Transcriptomic Insight into the Melon Morphology of Toothed Whales for Aquatic Molecular Developments
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. RNA Isolation, Library Preparation, and Sequencing
2.3. Bioinformatic Analysis
3. Results
3.1. Gene Clustering and Functional Enrichments
3.2. Differentially Expressed Genes (DEGs) and Functional Enrichments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reidenberg, J.S. Anatomical adaptations of aquatic mammals. Anat. Rec. 2007, 290, 507–513. [Google Scholar] [CrossRef]
- Uhen, M.D. The Origin(s) of Whales. Annu. Rev. Earth Planet. Sci. 2010, 38, 189–219. [Google Scholar] [CrossRef]
- Geisler, J.H.; McGowen, M.R.; Yang, G.; Gatesy, J. A supermatrix analysis of genomic, morphological, and paleontological data from crown Cetacea. BMC Evol. Biol. 2011, 11, 112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hooker, S.K. Toothed Whales, Overview. In Encyclopedia of Marine Mammals; Academic Press 30 Corporate Drive: Burlington, MA, USA, 2009; ISBN 9780123735539. [Google Scholar]
- Lonati, G.L.; Westgate, A.J.; Pabst, D.A.; Koopman, H.N. Nitrogen solubility in odontocete blubber and mandibular fats in relation to lipid composition. J. Exp. Biol. 2015, 218, 2620–2630. [Google Scholar] [CrossRef] [Green Version]
- Harper, C.J.; McLellan, W.A.; Rommel, S.A.; Gay, D.M.; Dillaman, R.M.; Pabst, D.A. Morphology of the melon and its tendinous connections to the facial muscles in bottlenose dolphins (Tursiops truncatus). J. Morphol. 2008, 269, 820–839. [Google Scholar] [CrossRef] [Green Version]
- Harper, C. Morphology of the Melon and Its Tendinous Connections to the Facial Muscles in Bottlenose Dolphins (Tursiops Truncatus). Master’s Thesis, University of North Carolina Wilmington, Wilmington, NC, USA, 2007. Available online: https://core.ac.uk/download/pdf/149230312.pdf (accessed on 1 December 2021).
- Mckenna, M.F.; Cranford, T.W.; Berta, A.; Pyenson, N.D. Morphology of the odontocete melon and its implications for acoustic function. Mar. Mammal Sci. 2012, 28, 690–713. [Google Scholar] [CrossRef]
- Karol, R.; Litchfield, C.; Caldwell, D.K.; Caldwell, M.C. Compositional topography of melon and spermaceti organ lipids in the pygmy sperm whale Kogia breviceps: Implications for echolocation. Mar. Biol. 1978, 47, 115–123. [Google Scholar] [CrossRef]
- Litchfield, C.; Ackman, R.G.; Sipos, J.C.; Eaton, C.A. Isovaleroyl triglycerides from the blubber and melon oils of the beluga whale (Delphinapterus leucas). Lipids 1971, 6, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Duggan, Z.P.Z.; Koopman, H.N.; Budge, S.M. Distribution and development of the highly specialized lipids in the sound reception systems of dolphins. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2009, 179, 783–798. [Google Scholar] [CrossRef]
- Jung, J.L.; Simon, G.; Alfonsi, E.; Thoraval, D.; Kervarec, N.; Ben Salem, D.; Hassani, S.; Domergue, F. Qualitative and quantitative study of the highly specialized lipid tissues of cetaceans using HR-MAS NMR and classical GC. PLoS ONE 2017, 12, e0180597. [Google Scholar] [CrossRef] [PubMed]
- Koopman, H.N. Function and evolution of specialized endogenous lipids in toothed whales. J. Exp. Biol. 2018, 221, jeb161471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivarson, E.; Iven, T.; Sturtevant, D.; Ahlman, A.; Cai, Y.; Chapman, K.; Feussner, I.; Zhu, L.H. Production of wax esters in the wild oil species Lepidium campestre. Ind. Crops Prod. 2017, 108, 535–542. [Google Scholar] [CrossRef]
- Morii, H.; Kaneda, T. Biosynthesis of branched-chain fatty acids from branched-chain amino acids in subcutaneous tissue of the marine little toothed whale, Stenella caeruleo-alba. Comp. Biochem. Physiol.—Part B Biochem. 1982, 71, 357–365. [Google Scholar] [CrossRef]
- Tandon, P.; Wafer, R.; Minchin, J.E.N. Adipose morphology and metabolic disease. J. Exp. Biol. 2018, 221, jeb164970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uslu, V.V.; Petretich, M.; Ruf, S.; Langenfeld, K.; Fonseca, N.A.; Marioni, J.C.; Spitz, F. Long-range enhancers regulating Myc expression are required for normal facial morphogenesis. Nat. Genet. 2014, 46, 753–758. [Google Scholar] [CrossRef] [PubMed]
- Viguerie, N.; Montastier, E.; Maoret, J.J.; Roussel, B.; Combes, M.; Valle, C.; Villa-Vialaneix, N.; Iacovoni, J.S.; Martinez, J.A.; Holst, C.; et al. Determinants of Human Adipose Tissue Gene Expression: Impact of Diet, Sex, Metabolic Status, and Cis Genetic Regulation. PLoS Genet. 2012, 8, e1002959. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.C.; Aizen, J.; Fitzgibbon, Q.P.; Elizur, A.; Ventura, T. Applying the power of transcriptomics: Understanding male sexual development in decapod crustacea. Integr. Comp. Biol. 2016, 56, 1144–1156. [Google Scholar] [CrossRef] [PubMed]
- Dodson, M.V.; Ramayo-Caldas, Y.; Puig-Oliveras, A.; Estellé, J.; Castelló, A.; Alves, E.; Pena, R.N.; Ballester, M.; Folch, J.M.; Dodson, M.; et al. Lipid metabolism, adipocyte depot physiology and utilization of meat animals as experimental models for metabolic research. Int. J. Biol. Sci. 2010, 14, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Khudyakov, J.I.; Champagne, C.D.; Meneghetti, L.M.; Crocker, D.E. Blubber transcriptome response to acute stress axis activation involves transient changes in adipogenesis and lipolysis in a fasting-adapted marine mammal. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Pintado, T.; Delgado-Pando, G. Towards more sustainable meat products: Extenders as a way of reducing meat content. Foods 2020, 9, 1044. [Google Scholar] [CrossRef]
- Russo, G.L.; Langellotti, A.L.; Oliviero, M.; Sacchi, R.; Masi, P. Sustainable production of food grade omega-3 oil using aquatic protists: Reliability and future horizons. New Biotechnol. 2021, 62, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Wendel, A.A.; Lewin, T.M.; Coleman, R.A. GPATs: Rate limiting enzymes of TAG biosynthesis. North 2010, 1791, 501–506. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, D.; Yin, Y.; Ji, M.; Xu, K.; Huang, X.; Peng, Y.; Zhang, J. Comprehensive transcriptomic view of the role of the LGALS12 gene in porcine subcutaneous and intramuscular adipocytes. BMC Genom. 2019, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hua, N.; Takahashi, H.; Yee, G.M.; Kitajima, Y.; Katagiri, S.; Kojima, M.; Anzai, K.; Eguchi, Y.; Hamilton, J.A. Influence of muscle fiber type composition on early fat accumulation under high-fat diet challenge. PLoS ONE 2017, 12, e0182430. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Mo, D.; Li, A.; Gong, W.; Xiao, S.; Zhang, Y.; Qin, L.; Niu, Y.; Guo, Y.; Liu, X.; et al. Comparative analyses by sequencing of transcriptomes during skeletal muscle development between pig breeds differing in muscle growth rate and fatness. PLoS ONE 2011, 6, e19774. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Viaud, K.A.; Lawley, C.T.; Vergara, M.M.; Ben-Zvi, G.; Biniashvili, T.; Baruch, K.; St Leger, J.; Le, J.; Natarajan, A.; Rivera, M.; et al. New de novo assembly of the Atlantic bottlenose dolphin (Tursiops truncatus) improves genome completeness and provides haplotype phasing. Gigascience 2019, 8, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frazee, A.C.; Pertea, G.; Jaffe, A.E.; Langmead, B.; Salzberg, S.L.; Leek, J.T. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 2015, 33, 243–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics 2018, 19, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemmrich, K.; Denecke, B.; Paul, N.E.; Hoffmeister, D.; Pallua, N. RNA isolation from adipose tissue: An optimized procedure for high RNA yield and integrity. Lab. Med. 2010, 41, 104–106. [Google Scholar] [CrossRef] [Green Version]
- McGowen, M.R. Toward the resolution of an explosive radiation--A multilocus phylogeny of oceanic dolphins (Delphinidae). Mol. Phylogenet. Evol. 2011, 60, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Senevirathna, J.D.M.; Yonezawa, R.; Saka, T.; Igarashi, Y.; Yoshitake, K.; Kinoshita, S.; Funasaka, N.; Asakawa, S. Another polymorphic mitochondrial genome of Grampus griseus and phylogeny of family Delphinidae. Mitochondrial DNA Part B 2021, 6, 2569–2571. [Google Scholar] [CrossRef]
- Vincelli, P. Genetic engineering and sustainable crop disease management: Opportunities for case-by-case decision-making. Sustain. 2016, 8, 495. [Google Scholar] [CrossRef] [Green Version]
- Small, B. Sustainable development and technology: Genetic engineering, social sustainability and empirical ethics. Int. J. Sustain. Dev. 2007, 10, 402–435. [Google Scholar] [CrossRef]
- Jensen, P.; Andersson, L. Genomics meets ethology: A new route to understanding domestication, behavior, and sustainability in animal breeding. Ambio 2005, 34, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016, 2016, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hogan, B.L.M. Morphogenesis. Cell 1999, 96, 225–233. [Google Scholar] [CrossRef] [Green Version]
- Wiesen, M.H.J.; Bogdanovich, S.; Agarkova, I.; Perriard, J.C.; Khurana, T.S. Identification and characterization of layer-specific differences in extraocular muscle M-bands. Investig. Ophthalmol. Vis. Sci. 2007, 48, 1119–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bottje, W.; Kong, B.W.; Reverter, A.; Waardenberg, A.J.; Lassiter, K.; Hudson, N.J. Progesterone signalling in broiler skeletal muscle is associated with divergent feed efficiency. BMC Syst. Biol. 2017, 11, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kepser, L.J.; Damar, F.; De Cicco, T.; Chaponnier, C.; Prószynski, T.J.; Pagenstecher, A.; Rust, M.B. CAP2 deficiency delays myofibril actin cytoskeleton differentiation and disturbs skeletal muscle architecture and function. Proc. Natl. Acad. Sci. USA 2019, 116, 8397–8402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Zhang, S.; Li, X.; Yao, J.; Ling, L.; Huang, X.; Hu, C.; Zhang, Y.; Sun, X.; Qin, B.; et al. Integrative transcriptomics and proteomic analysis of extraocular muscles from patients with thyroid-associated ophthalmopathy. Exp. Eye Res. 2020, 193, 107962. [Google Scholar] [CrossRef] [PubMed]
- Shapshak, P. Molecule of the month. Drug News Perspect. 2005, 18, 523. [Google Scholar] [CrossRef]
- Nelson, B.R.; Makarewich, C.A.; Anderson, D.M.; Winders, B.R.; Troupes, C.D.; Wu, F.; Reese, A.L.; McAnally, J.R.; Chen, X.; Kavalali, E.T.; et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016, 351, 271–275. [Google Scholar] [CrossRef] [Green Version]
- Jensen, J.H.; Conley, L.N.; Hedegaard, J.; Nielsen, M.; Young, J.F.; Oksbjerg, N.; Hornshøj, H.; Bendixen, C.; Thomsen, B. Gene expression profiling of porcine skeletal muscle in the early recovery phase following acute physical activity. Exp. Physiol. 2012, 97, 833–848. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Wang, G.; Liao, J.; Chen, X. A functional mutation in the AMPD1 promoter region affects promoter activity and breast meat freshness in chicken. Anim. Genet. 2021, 52, 121–125. [Google Scholar] [CrossRef]
- Azeez, O.I.; Meintjes, R.; Chamunorwa, J.P. Fat body, fat pad and adipose tissues in invertebrates and vertebrates: The nexus. Lipids Health Dis. 2014, 13, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Miettinen, S.; Sarkanen, J.R.; Ashammakhi, N. Adipose Tissue and Adipocyte Differentiation: Molecular and Cellular Aspects and Tissue Engineering Applications. Top. Tissue Eng. 2008, 4, 1–26. [Google Scholar]
- Luo, L.; Liu, M. Adipose tissue in control of metabolism. J. Endocrinol. 2016, 231, R77–R99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serão, N.V.L.; Veroneze, R.; Ribeiro, A.M.F.; Verardo, L.L.; Braccini Neto, J.; Gasparino, E.; Campos, C.F.; Lopes, P.S.; Guimarães, S.E.F. Candidate gene expression and intramuscular fat content in pigs. J. Anim. Breed. Genet. 2011, 128, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Qi, K.; Men, X.; Wu, J.; Xu, Z. Rearing pattern alters porcine myofiber type, fat deposition, associated microbial communities and functional capacity. BMC Microbiol. 2019, 19, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Wasylenko, T.M.; Ahn, W.S.; Stephanopoulos, G. The oxidative pentose phosphate pathway is the primary source of NADPH for lipid overproduction from glucose in Yarrowia lipolytica. Metab. Eng. 2015, 30, 27–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Tang, X.; Luan, X.; Chen, H.; Chen, Y.Q.; Chen, W.; Song, Y.; Ratledge, C. Role of pentose phosphate pathway in lipid accumulation of oleaginous fungus Mucor circinelloides. RSC Adv. 2015, 5, 97658–97664. [Google Scholar] [CrossRef]
- Patra, K.C.; Hay, N. The pentose phosphate pathway and cancer. Trends Biochem. Sci. 2014, 39, 347–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watts, J.L.; Ristow, M. Lipid and carbohydrate metabolism in Caenorhabditis elegans. Genetics 2017, 207, 413–446. [Google Scholar] [CrossRef] [PubMed]
- Jeukendrup, A.E.; Saris, W.H.M.; Wagenmakers, A.J.M. Fat Metabolism During Exercise: A Review. Part I: Fatty Acid Mobilization and Muscle Metabolism. Int. J. Sports Med. 2007, 19, 231–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinn, L.S. Interleukin-15: A muscle-derived cytokine regulating fat-to-lean body composition. J. Anim. Sci. 2008, 86, 75–83. [Google Scholar] [CrossRef]
- Volek, J.S.; Noakes, T.; Phinney, S.D. Rethinking fat as a fuel for endurance exercise. Eur. J. Sport Sci. 2015, 15, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Slomovic, S.; Pardee, K.; Collins, J.J. Synthetic biology devices for in vitro and in vivo diagnostics. Proc. Natl. Acad. Sci. USA 2015, 112, 14429–14435. [Google Scholar] [CrossRef] [Green Version]
- Hanigan, M.D.; Daley, V.L. Use of Mechanistic Nutrition Models to Identify Sustainable Food Animal Production. Annu. Rev. Anim. Biosci. 2020, 8, 355–376. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Cai, Z.; Xie, Y.; Ma, A.; Zhang, H.; Rao, P.; Wang, Q. Synthesis, physicochemical properties, and health aspects of structured lipids: A review. Compr. Rev. Food Sci. Food Saf. 2020, 19, 759–800. [Google Scholar] [CrossRef] [PubMed]
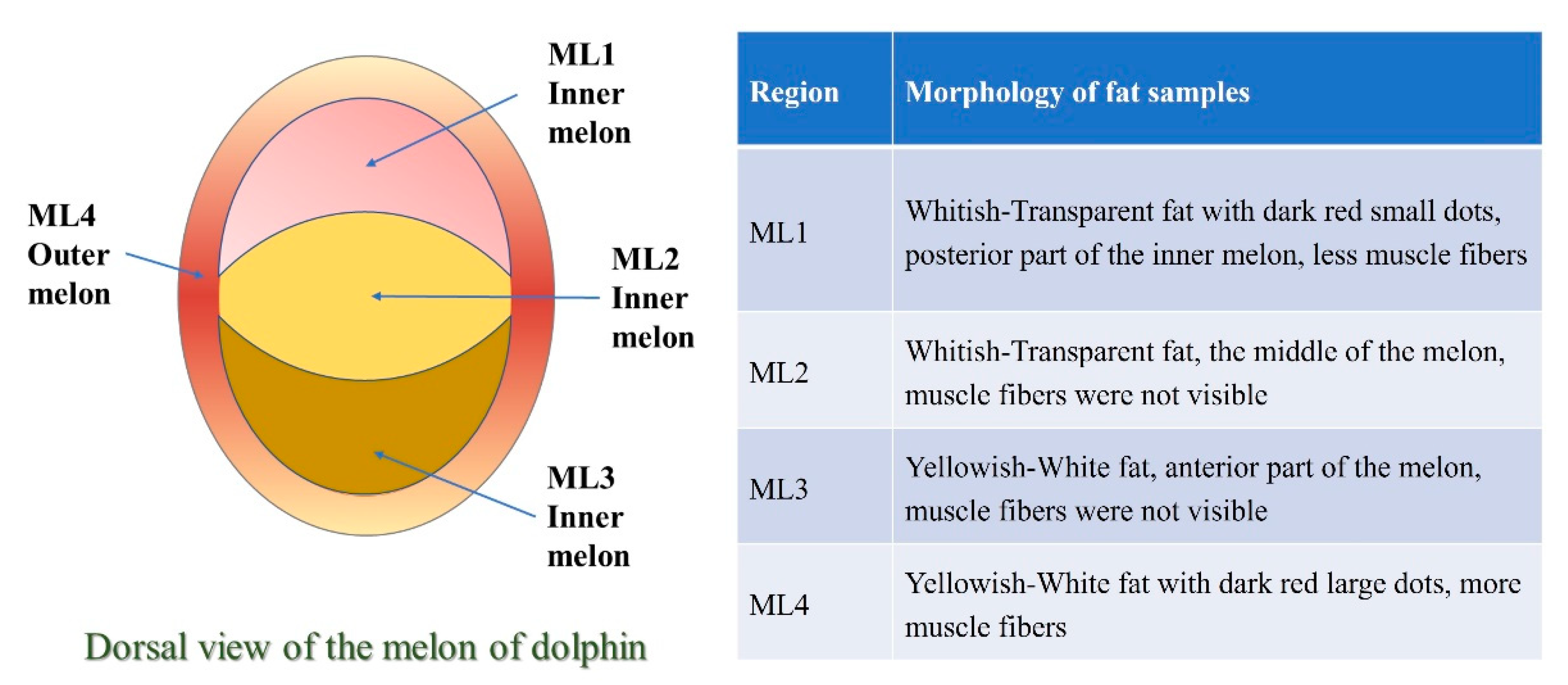

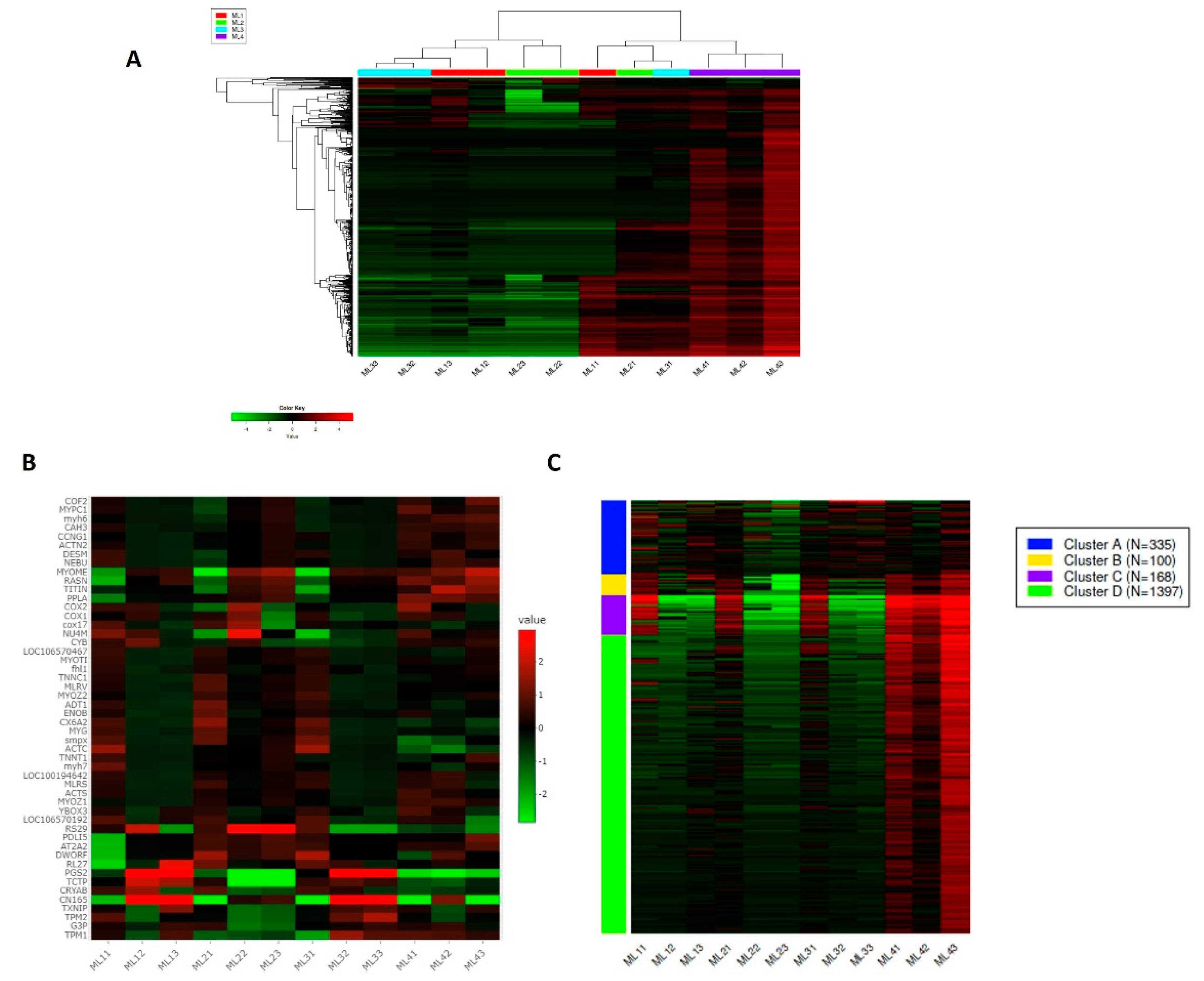


| Comparison/Direction | DEGs |
|---|---|
| ML2 up—ML1 down | AT2A2, DWORF |
| ML2 down—ML1 up | CN165 |
| ML3 up—ML1 down | AT2A2, DWORF |
| ML4 up—ML1 down | ASB5, AT1B1, AT2A2, AT5F1, BTBD1, DWORF, COX3, SERC1, SMYD2, NAA50, AATM, AMPD1, ACADL, DDT4L, CAP2, CATA, GLYG, NRAP, LDB3, GNAS3, IF4G2, LRC39, M3K20, MYH13, MYOM2, MYOM3, MYOME, NF2L1, NNTM, NUD4B, OBSCN, PDLI5, PFKAM, PGM1, PPLA, RASN |
| ML3 up—ML2 down | CN165 |
| ML4 up—ML2 down | CN165 |
| ML4 up—ML3 down | NAA50, IF4G2, MYH13, MYOME |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Senevirathna, J.D.M.; Yonezawa, R.; Saka, T.; Igarashi, Y.; Funasaka, N.; Yoshitake, K.; Kinoshita, S.; Asakawa, S. Transcriptomic Insight into the Melon Morphology of Toothed Whales for Aquatic Molecular Developments. Sustainability 2021, 13, 13997. https://doi.org/10.3390/su132413997
Senevirathna JDM, Yonezawa R, Saka T, Igarashi Y, Funasaka N, Yoshitake K, Kinoshita S, Asakawa S. Transcriptomic Insight into the Melon Morphology of Toothed Whales for Aquatic Molecular Developments. Sustainability. 2021; 13(24):13997. https://doi.org/10.3390/su132413997
Chicago/Turabian StyleSenevirathna, Jayan Duminda Mahesh, Ryo Yonezawa, Taiki Saka, Yoji Igarashi, Noriko Funasaka, Kazutoshi Yoshitake, Shigeharu Kinoshita, and Shuichi Asakawa. 2021. "Transcriptomic Insight into the Melon Morphology of Toothed Whales for Aquatic Molecular Developments" Sustainability 13, no. 24: 13997. https://doi.org/10.3390/su132413997
APA StyleSenevirathna, J. D. M., Yonezawa, R., Saka, T., Igarashi, Y., Funasaka, N., Yoshitake, K., Kinoshita, S., & Asakawa, S. (2021). Transcriptomic Insight into the Melon Morphology of Toothed Whales for Aquatic Molecular Developments. Sustainability, 13(24), 13997. https://doi.org/10.3390/su132413997






