Sponge–Microbial Symbiosis and Marine Extremozymes: Current Issues and Prospects
Abstract
:1. Introduction
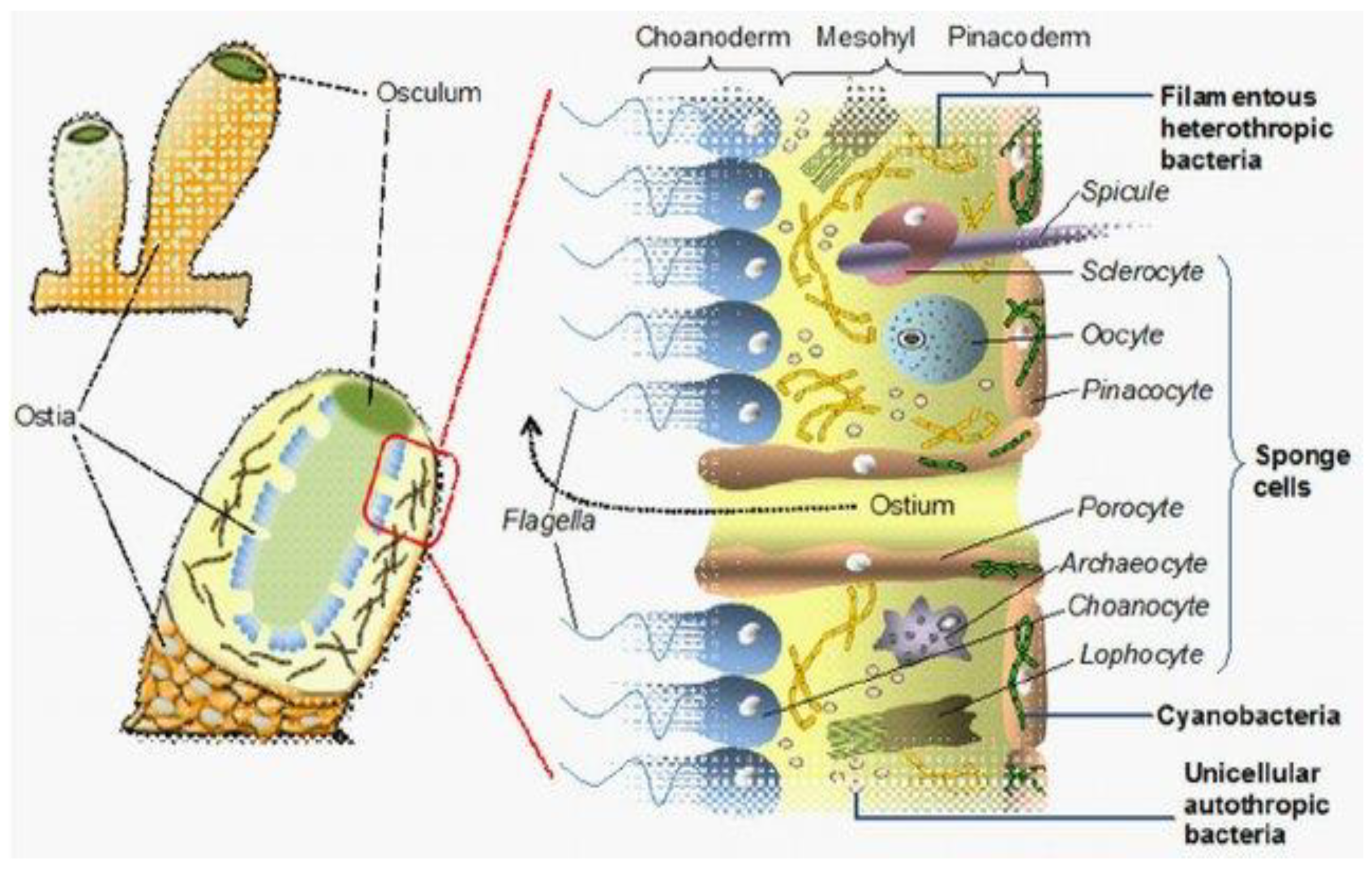
2. Microbial Biodiversity and Marine Sponge Symbiosis
3. Benefits of Enzyme Production from Marine Extremophiles
4. Marine Extremozymes and Biotechnological Applications
5. Current Issues on Sponge–Microbial Symbiosis and Marine Extremozyme Application
5.1. Research
- a.
- The possibility of distortion or destruction of microbial interactions in their natural environment by culture media. The rapidly growing microbes may produce inhibitory compounds that will in turn inactivate the very slow growers.
- b.
- A lack of knowledge of organic substrates that are metabolized by the marine microorganism in their natural habitat may have resulted in the wrong concentration or absence of nutrients in the available culture media.
- c.
- The possibility of virus infection may prevent bacterial growth in medium; this could be due to infection with bacteriophages or starvation of bacterial cells due to changes to the lytic cycle of temperate phages during the supply of nutrients.
- d.
- Difficulty in the detection of growth may also be associated with high concentrations of substrate in the medium. This condition may be toxic to bacteria that have evolved under oligotrophic conditions.
- e.
- Poor cell density detection methods have resulted in negligence regarding the invisible first round of culture in liquid media [49].
5.2. Industry and Bioremediation
6. Prospects
6.1. Cost Effectiveness
6.2. Species Diversity Approach
6.3. Integrative Bioinformatics Software
6.4. Government and Industries
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Otaiku, A.A. Bioremediation of Hydrocarbon Production Wastes using Environmental Biotechnology Inoculants (OBD-Plus®) Obagi, Niger Delta, Nigeria: Findings. (MSc); Enugu State University of Science and Technology Nigeria: Enugu, Nigeria, 2007. [Google Scholar]
- Baharum, S.; Beng, E.; Mokhtar, M. Marine microorganisms: Potential application and challenges. J. Biol. Sci. 2010, 10, 555–564. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Kim, S.-K. Research and application of marine microbial enzymes: Status and prospects. Mar. Drugs 2010, 8, 1920–1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uria, A.R. Capturing natural product biosynthetic pathways from uncultivated symbiotic bacteria of marine sponges through metagenome mining: A mini-review. Squalen Bull. Mar. Fish. Postharvest Biotechnol. 2015, 10, 35–49. [Google Scholar] [CrossRef] [Green Version]
- Mohapatra, B.; Bapuji, M. Characterization of urethanase from Micrococcus species associated with the marine sponge (Spirasfrella species). Lett. Appl. Microbiol. 1997, 25, 393–396. [Google Scholar] [CrossRef]
- Lee, Y.-K.; Lee, J.-H.; Lee, H.-K. Microbial symbiosis in marine sponges. J. Microbiol. 2001, 39, 254–264. [Google Scholar]
- Robbins, S.; Song, W.; Engelberts, J.; Glasl, B.; Slaby, B.M.; Boyd, J.; Marangon, E.; Botté, E.; Laffy, P.; Thomas, T.; et al. A genomic view of the microbiome of coral reef demosponges. ISME J. 2021, 15, 1641–1654. [Google Scholar] [CrossRef]
- Merino, N.; Aronson, H.S.; Bojanova, D.P.; Feyhl-Buska, J.; Wong, M.L.; Zhang, S.; Giovannelli, D. Living at the extremes: Extremophiles and the limits of life in a planetary context. Front. Microbiol. 2019, 10, 780. [Google Scholar] [CrossRef] [Green Version]
- Di Donato, P.; Buono, A.; Poli, A.; Finore, I.; Abbamondi, G.R.; Nicolaus, B.; Lama, L. Exploring marine environments for the identification of extremophiles and their enzymes for sustainable and green bioprocesses. Sustainability 2018, 11, 149. [Google Scholar] [CrossRef] [Green Version]
- Wang, G. Diversity and biotechnological potential of the sponge-associated microbial consortia. J. Ind. Microbiol. Biotechnol. 2006, 33, 545. [Google Scholar] [CrossRef]
- Rohban, R.; Amoozegar, M.A.; Ventosa, A. Screening and isolation of halophilic bacteria producing extracellular hydrolyses from Howz Soltan Lake, Iran. J. Ind. Microbiol. Biotechnol. 2009, 36, 333–340. [Google Scholar] [CrossRef]
- Gurung, N.; Ray, S.; Bose, S.; Rai, V. A broader view: Microbial enzymes and their relevance in industries, medicine, and beyond. BioMed Res. Int. 2013, 2013, 329121. [Google Scholar] [CrossRef] [Green Version]
- Raveendran, S.; Parameswaran, B.; Ummalyma, S.B.; Abraham, A.; Mathew, A.K.; Madhavan, A.; Rebello, S.; Pandey, A. Applications of microbial enzymes in food industry. Food Technol. Biotechnol. 2018, 56, 16. [Google Scholar] [CrossRef]
- Anbu, P.; Gopinath, S.C.; Chaulagain, B.P.; Lakshmipriya, T. Microbial enzymes and their applications in industries and medicine 2016. BioMed Res. Int. 2017, 2017, 2195808. [Google Scholar] [CrossRef]
- Anbu, P.; Gopinath, S.C.; Chaulagain, B.P.; Tang, T.-H.; Citartan, M. Microbial enzymes and their applications in industries and medicine 2014. BioMed Res. Int. 2015, 2015, 816419. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Kumar, M.; Mittal, A.; Mehta, P.K. Microbial enzymes: Industrial progress in 21st century. 3 Biotech 2016, 6, 174. [Google Scholar] [CrossRef] [Green Version]
- Oyewole, O.A.; Oyeleke, S.B.; Dauda, B.; Emiade, S. Production of amylase and protease enzymes by Aspergillus niger and Penicillium frequestans isolated from abattoir effluent. Microbiol. J. 2011, 1, 174–180. [Google Scholar] [CrossRef] [Green Version]
- Suriya, J.; Bharathiraja, S.; Krishnan, M.; Manivasagan, P.; Kim, S.-K. Marine microbial amylases: Properties and applications. Adv. Food Nutr. Res. 2016, 79, 161–177. [Google Scholar]
- Lilja, T. Isolating Microorganisms from Marine and Marine-Associated Samples—A Targeted Search for Novel Natural Antibiotics; Independent Project in Biology; Swedish University of Agricultural Science: Upsala, Sweden, 2013. [Google Scholar]
- Gomes, J.; Steiner, W. The biocatalytic potential of extremophiles and extremozymes. Food Technol. Biotechnol. 2004, 42, 223–225. [Google Scholar]
- Zhu, D.; Adebisi, W.A.; Ahmad, F.; Sethupathy, S.; Danso, B.; Sun, J. Recent development of extremophilic bacteria and their application in biorefinery. Front. Bioeng. Biotechnol. 2020, 8, 483. [Google Scholar] [CrossRef]
- De Lourdes Moreno, M.; Pérez, D.; García, M.T.; Mellado, E. Halophilic Bacteria as a source of novel hydrolytic enzymes. Life 2013, 3, 38–51. [Google Scholar] [CrossRef] [Green Version]
- Calimlioglu, B.; Arga, K.Y. Proteins from Halophilic Bacteria: Purification and Their Applications; iConcept Press Ltd.: Hong Kong, China, 2014. [Google Scholar]
- Das, S.; Lyla, P.; Khan, S.A. Marine microbial diversity and ecology: Importance and future perspectives. Curr. Sci. 2006, 90, 1325–1335. [Google Scholar]
- Folkers, M.; Rombouts, T. Sponges revealed: A synthesis of their overlooked ecological functions within aquatic ecosystems. In YOUMARES 9—The Oceans: Our Research, Our Future; Springer: Cham, Switzerland, 2020; pp. 181–193. [Google Scholar]
- Orcutt, B.N.; Sylvan, J.B.; Knab, N.J.; Edwards, K.J. Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol. Mol. Biol. Rev. 2011, 75, 361–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akpiri, R.U. The Development of Sea Sponges (Hymeniacidon perlevis and Amorphinopsis sp.) as Novel Models for Genotoxicity Assessment and Environmental Monitoring of Pollutants in the Aquatic Environment. Ph.D. Thesis, University of Birmingham, Birmingham, UK, 2019. [Google Scholar]
- Carter, A. Coral’s indispensable bacterial buddies. Oceanus 2013, 50, 6. [Google Scholar]
- Hudspith, M.; Rix, L.; Achlatis, M.; Bougoure, J.; Guagliardo, P.; Clode, P.L.; Webster, N.S.; Muyzer, G.; Pernice, M.; de Goeij, J.M. Subcellular view of host–microbiome nutrient exchange in sponges: Insights into the ecological success of an early metazoan–microbe symbiosis. Microbiome 2021, 9, 44. [Google Scholar] [CrossRef]
- Pereira, M.G.; Vici, A.C.; Facchini, F.D.A.; Tristao, A.P.; Cursino-Santos, J.R.; Sanches, P.R.; Jorge, J.A.; de Lourdes Teixeira de Moraes Polizeli, M. Screening of filamentous fungi for lipase production: Hypocrea pseudokoningii a new producer with a high biotechnological potential. Biocatal. Biotransform. 2014, 32, 74–83. [Google Scholar] [CrossRef]
- Prabhu, R.; Bhise, K.; Patravale, V. Marine enzymes in cancer: A new paradigm. In Advances in Food and Nutrition Research; Elsevier: Amsterdam, The Netherlands, 2017; Volume 80, pp. 1–14. [Google Scholar]
- Andualema, B.; Gessesse, A. Microbial lipases and their industrial applications. Biotechnology 2012, 11, 100. [Google Scholar] [CrossRef] [Green Version]
- Rao, T.E.; Imchen, M.; Kumavath, R. Marine enzymes: Production and applications for human health. In Advances in Food and Nutrition Research; Elsevier: Amsterdam, The Netherlands, 2017; Volume 80, pp. 149–163. [Google Scholar]
- Parte, S.; Sirisha, V.; D’Souza, J. Biotechnological applications of marine enzymes from algae, bacteria, fungi, and sponges. In Advances in Food and Nutrition Research; Elsevier: Amsterdam, The Netherlands, 2017; Volume 80, pp. 75–106. [Google Scholar]
- Whitehurst, R.J.; Van Oort, M. Enzymes in Food Technology; Wiley-Blackwell: Singapore, 2010; Volume 388. [Google Scholar]
- Sysoev, M.; Grötzinger, S.W.; Renn, D.; Eppinger, J.; Rueping, M.; Karan, R. Bioprospecting of novel extremozymes from prokaryotes—The advent of culture-independent methods. Front. Microbiol. 2021, 12, 630013. [Google Scholar] [CrossRef]
- Abe, T.; Sahin, F.P.; Akiyama, K.; Naito, T.; Kishigami, M.; Miyamoto, K.; Sakakibara, Y.; Uemura, D. Construction of a metagenomic library for the marine sponge Halichondria okadai. Biosci. Biotechnol. Biochem. 2012, 76, 633–639. [Google Scholar] [CrossRef]
- Qin, Y.; Huang, Z.; Liu, Z. A novel cold-active and salt-tolerant α-amylase from marine bacterium Zunongwangia profunda: Molecular cloning, heterologous expression and biochemical characterization. Extremophiles 2014, 18, 271–281. [Google Scholar] [CrossRef]
- Mohriak, W.; Mello, M.; Dewey, J.; Maxwell, J.R. Petroleum Geology of the Campos Basin, Offshore Brazil; Geological Society: London, UK, 1990; Volume 50, pp. 119–141. [Google Scholar]
- Hasan, F.; Shah, A.A.; Javed, S.; Hameed, A. Enzymes used in detergents: Lipases. Afr. J. Biotechnol. 2010, 9, 4836–4844. [Google Scholar]
- Mienda, B.S.; Yahya, A.; Galadima, I.; Shamsir, M. An overview of microbial proteases for industrial applications. Res. J. Pharm. Biol. Chem. Sci. 2014, 5, 388–396. [Google Scholar]
- Bhatnagar, I.; Kim, S.-K. Immense essence of excellence: Marine microbial bioactive compounds. Mar. Drugs 2010, 8, 2673–2701. [Google Scholar] [CrossRef] [Green Version]
- Jin, M.; Gai, Y.; Guo, X.; Hou, Y.; Zeng, R. Properties and applications of extremozymes from deep-sea extremophilic microorganisms: A mini review. Mar. Drugs 2019, 17, 656. [Google Scholar] [CrossRef] [Green Version]
- Damare, S.; Raghukumar, C.; Raghukumar, S. Fungi in deep-sea sediments of the Central Indian Basin. Deep. Sea Res. Part I Oceanogr. Res. Pap. 2006, 53, 14–27. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.K.; Fuerst, J.A. Diversity of polyketide synthase genes from bacteria associated with the marine sponge Pseudoceratina clavata: Culture-dependent and culture-independent approaches. Environ. Microbiol. 2006, 8, 1460–1470. [Google Scholar] [CrossRef]
- Dias, R.; Silva, L.C.; Eller, M.R.; Oliveira, V.M.; De Paula, S.; Silva, C.C. Metagenomics: Library construction and screening methods. V Metagenom. Methods Appl. Perspect 2014, 5, 28–34. [Google Scholar]
- Teeling, H.; Glöckner, F.O. Current opportunities and challenges in microbial metagenome analysis—A bioinformatic perspective. Brief. Bioinform. 2012, 13, 728–742. [Google Scholar] [CrossRef]
- Kumar, S.; Tamura, K.; Jakobsen, I.B.; Nei, M. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 2001, 17, 1244–1245. [Google Scholar] [CrossRef] [Green Version]
- Joint, I.; Mühling, M.; Querellou, J. Culturing marine bacteria—An essential prerequisite for biodiscovery. Microb. Biotechnol. 2010, 3, 564–575. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Liu, W. Scientists Develop Enzyme Produced from Agricultural Waste for Use as Laundry Detergent. 2020. Available online: https://www.eurekalert.org/news-releases/836376 (accessed on 30 July 2021).
- Ezeonu, C.S.; Tagbo, R.; Anike, E.N.; Oje, O.A.; Onwurah, I.N. Biotechnological tools for environmental sustainability: Prospects and challenges for environments in Nigeria—A standard review. Biotechnol. Res. Int. 2012, 2012, 450802. [Google Scholar] [CrossRef]
- Wu, S.; Snajdrova, R.; Moore, J.C.; Baldenius, K.; Bornscheuer, U.T. Biocatalysis: Enzymatic synthesis for industrial applications. Angew. Chem. Int. Ed. 2021, 60, 88–119. [Google Scholar] [CrossRef] [PubMed]
- Sivaperumal, P.; Kamala, K.; Rajaram, R. Bioremediation of industrial waste through enzyme producing marine microorganisms. In Advances in Food and Nutrition Research; Elsevier: Amsterdam, The Netherlands, 2017; Volume 80, pp. 165–179. [Google Scholar]
- Jenab, K.; Moghimi, H.; Hamedi, J. Bioremediation of Crude Oil by Halophilic Funges Engyodontium album in Saline Culture. In Proceedings of the International and Iranian Congress of Microbiology, Tehran, Iran, 25–27 August 2015. [Google Scholar]
- Bano, A.; Hussain, J.; Akbar, A.; Mehmood, K.; Anwar, M.; Hasni, M.S.; Ullah, S.; Sajid, S.; Ali, I. Biosorption of heavy metals by obligate halophilic fungi. Chemosphere 2018, 199, 218–222. [Google Scholar] [CrossRef] [PubMed]

| Extremophilic Microbes | Marine Habitat | Extreme Characteristics | References |
|---|---|---|---|
| Thermococcus piezophilus CDGS Pyrococcus, Thermotoga, Thermococcus, Archeoaglobus, Methanococcus, Pyrodictium, Aquifex | Deep-sea hydrothermal vent Shallow-water and deep-sea hydrothermal vents | [8] [9] | |
| Planococcus halocryophilus Or1 | Sea ice core | Halo-psychrophilic | [8] |
| Halarsenatibacter silvermanii SLAS-1 | Soda lake | Halo-alkaliphilic | [8] |
| Cyanobacteria, Ceratoporella nicholsoni, Rhopaloides odorabile, Aplysina aerophoba Cytophaga, and Desulfovibrio Micrococcus species | Marine sponges Marine sponges | [10] [5] | |
| Salicola, Halovibrio, Halomonas, Bacillus, Oceanobacillus, Thalasobacillus, Virgibacillus, Gracilibacillus, Halobacillus, Piscibacillus and Salinicoccus | Howz Soltan Lake | Halophilic | [11] |
| Extremozymes | Extremophilic Property | Sources | References |
|---|---|---|---|
| Amylase | Thermophilic | Bacillius, Clostridia, Fervidobacterium | [20] |
| β-Galactosidase | Psychrophilic | Alteromonas sp. ML117 | [21] |
| Lipase | Halophilic | Salicola marasensis Marinobacter lipolyticu | [22,23] |
| Esterase | Psychrophilic | Acinetobacter sp. | [20] |
| α-Amylase | Thermophilic | Thermophilic Anoxybacillus sp. | [21] |
| α-Amylase | Thermophilic | Bacillus mojavensis SO-10 | [21] |
| Protease | Halophilic | Salicola marasensis | [21] |
| α-Amylase | Halophilic | Zunongwangia profunda |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nnaji, P.T.; Morse, H.R.; Adukwu, E.; Chidugu-Ogborigbo, R.U. Sponge–Microbial Symbiosis and Marine Extremozymes: Current Issues and Prospects. Sustainability 2022, 14, 6984. https://doi.org/10.3390/su14126984
Nnaji PT, Morse HR, Adukwu E, Chidugu-Ogborigbo RU. Sponge–Microbial Symbiosis and Marine Extremozymes: Current Issues and Prospects. Sustainability. 2022; 14(12):6984. https://doi.org/10.3390/su14126984
Chicago/Turabian StyleNnaji, Praise Tochukwu, H. Ruth Morse, Emmanuel Adukwu, and Rachael U. Chidugu-Ogborigbo. 2022. "Sponge–Microbial Symbiosis and Marine Extremozymes: Current Issues and Prospects" Sustainability 14, no. 12: 6984. https://doi.org/10.3390/su14126984






