology | Lolium perenne | 6.19% TPH | 750 g soil/20–30% moisture content/162 days. | The results show that the combination of ryegrass with mixed microbial strains gave the best result with a degradation rate of 58%. | [129] |
| Medicaga sativa | 30% (40% TPH oily sludge)+70% non-pollution soil. | 1 kg soil/N:P 10:1/75–80% moisture content/60 days. | Consortium degraded more than 63% TPH. | [130] | |
| Medicaga sativa/vicia faba/Lolium perenne | 1.13% TPH. | 2 kg soil/18 months. | The TPH degradation in the soil cultivated with broad beans and alfalfa was 36.6% and 35.8%, respectively, compared with 24% degradation in case of ryegrass. | [131] | |
| biopiles (bark chips) | 700 mg kg−1 TPH | soil to bulking agent was approximately 1:3/15–20 °C/5 months. | The TPH content in the pile with oil-contaminated soil decreased with 71%. | [132] | |
| biopiles (peanut hull powder) | 29,500 mg kg−1 TPH | 5 kg of soil/15% w/w peanut hull powder/18–20% moisture content/C:N:P 100:10:1/25–30 °C/12 weeks. | Biodegradation was enhanced with free-living bacterial culture and biocarrier with a TPH removal ranging from 26% to 61%. | [133] | |
| biopiles (food waste) | 2% diesel oil | soil [77% (w/w)] and food waste [23% (w/w)]/C:N 11:1/13 days. | 84% of the TPH was degraded, compared with 48% of removal ratio in control reactor without inoculum. | [134] | |
| earthworms (Eisenia fetida/Allolobophora chlorotica/Lumbricus terrestris) | 10,000 mg kg−1 TPH | 1000 g soil/ten adult worms per container/28 days. | The TPH concentration decreased by 30–42% in samples with L. terrestris, by 31–37% in samples with E. fetida, and by 17–18% in samples with A. chlorotica. | [135] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sui, X.; Wang, X.; Li, Y.; Ji, H. Remediation of Petroleum-Contaminated Soils with Microbial and Microbial Combined Methods: Advances, Mechanisms, and Challenges. Sustainability 2021, 13, 9267. https://doi.org/10.3390/su13169267
Sui X, Wang X, Li Y, Ji H. Remediation of Petroleum-Contaminated Soils with Microbial and Microbial Combined Methods: Advances, Mechanisms, and Challenges. Sustainability. 2021; 13(16):9267. https://doi.org/10.3390/su13169267
Chicago/Turabian StyleSui, Xin, Xuemei Wang, Yuhuan Li, and Hongbing Ji. 2021. "Remediation of Petroleum-Contaminated Soils with Microbial and Microbial Combined Methods: Advances, Mechanisms, and Challenges" Sustainability 13, no. 16: 9267. https://doi.org/10.3390/su13169267
APA StyleSui, X., Wang, X., Li, Y., & Ji, H. (2021). Remediation of Petroleum-Contaminated Soils with Microbial and Microbial Combined Methods: Advances, Mechanisms, and Challenges. Sustainability, 13(16), 9267. https://doi.org/10.3390/su13169267






