Planifilum fulgidum Is the Dominant Functional Microorganism in Compost Containing Spent Mushroom Substrate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Compost Sampling
2.2. Analysis of Physicochemical Properties
2.3. Enzyme Activity Assay
2.4. DNA Extraction and Amplicon Sequencing
2.5. Metaproteomics and Bioinformatics Analyses
2.6. Statistical Analysis
3. Results and Discussion
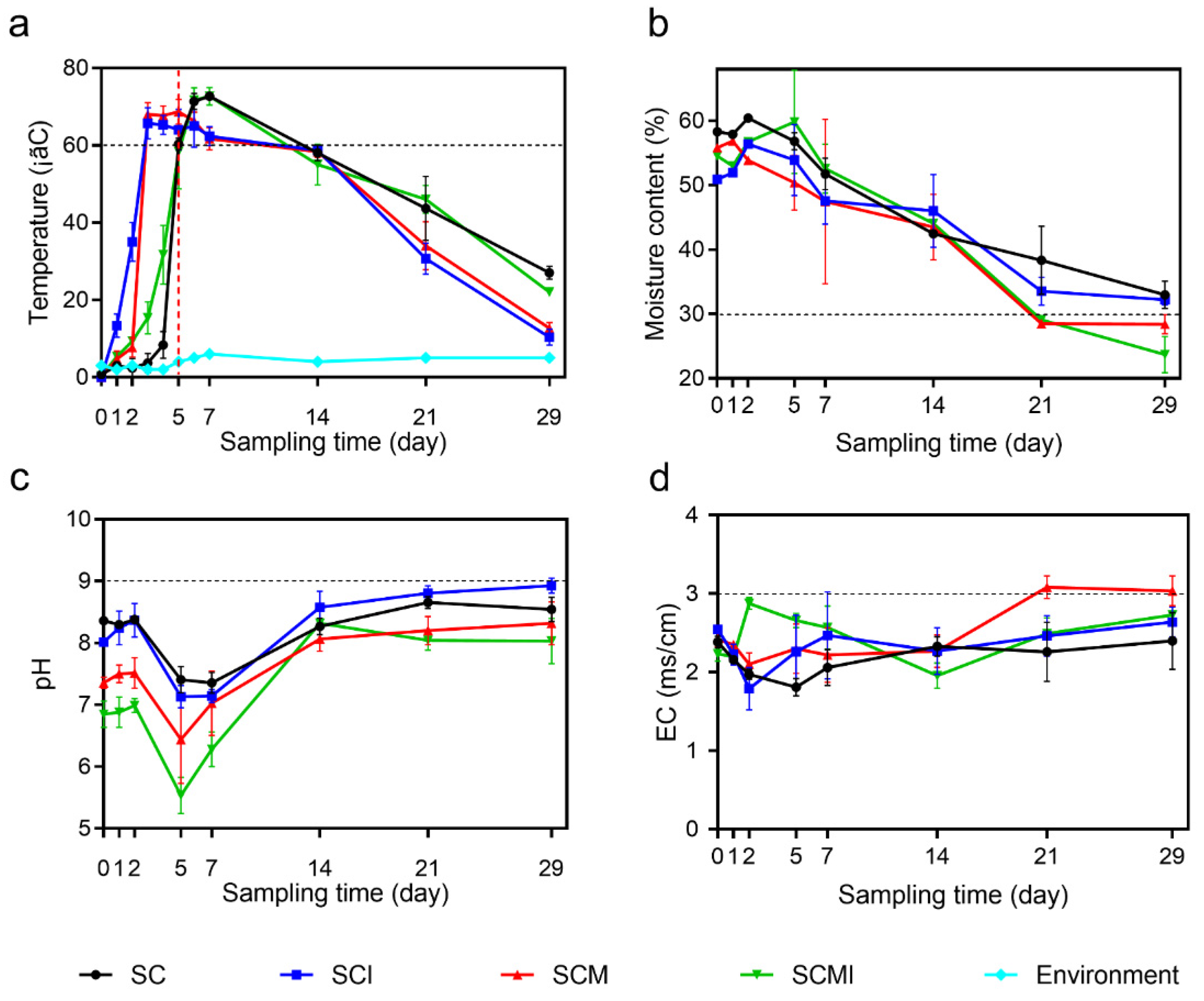
3.1. Physicochemical Properties of the Four Composting Systems
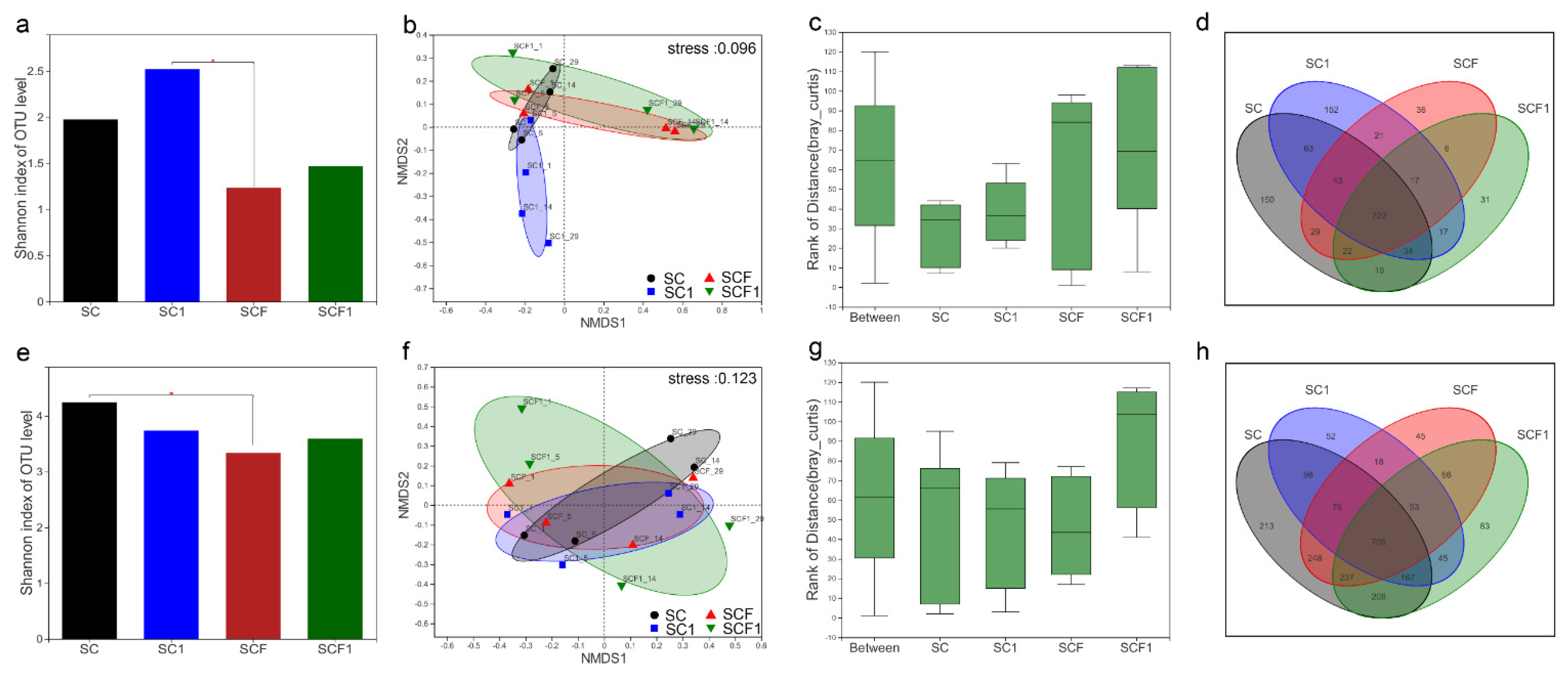
3.2. Characterization of the Microbial Communities
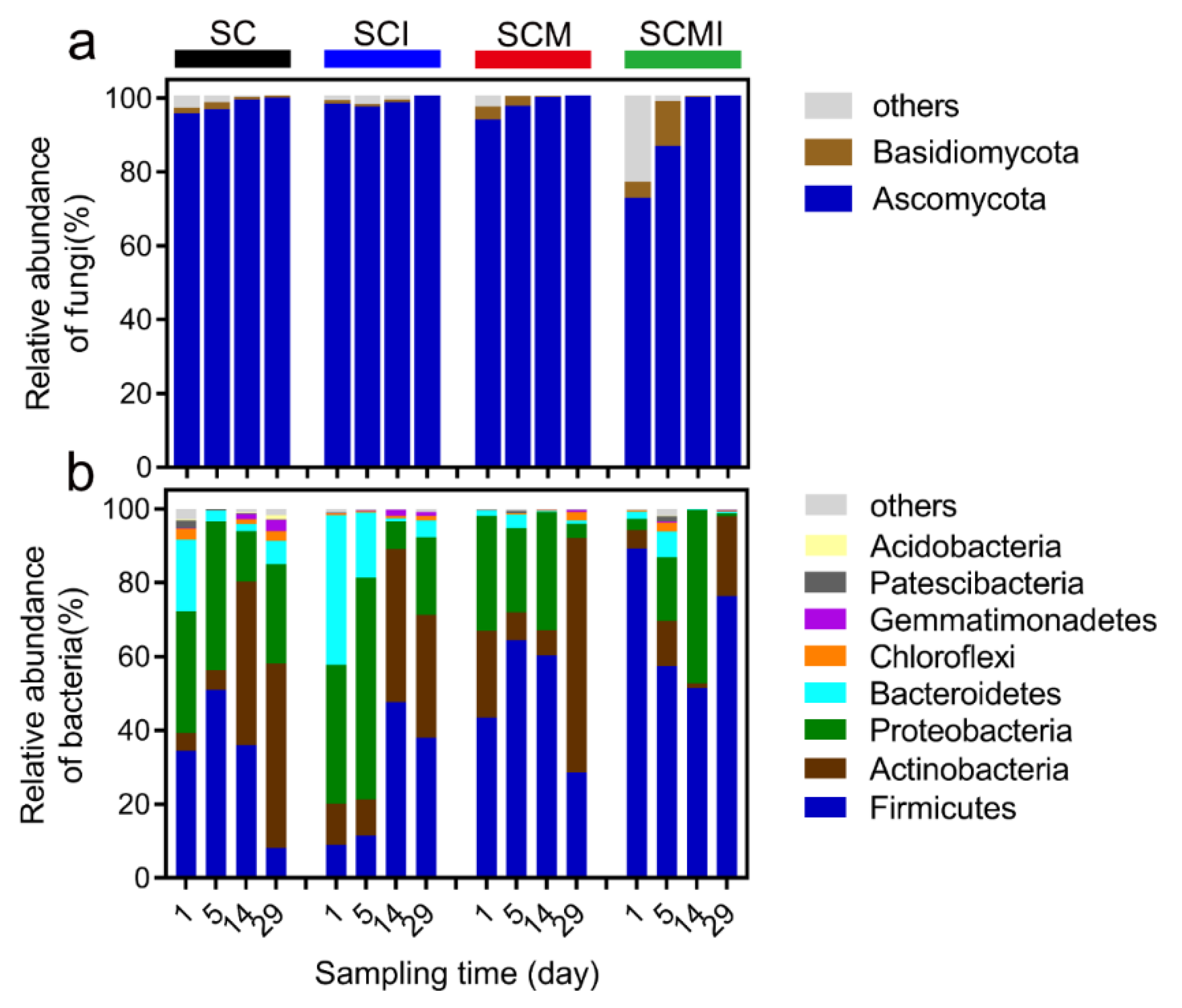
3.3. Composition of the Fungal Communities
3.4. Composition of the Bacterial Communities
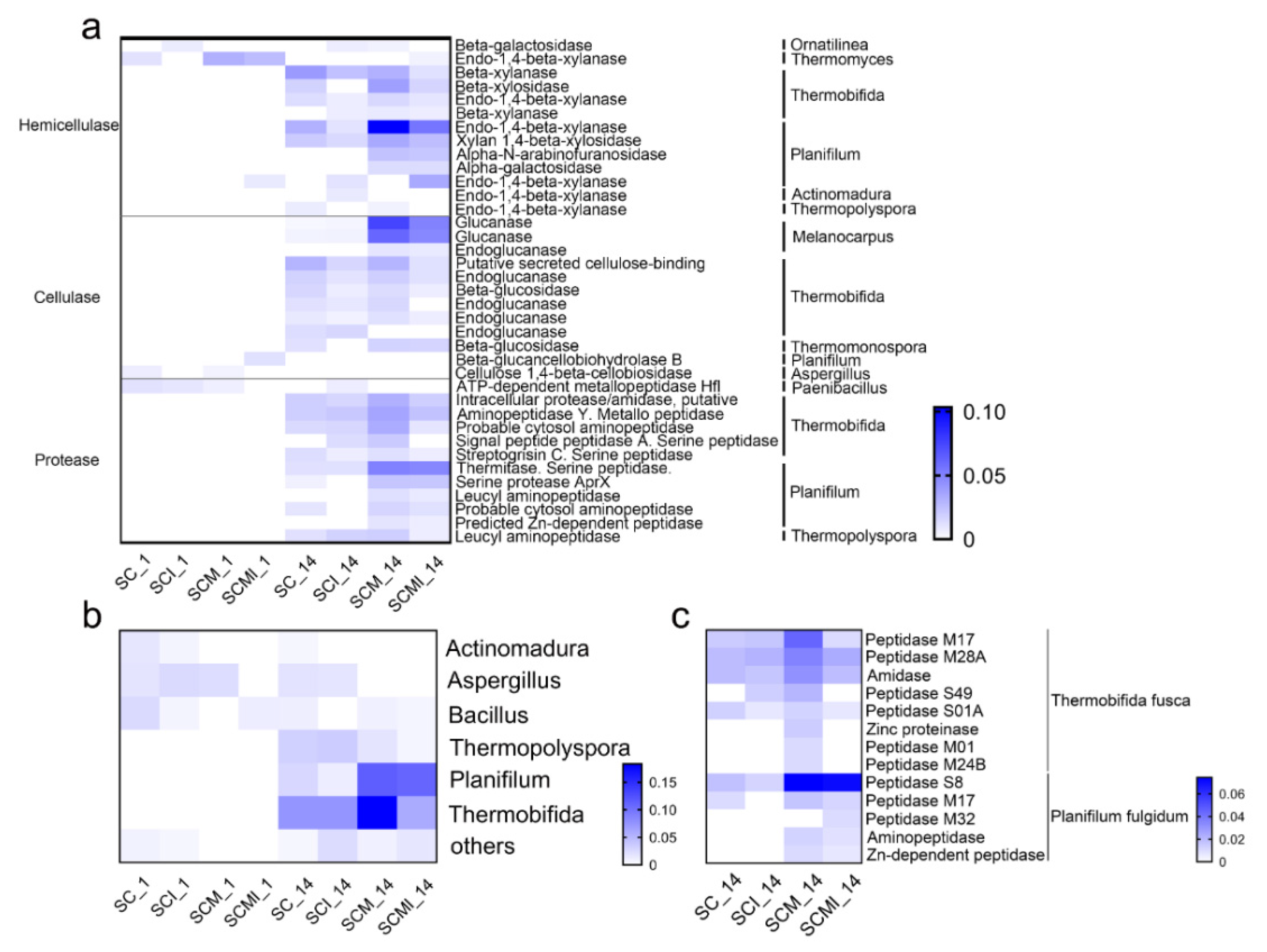
3.5. Metaproteome Analysis of the Dominant Microbial Communities
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, H.J.; Sun, L.F.; Zhang, Y.F.; Zhang, X.L.; Qiao, J.J. Conversion of spent mushroom substrate to biofertilizer using a stress-tolerant phosphate-solubilizing Pichia. farinose. FL7. Bioresour. Technol. 2012, 111, 410–416. [Google Scholar] [CrossRef]
- Finney, K.N.; Ryu, C.; Sharifi, V.N.; Swithenbank, J. The reuse of spent mushroom compost and coal tailings for energy recovery: Comparison of thermal treatment technologies. Bioresour. Technol. 2009, 100, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Wang, X.; Zhen, L.; Gu, J.; Zhang, K.; Wang, Q.; Ma, J.; Peng, H.; Lei, L.; Zhao, W. Effects of inoculating with lignocellulose-degrading consortium on cellulose-degrading genes and fungal community during co-composting of spent mushroom substrate with swine manure. Bioresour. Technol. 2019, 291, 121876. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.L.; Liu, J.Y.; Chen, J.C.; Xie, W.M.; Kuo, J.H.; Lu, X.W.; Chang, K.L.; Wen, S.T.; Sun, G.; Cai, H.M. Combustion behaviors of spent mushroom substrate using TG-MS and TG-FTIR: Thermal conversion, kinetic, thermodynamic and emission analyses. Bioresour. Technol. 2018, 266, 389–397. [Google Scholar] [CrossRef]
- Fang, W.; Zhang, P.Y.; Gou, X.Y.; Zhang, H.B.; Wu, Y.; Ye, J.; Zeng, G.M. Volatile fatty acid production from spent mushroom compost: Effect of total solid content. Int. Biodeterior Biodegrad. 2016, 113, 217–221. [Google Scholar] [CrossRef]
- Lou, Z.M.; Zhu, J.; Wang, Z.X.; Baig, S.A.; Fang, L.; Hu, B.L.; Xu, X.H. Release characteristics and control of nitrogen, phosphate, organic matter from spent mushroom compost amended soil in a column experiment. Process. Saf. Environ. Prot. 2015, 98, 417–423. [Google Scholar] [CrossRef]
- Duan, Y.; Awasthi, S.K.; Liu, T.; Verma, S.; Wang, Q.; Chen, H.; Ren, X.; Zhang, Z.; Awasthi, M.K. Positive impact of biochar alone and combined with bacterial consortium amendment on improvement of bacterial community during cow manure composting. Bioresour. Technol. 2019, 280, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Wang, X.; Cong, C.; Li, J.; Xu, Y.; Li, X.; Hou, F.; Wu, Y.; Wang, L. Effect of inoculating microorganisms in chicken manure composting with maize straw. Bioresour. Technol. 2020, 301, 122730. [Google Scholar] [CrossRef]
- Ren, G.M.; Xu, X.H.; Qu, J.J.; Zhu, L.P.; Wang, T.T. Evaluation of microbial population dynamics in the co-composting of cow manure and rice straw using high throughput sequencing analysis. World J. Microb. Biot. 2016, 32, 1–11. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, J.; Sui, Q.; Wan, H.; Tong, J.; Chen, M.; Wei, Y.; Wei, D. Effect of red mud addition on tetracycline and copper resistance genes and microbial community during the full scale swine manure composting. Bioresour. Technol. 2016, 216, 1049–1057. [Google Scholar] [CrossRef]
- Meng, L.; Li, W.; Zhang, S.; Wu, C.; Lv, L. Feasibility of co-composting of sewage sludge, spent mushroom substrate and wheat straw. Bioresour. Technol. 2017, 226, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Uppal, H.S.; Singh, R.; Beri, S.; Mohan, K.S.; Gupta, V.C.; Adholeya, A. Co-composting of physic nut (Jatropha curcas) deoiled cake with rice straw and different animal dung. Bioresour. Technol. 2011, 102, 6541–6546. [Google Scholar] [CrossRef] [PubMed]
- Paredes, C.; Bernal, M.P.; Cegarra, J.; Roig, A. Bio-degradation of olive mill wastewater sludge by its co-composting with agricultural wastes. Bioresour. Technol. 2002, 85, 1–8. [Google Scholar] [CrossRef]
- Zhang, L.L.; Ma, H.X.; Zhang, H.Q.; Xun, L.Y.; Chen, G.J.; Wang, L.S. Thermomyces lanuginosus is the dominant fungus in maize straw composts. Bioresour. Technol. 2015, 197, 266–275. [Google Scholar] [CrossRef]
- Fendrihan, S.; Pop, C.E. Biotechnological potential of plant associated microorganisms. Rom. Biotechnol. Lett. 2021, 26, 2700–2706. [Google Scholar] [CrossRef]
- Lyu, D.; Backer, R.; Subramanian, S.; Smith, D.L. Phytomicrobiome Coordination Signals Hold Potential for Climate Change-Resilient Agriculture. Front. Plant. Sci. 2020, 11, 634. [Google Scholar] [CrossRef] [PubMed]
- Shubina, V.; Birsa, M.; Burtseva, S. Antifungal Activity of Bacteria of the Genus Bacillus and Streptomyces isolated from the Soil of the Republic of Moldova. Mikrobiolohichnyi Zhurnal 2018, 80, 41–53. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Wang, L.; Zhu, K.; Hou, S.; Chen, L.; Mi, D.; Gui, Y.; Qi, Y.; Jiang, C.; Guo, J.H. Plant Root Exudates Are Involved in Bacillus cereus AR156 Mediated Biocontrol Against Ralstonia solanacearum. Front. Microbiol. 2019, 10, 98. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.; Liu, B.; Xi, C.; Luo, X.; Yuan, X.; Wang, X.; Zhu, W.; Wang, H.; Cui, Z. Effect of pig manure on the chemical composition and microbial diversity during co-composting with spent mushroom substrate and rice husks. Bioresour. Technol. 2018, 251, 22–30. [Google Scholar] [CrossRef]
- Wu, D.; Wei, Z.; Gao, X.; Wu, J.; Chen, X.; Zhao, Y.; Jia, L.; Wen, D. Reconstruction of core microbes based on producing lignocellulolytic enzymes causing by bacterial inoculation during rice straw composting. Bioresour. Technol. 2020, 315, 123849. [Google Scholar] [CrossRef]
- Xiao, Z.; Lin, M.H.; Fan, J.L.; Chen, Y.X.; Zhao, C.; Liu, B. Anaerobic digestion of spent mushroom substrate under thermophilic conditions: Performance and microbial community analysis. Appl. Microbiol. Biot. 2018, 102, 499–507. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, H.; Wang, Z.; Chen, G.; Wang, L. Dynamic changes of the dominant functioning microbial community in the compost of a 90-m(3) aerobic solid state fermentor revealed by integrated meta-omics. Bioresour. Technol. 2016, 203, 1–10. [Google Scholar] [CrossRef]
- Chi, C.P.; Chu, S.; Wang, B.; Zhang, D.; Zhi, Y.; Yang, X.; Zhou, P. Dynamic bacterial assembly driven by Streptomyces. griseorubens. JSD-1 inoculants correspond to composting performance in swine manure and rice straw co-composting. Bioresour. Technol. 2020, 313, 123692. [Google Scholar] [CrossRef]
- Yang, X.; Hu, Q.; Han, Z.; Ruan, X.; Jiang, S.; Chai, J.; Zheng, R. Effects of exogenous microbial inoculum on the structure and dynamics of bacterial communities in swine carcass composting. Can. J. Microbiol. 2018, 64, 1042–1053. [Google Scholar] [CrossRef]
- Wei, Y.; Wu, D.; Wei, D.; Zhao, Y.; Wu, J.; Xie, X.; Zhang, R.; Wei, Z. Improved lignocellulose-degrading performance during straw composting from diverse sources with actinomycetes inoculation by regulating the key enzyme activities. Bioresour. Technol. 2019, 271, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Lv, Z.; Sun, H.; Li, R.; Zhai, B.; Wang, Z.; Awasthi, M.K.; Wang, Q.; Zhou, L. Improvement of biochar and bacterial powder addition on gaseous emission and bacterial community in pig manure compost. Bioresour. Technol. 2018, 258, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, Y.; Chen, Y.; Lu, Q.; Li, M.; Wang, X.; Wei, Y.; Xie, X.; Wei, Z. A regulating method for reducing nitrogen loss based on enriched ammonia-oxidizing bacteria during composting. Bioresour. Technol. 2016, 221, 276–283. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, W.; Chen, L.; Meng, L.; Zheng, Z. Effect of enriched thermotolerant nitrifying bacteria inoculation on reducing nitrogen loss during sewage sludge composting. Bioresour. Technol. 2020, 311, 123461. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Liu, X.; Huang, Y.; Huang, H. Inoculation with nitrogen turnover bacterial agent appropriately increasing nitrogen and promoting maturity in pig manure composting. Waste Manag. 2015, 39, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Ballardo, C.; Vargas-Garcia, M.D.C.; Sanchez, A.; Barrena, R.; Artola, A. Adding value to home compost: Biopesticide properties through Bacillus. thuringiensis. inoculation. Waste Manag. 2020, 106, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.K.; Richard, T.L.; Glanville, T.D. Laboratory determination of compost physical parameters for modeling of airflow characteristics. Waste Manag. 2008, 28, 660–670. [Google Scholar] [CrossRef] [PubMed]
- Kwaansa-Ansah, E.E.; Voegborlo, R.B.; Adimado, A.A.; Ephraim, J.H.; Nriagu, J.O. Effect of pH, sulphate concentration and total organic carbon on mercury accumulation in sediments in the Volta Lake at Yeji, Ghana. Bull. Environ. Contam. Toxicol. 2012, 88, 418–421. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sun, X. Changes in physical, chemical, and microbiological properties during the two-stage co-composting of green waste with spent mushroom compost and biochar. Bioresour. Technol. 2014, 171, 274–284. [Google Scholar] [CrossRef]
- Xing, S.; Li, G.; Sun, X.; Ma, S.; Chen, G.; Wang, L.; Gao, P. Dynamic changes in xylanases and beta-1,4-endoglucanases secreted by Aspergillus. niger. An-76 in response to hydrolysates of lignocellulose polysaccharide. Appl. Biochem. Biotechnol. 2013, 171, 832–846. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, N.; Yang, F.; Li, J.; Wang, L.; Chen, G.; Gao, P. In situ demonstration and quantitative analysis of the intrinsic properties of glycoside hydrolases. Electrophoresis 2012, 33, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Tsujii, M.; Kawano, S.; DuBois, R.N. Cyclooxygenase-2 expression in human colon cancer cells increases metastatic potential. Proc. Natl. Acad. Sci. USA 1997, 94, 3336–3340. [Google Scholar] [CrossRef] [Green Version]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.L.; Li, L.J.; Sha, G.M.; Liu, C.X.; Wang, Z.H.; Wang, L.S. Aerobic composting as an effective cow manure management strategy for reducing the dissemination of antibiotic resistance genes: An integrated meta-omics study. J. Hazard. Mater. 2020, 386. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, X.Y.; Tian, Y.; Gong, X.Q. Effects of brown sugar and calcium superphosphate on the secondary fermentation of green waste. Bioresour. Technol. 2013, 131, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.P.; Chen, Y.X.; Shaaban, M.; Zhu, D.W.; Hu, C.X.; Chen, Z.B.; Wang, Y. Evaluation of microbial inoculants pretreatment in straw and manure co-composting process enhancement. J. Clean. Prod. 2019, 239, 118078. [Google Scholar] [CrossRef]
- Xie, X.Y.; Zhao, Y.; Sun, Q.H.; Wang, X.Q.; Cui, H.Y.; Zhang, X.; Li, Y.J.; Wei, Z.M. A novel method for contributing to composting start-up at low temperature by inoculating cold-adapted microbial consortium. Bioresour. Technol. 2017, 238, 39–47. [Google Scholar] [CrossRef]
- Li, S.Y.; Li, D.Y.; Li, J.J.; Li, Y.Y.; Li, G.X.; Zang, B.; Li, Y. Effect of spent mushroom substrate as a bulking agent on gaseous emissions and compost quality during pig manure composting. Environ. Sci. Pollut. R. 2018, 25, 12398–12406. [Google Scholar] [CrossRef]
- Gong, X.; Li, S.; Carson, M.A.; Chang, S.X.; Wu, Q.; Wang, L.; An, Z.; Sun, X. Spent mushroom substrate and cattle manure amendments enhance the transformation of garden waste into vermicomposts using the earthworm Eisenia fetida. J. Environ. Manag. 2019, 248, 109263. [Google Scholar] [CrossRef]
- Hanc, A.; Chadimova, Z. Nutrient recovery from apple pomace waste by vermicomposting technology. Bioresour. Technol. 2014, 168, 240–244. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, X. Influence of bulking agents on physical, chemical, and microbiological properties during the two-stage composting of green waste. Waste Manag. 2016, 48, 115–126. [Google Scholar] [CrossRef]
- Perez-Godinez, E.A.; Lagunes-Zarate, J.; Corona-Hernandez, J.; Barajas-Aceves, M. Growth and reproductive potential of Eisenia. foetida. (Sav) on various zoo animal dungs after two methods of pre-composting followed by vermicomposting. Waste Manag. 2017, 64, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Lasaridi, K.; Protopapa, I.; Kotsou, M.; Pilidis, G.; Manios, T.; Kyriacou, A. Quality assessment of composts in the Greek market: The need for standards and quality assurance. J. Environ. Manage. 2006, 80, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.C.; Li, Y.C.; Gao, Y.F.; Yin, M.Q.; Wu, Y.X.; Liu, L.L.; Du, N.; Liu, J.; Yu, X.N.; Wang, L.S. Insight into the effect of nitrogen-rich substrates on the community structure and the co-occurrence network of thermophiles during lignocellulose-based composting. Bioresour. Technol. 2021, 319. [Google Scholar] [CrossRef]
- Bello, A.; Han, Y.; Zhu, H.; Deng, L.; Yang, W.; Meng, Q.; Sun, Y.; Egbeagu, U.U.; Sheng, S.; Wu, X. Microbial community composition, co-occurrence network pattern and nitrogen transformation genera response to biochar addition in cattle manure-maize straw composting. Sci. Total Environ. 2020, 721, 137759. [Google Scholar] [CrossRef]
- Zhang, J.; Zeng, G.; Chen, Y.; Yu, M.; Yu, Z.; Li, H.; Yu, Y.; Huang, H. Effects of physico-chemical parameters on the bacterial and fungal communities during agricultural waste composting. Bioresour. Technol. 2011, 102, 2950–2956. [Google Scholar] [CrossRef] [PubMed]
- Steger, K.; Jarvis, A.; Vasara, T.; Romantschuk, M.; Sundh, I. Effects of differing temperature management on development of Actinobacteria populations during composting. Res. Microbiol. 2007, 158, 617–624. [Google Scholar] [CrossRef]
- Nakasaki, K.; Hirai, H. Temperature control strategy to enhance the activity of yeast inoculated into compost raw material for accelerated composting. Waste Manag. 2017, 65, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Neher, D.A.; Weicht, T.R.; Bates, S.T.; Leff, J.W.; Fierer, N. Changes in bacterial and fungal communities across compost recipes, preparation methods, and composting times. PLoS. ONE 2013, 8, e79512. [Google Scholar]
- Jiang, X.; Deng, L.; Meng, Q.; Sun, Y.; Han, Y.; Wu, X.; Sheng, S.; Zhu, H.; Ayodeji, B.; Egbeagu, U.U. Fungal community succession under influence of biochar in cow manure composting. Environ. Sci. Pollut. Res. Int. 2020, 27, 9658–9668. [Google Scholar] [CrossRef]
- Tian, X.; Yang, T.; He, J.; Chu, Q.; Jia, X.; Huang, J. Fungal community and cellulose-degrading genes in the composting process of Chinese medicinal herbal residues. Bioresour. Technol. 2017, 241, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yin, X.; Mao, H.; Chu, C.; Tian, Y. Changes in structure and function of fungal community in cow manure composting. Bioresour. Technol. 2018, 255, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Narang, S.; Sahai, V.; Bisaria, V.S. Optimization of xylanase production by Melanocarpus albomyces IIS68 in solid state fermentation using response surface methodology. J. Biosci. Bioeng. 2001, 91, 425–427. [Google Scholar] [CrossRef]
- Li, P.; Liu, M.; Ma, X.; Wu, M.; Jiang, C.; Liu, K.; Liu, J.; Li, Z. Responses of microbial communities to a gradient of pig manure amendment in red paddy soils. Sci. Total Environ. 2020, 705, 135884. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Xu, X.; Zhang, W.; Men, M.; Xu, B.; Deng, L.; Bello, A.; Jiang, X.; Sheng, S.; Wu, X. Bacterial community succession in dairy manure composting with a static composting technique. Can. J. Microbiol. 2019, 65, 436–449. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Wei, D.; An, Z.; Zhang, C.; Jin, L.; Wang, L.; Li, Y.; Li, Q. Succession of the bacterial community structure and functional prediction in two composting systems viewed through metatranscriptomics. Bioresour. Technol. 2020, 313, 123688. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Pu, J.W.; Liu, X.M.; Wu, Y.Y. Characterization of the alkali-tolerant thermostable enzyme from Saccharomonospora Viridis and its application in pulp bleaching. Cell. Chem. Technol. 2008, 42, 363–369. [Google Scholar]
- Storey, S.; Chualain, D.N.; Doyle, O.; Clipson, N.; Doyle, E. Comparison of bacterial succession in green waste composts amended with inorganic fertiliser and wastewater treatment plant sludge. Bioresour. Technol. 2015, 179, 71–77. [Google Scholar] [CrossRef]
- Zhu, P.; Qin, H.; Zhang, H.; Luo, Y.; Ru, Y.; Li, J.; San, K.W.; Wang, L.; Yu, X.; Guo, W. Variations in antibiotic resistance genes and removal mechanisms induced by C/N ratio of substrate during composting. Sci. Total Environ. 2021, 798, 149288. [Google Scholar] [CrossRef]
- Shi, Z.; Han, C.; Zhang, X.; Tian, L.; Wang, L. Novel Synergistic Mechanism for Lignocellulose Degradation by a Thermophilic Filamentous Fungus and a Thermophilic Actinobacterium Based on Functional Proteomics. Front. Microbiol. 2020, 11, 539438. [Google Scholar] [CrossRef]
- Makarov, A.; Denisov, E.; Lange, O.; Horning, S. Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J. Am. Soc. Mass Spectrom. 2006, 17, 977–982. [Google Scholar] [CrossRef] [Green Version]
- Saykhedkar, S.; Ray, A.; Ayoubi-Canaan, P.; Hartson, S.D.; Prade, R.; Mort, A.J. A time course analysis of the extracellular proteome of Aspergillus nidulans growing on sorghum stover. Biotechnol. Biofuels 2012, 5, 52. [Google Scholar] [CrossRef] [Green Version]
- Wilson, D.B. Studies of Thermobifida fusca plant cell wall degrading enzymes. Chem. Rec. 2004, 4, 72–82. [Google Scholar] [CrossRef]
- Zhang, L.; Li, L.; Pan, X.; Shi, Z.; Feng, X.; Gong, B.; Li, J.; Wang, L. Enhanced Growth and Activities of the Dominant Functional Microbiota of Chicken Manure Composts in the Presence of Maize Straw. Front. Microbiol. 2018, 9, 1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| pile | Time | Fungi | Bacteria | ||||
|---|---|---|---|---|---|---|---|
| Seq-num | OTUs | Chao1 | Seq-num | OTUs | Chao1 | ||
| SC | 1 d | 71,756 | 448 | 517 | 50,990 | 1653 | 1808 |
| 5 d | 58,897 | 432 | 437 | 55,303 | 742 | 1119 | |
| 14 d | 60,693 | 269 | 286 | 46,654 | 660 | 905 | |
| 29 d | 61,284 | 147 | 155 | 52,092 | 943 | 1097 | |
| SCI | 1 d | 73,566 | 430 | 425 | 61,831 | 659 | 856 |
| 5 d | 74,619 | 375 | 385 | 56,385 | 750 | 995 | |
| 14 d | 74,243 | 274 | 284 | 59,454 | 584 | 647 | |
| 29 d | 74,055 | 222 | 217 | 71,910 | 801 | 883 | |
| SCM | 1 d | 34,452 | 245 | 289 | 72,962 | 584 | 893 |
| 5 d | 64,908 | 341 | 385 | 63,189 | 1222 | 1493 | |
| 14 d | 43,927 | 149 | 170 | 54,033 | 379 | 522 | |
| 29 d | 57,564 | 89 | 115 | 71,100 | 540 | 696 | |
| SCMI | 1 d | 34,607 | 203 | 205 | 71,239 | 777 | 907 |
| 5 d | 74,430 | 352 | 397 | 70,632 | 1552 | 1798 | |
| 14 d | 71,465 | 97 | 94 | 73,948 | 309 | 375 | |
| 29 d | 70,014 | 17 | 12 | 73,057 | 327 | 417 | |
| Total | 1,000,480 | 4090 | — | 1,004,779 | 12,482 | — | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Wang, W.; Li, Z.; Yang, C.; Liang, S.; Wang, L. Planifilum fulgidum Is the Dominant Functional Microorganism in Compost Containing Spent Mushroom Substrate. Sustainability 2021, 13, 10002. https://doi.org/10.3390/su131810002
Zhang H, Wang W, Li Z, Yang C, Liang S, Wang L. Planifilum fulgidum Is the Dominant Functional Microorganism in Compost Containing Spent Mushroom Substrate. Sustainability. 2021; 13(18):10002. https://doi.org/10.3390/su131810002
Chicago/Turabian StyleZhang, Hong, Wenying Wang, Zaixue Li, Chuanlun Yang, Shuang Liang, and Lushan Wang. 2021. "Planifilum fulgidum Is the Dominant Functional Microorganism in Compost Containing Spent Mushroom Substrate" Sustainability 13, no. 18: 10002. https://doi.org/10.3390/su131810002





