Integrating Transformer and GCN for COVID-19 Forecasting
Abstract
:1. Introduction
2. Literature Reviews
3. Data Reduction
3.1. Confirmed Cased and Deaths Datasets
- Date: Observation date in mm/dd/yyyy
- State: State of the USA
- Cases: Cumulative counts of coronavirus cases till that date
- Deaths: Cumulative counts of coronavirus deaths till that date
3.2. Vaccinations Dataset
- Date
- State Name
- Daily count of vaccinations
4. Architecture Design for Hybrid Models
4.1. Data Preprocessing
4.2. Model Theory
4.2.1. Encoder Structure
4.2.2. Decoder Structure
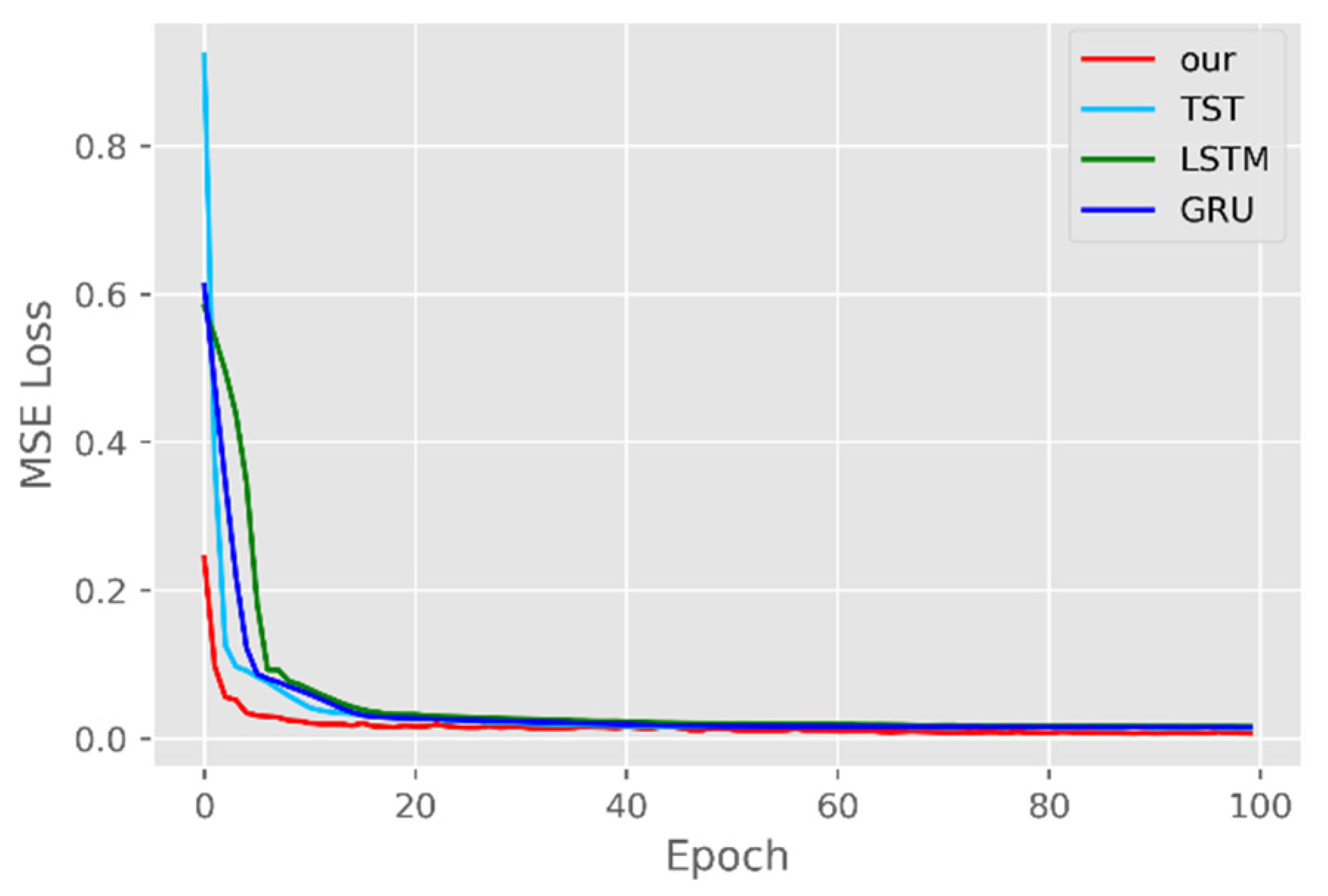
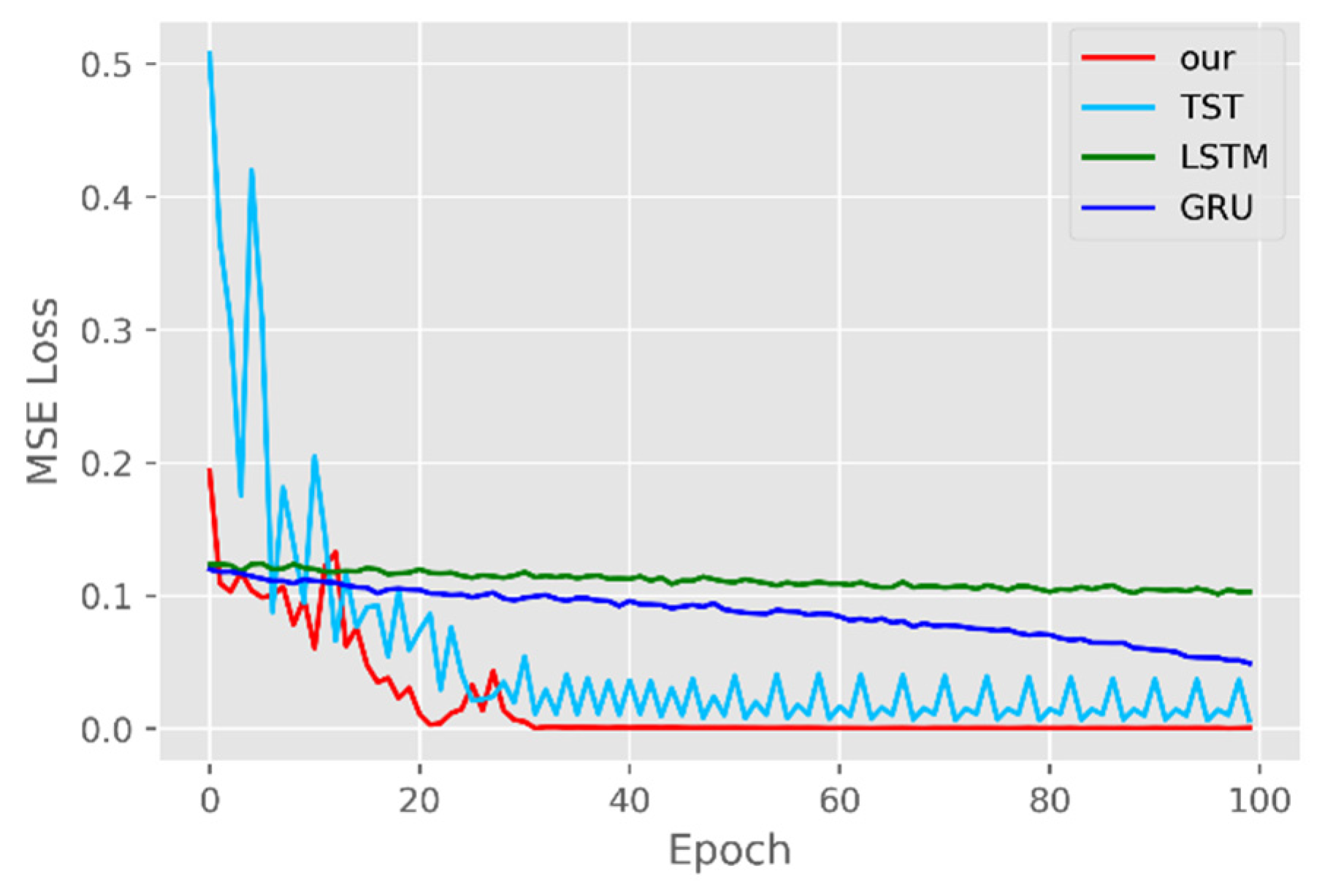
4.3. Training Schemes
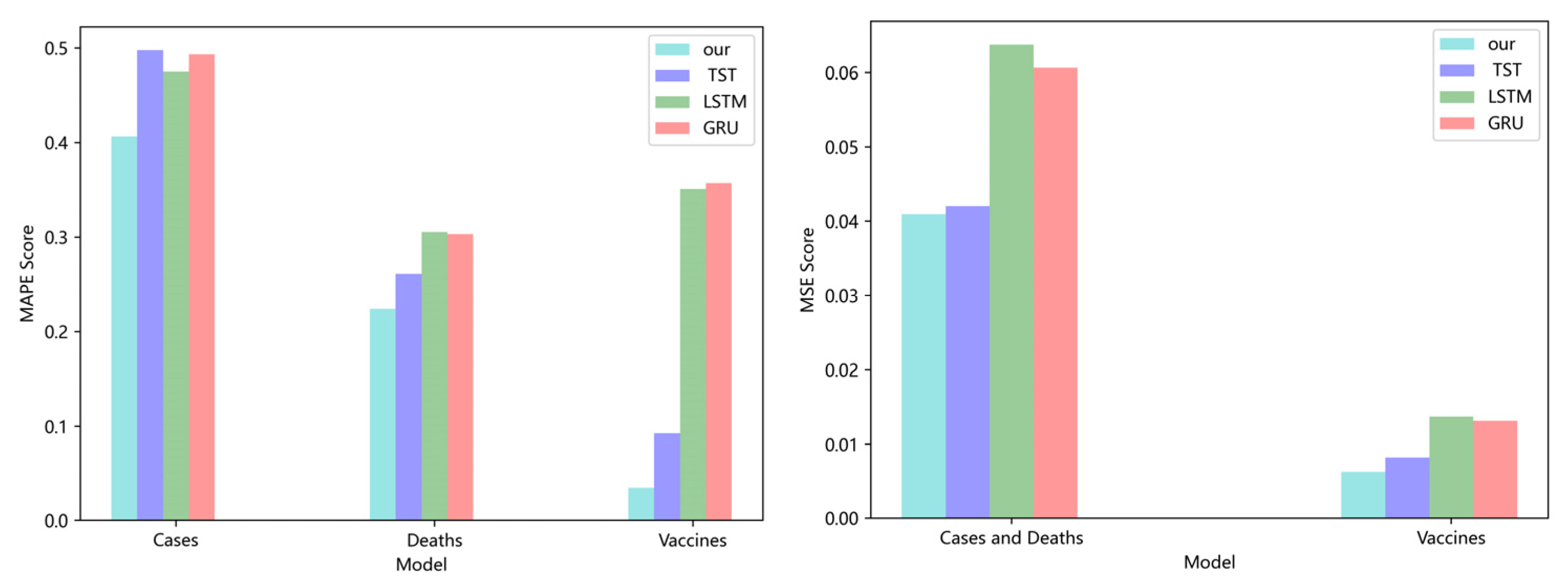
4.4. Prediction Accuracy Measurement
5. Results and Discussion
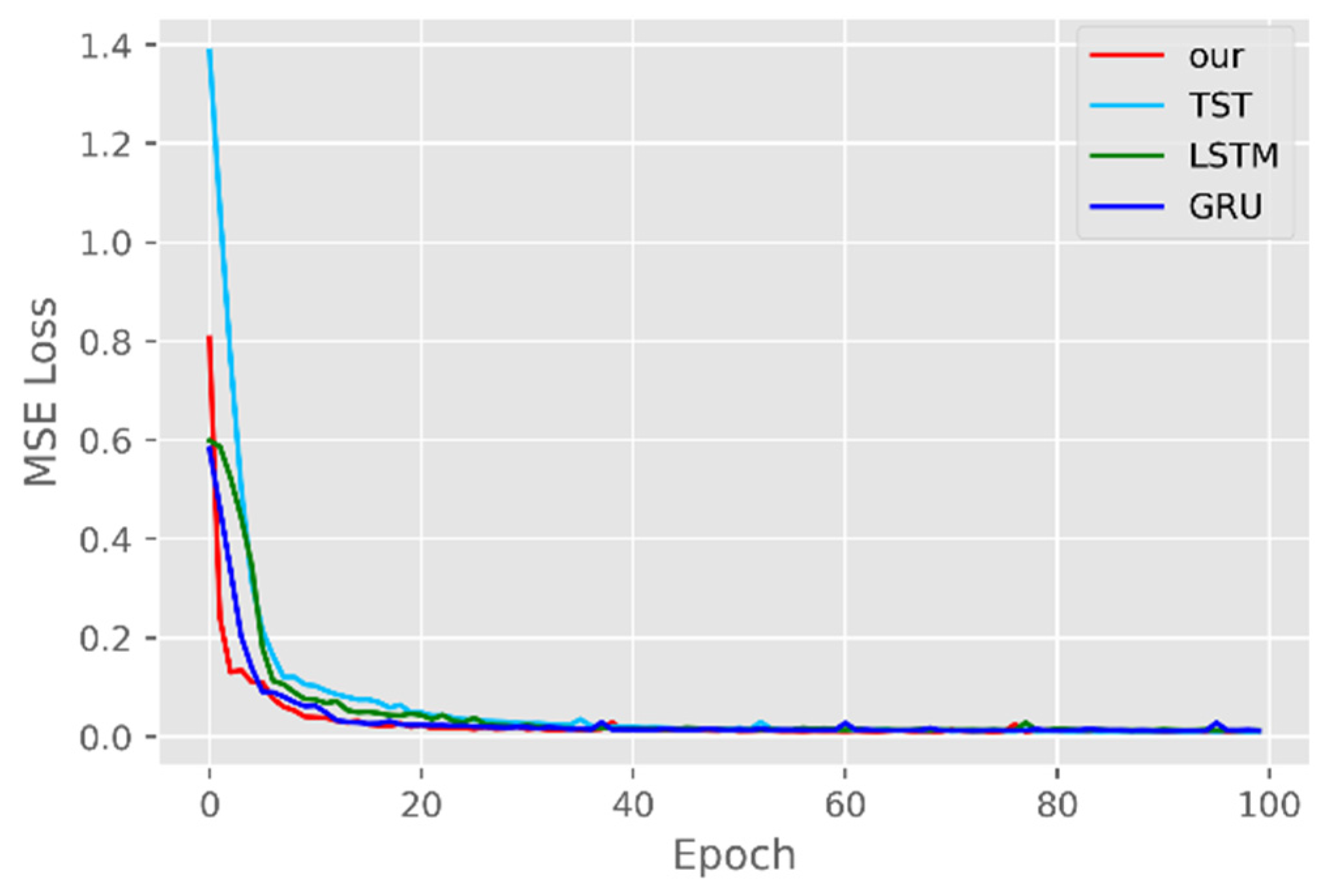
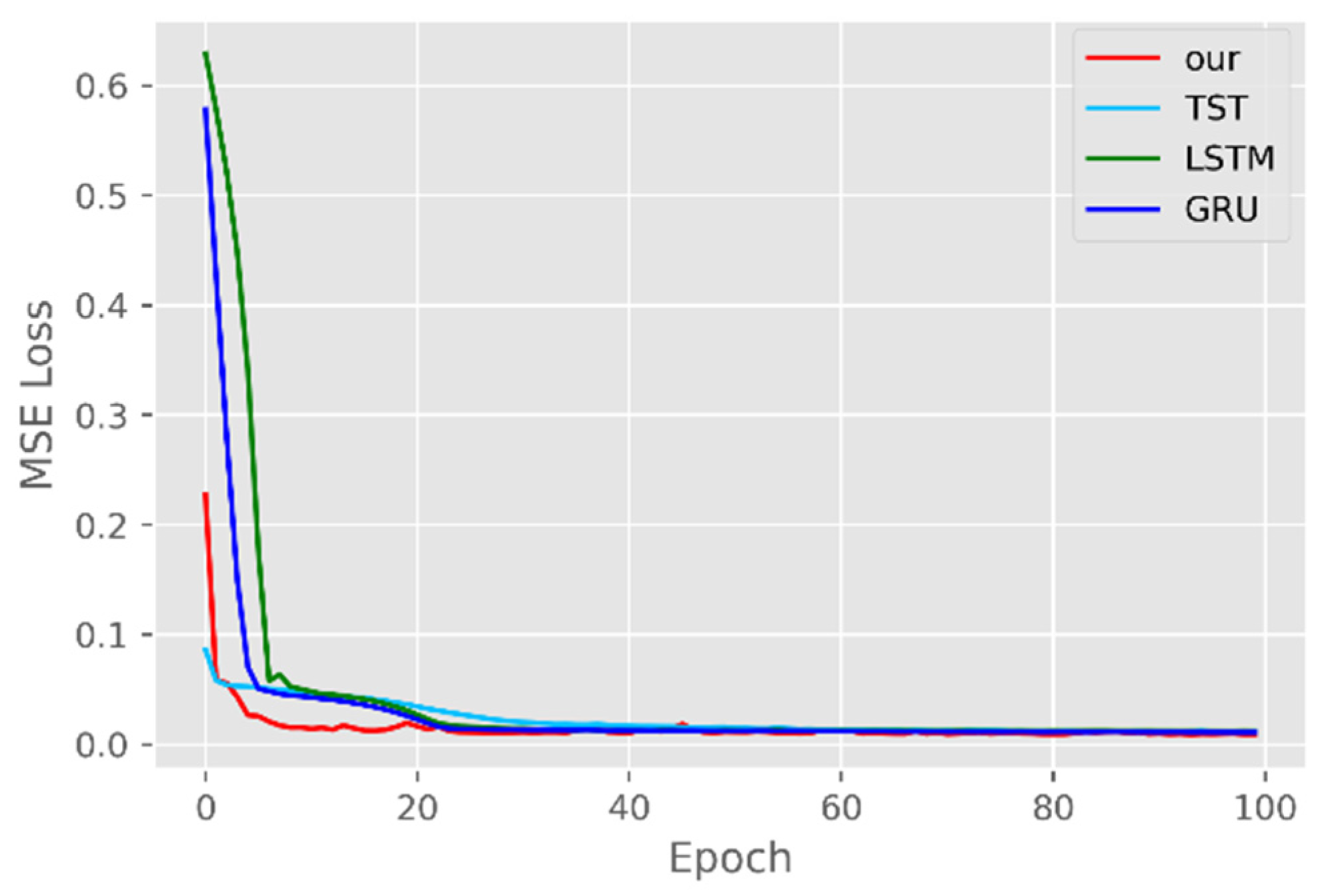
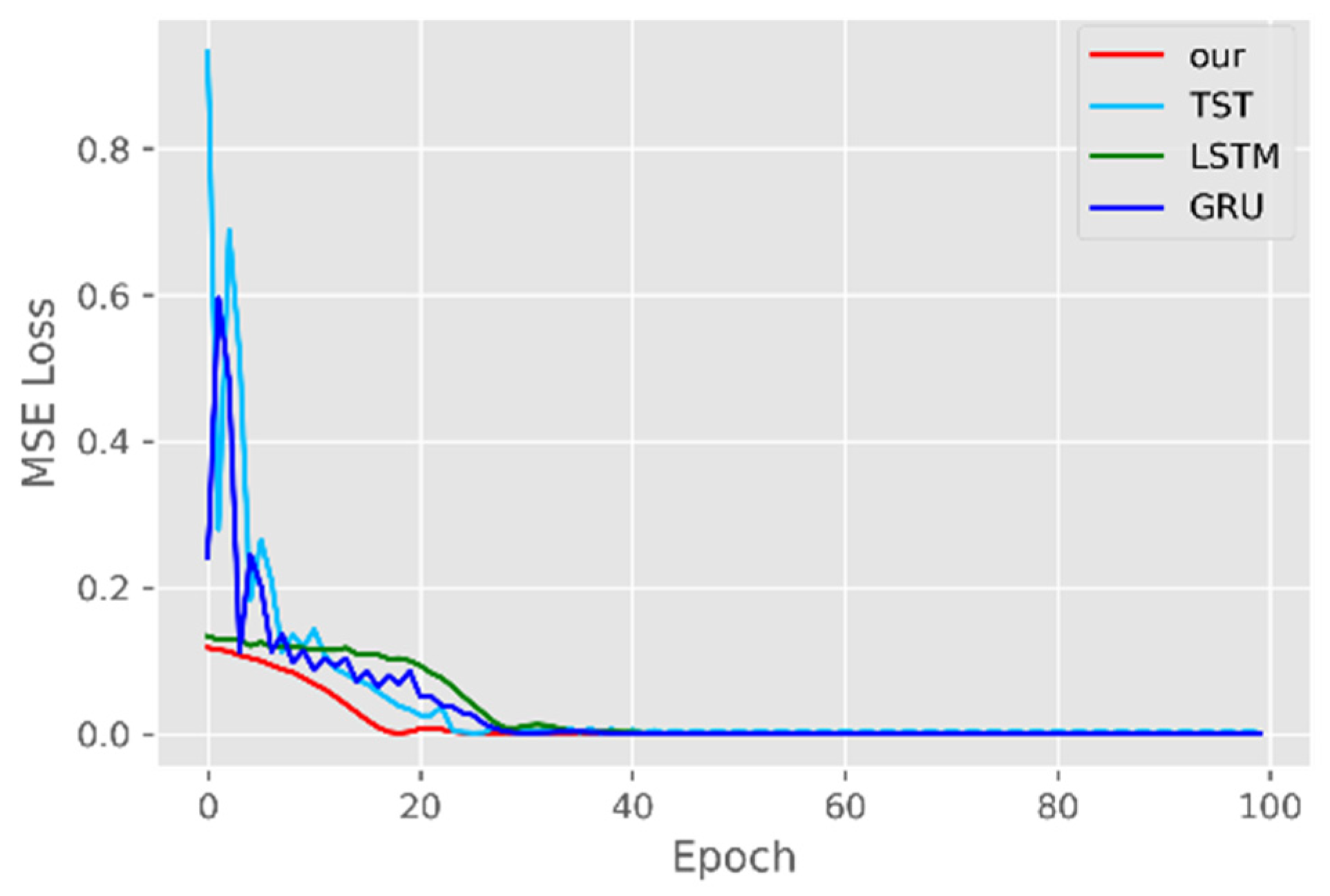
5.1. Comparison of Accuracy and Convergence of Models
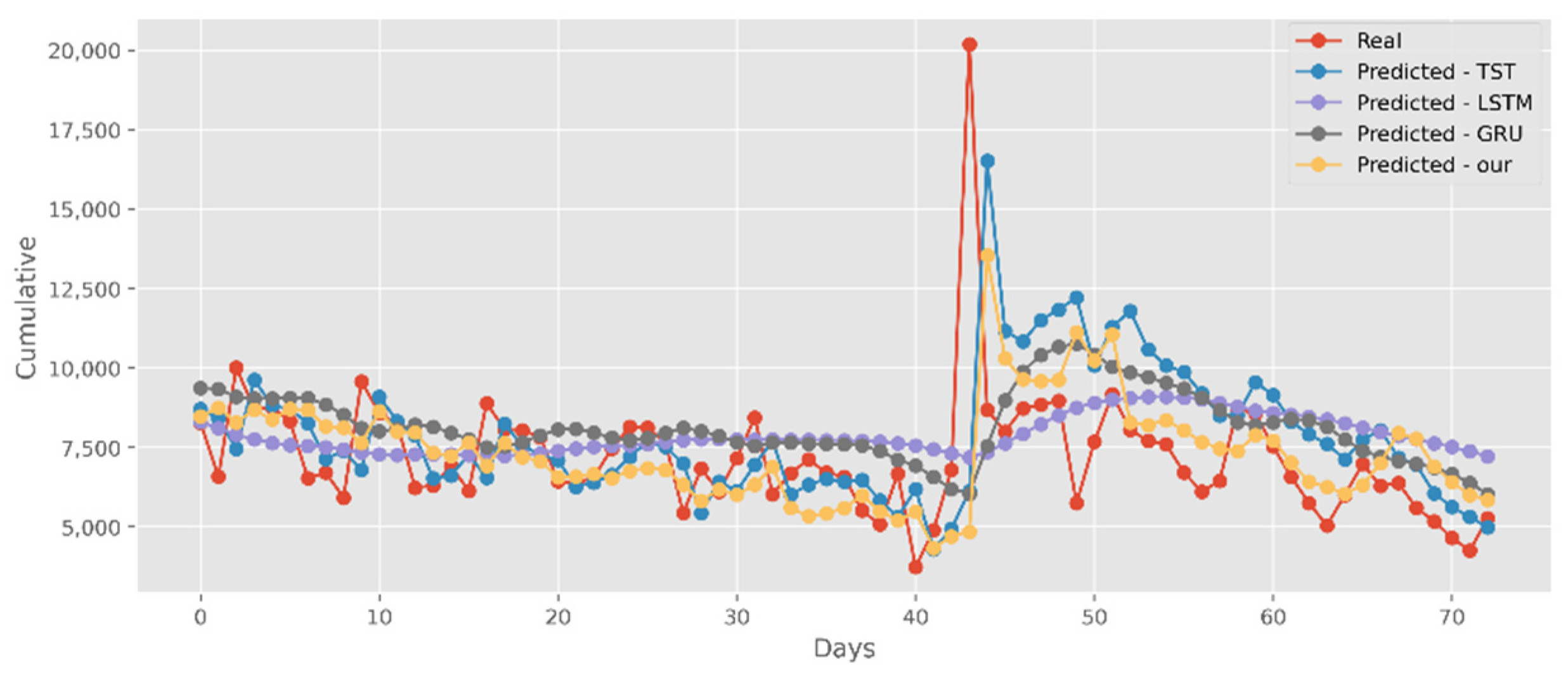
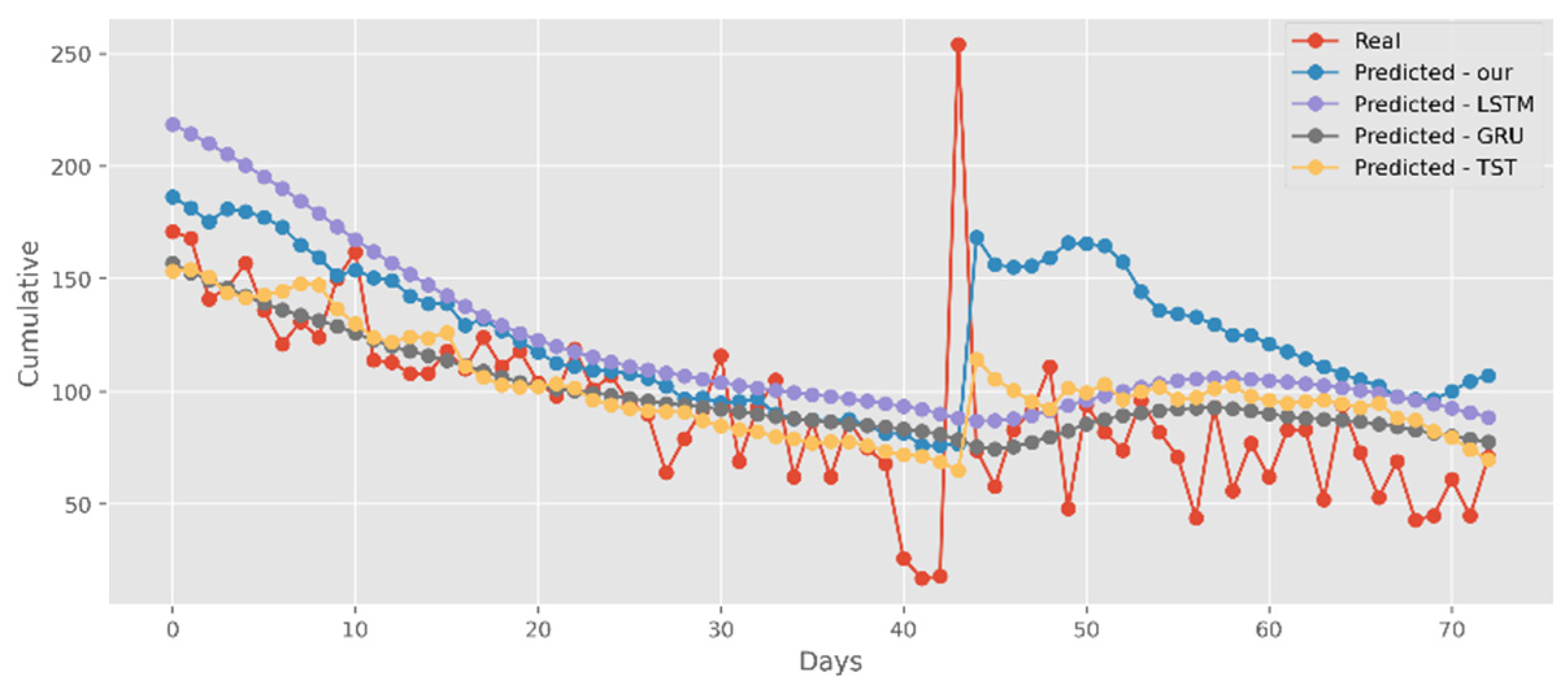
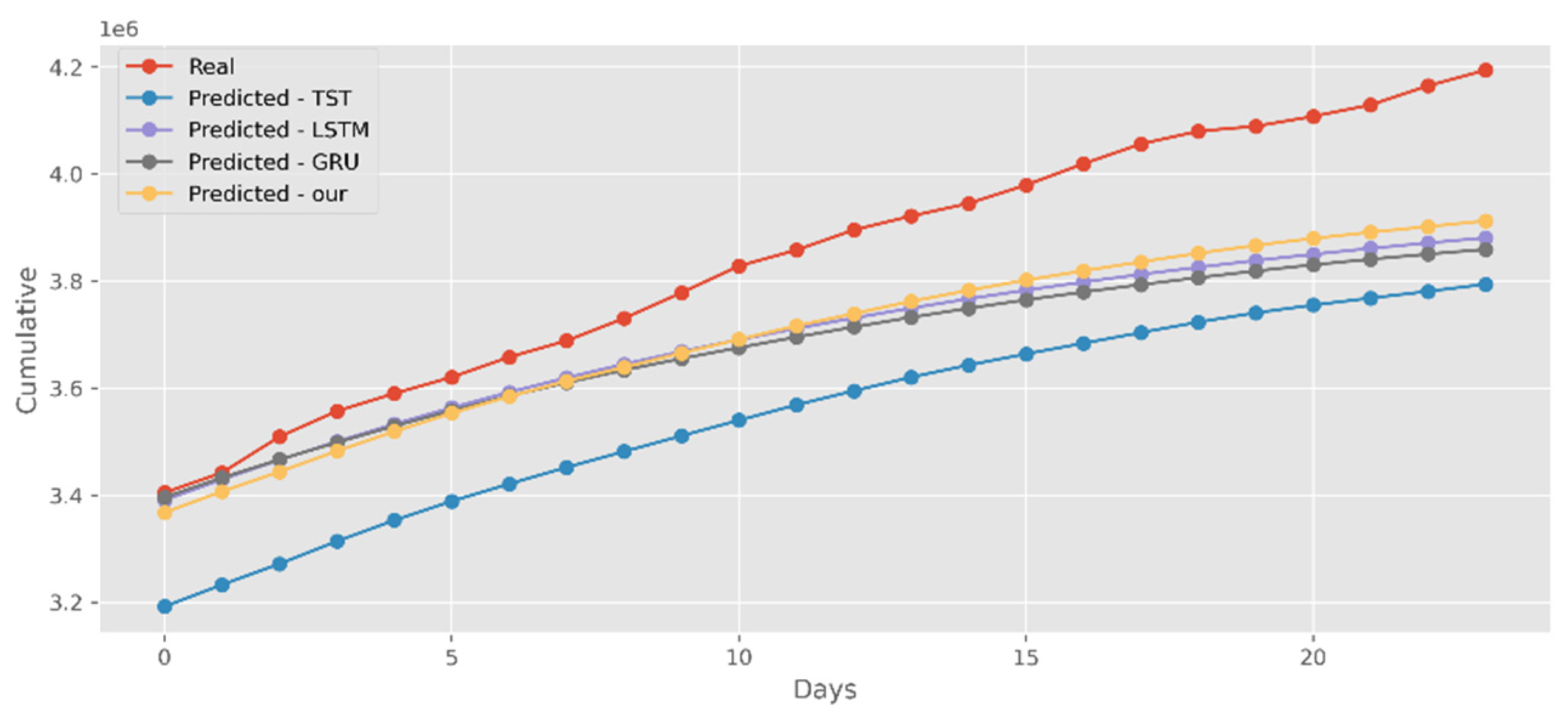
5.2. Forecasting the Number of Confirmed Cases
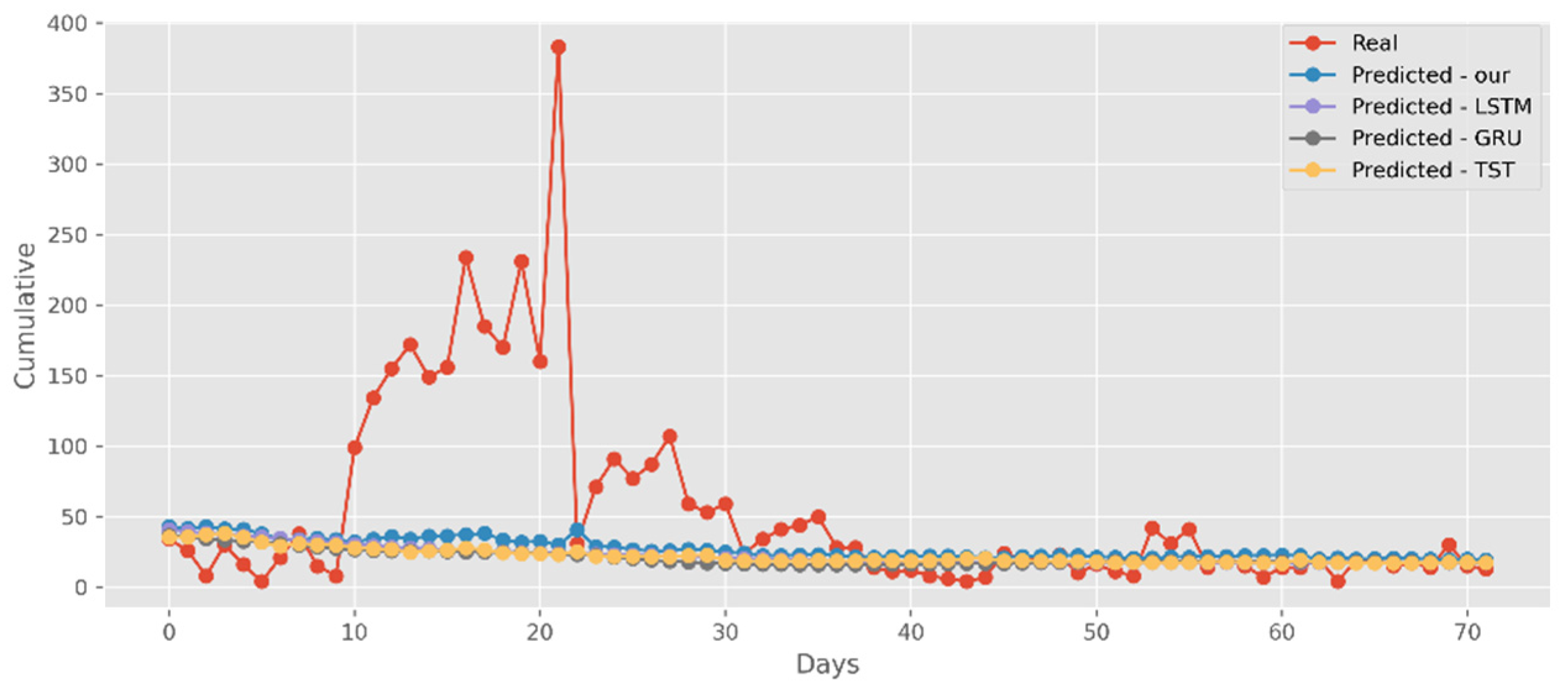
5.3. Forecasting the Number of Deaths
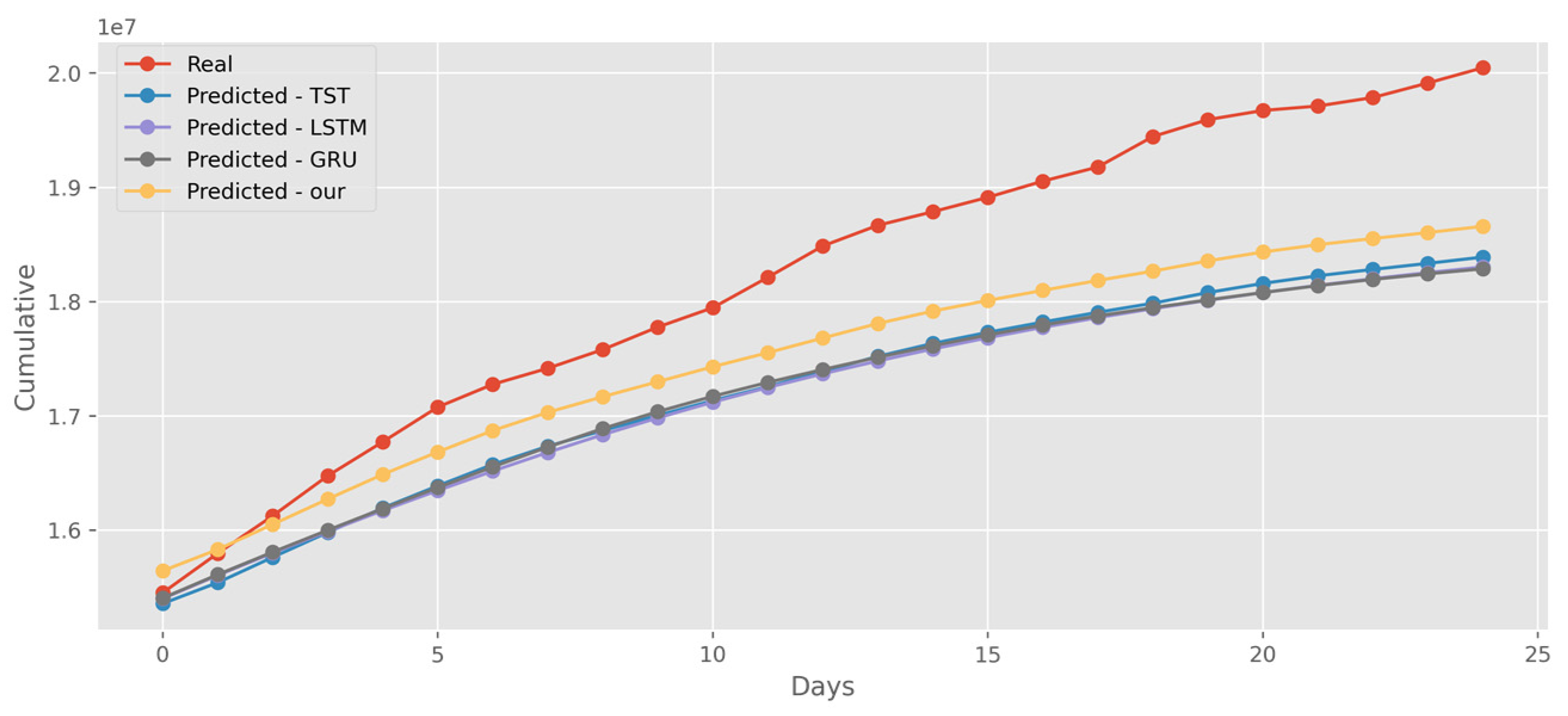
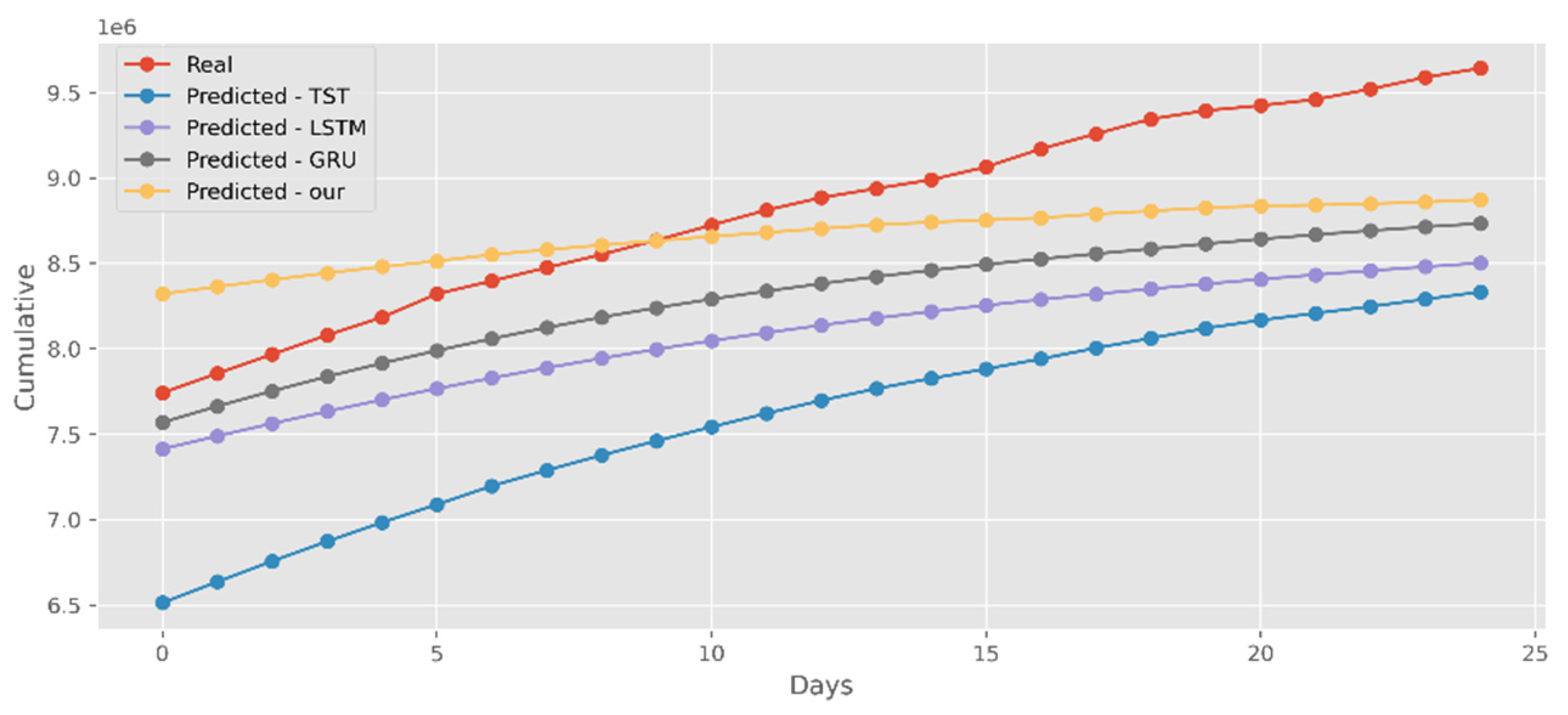
5.4. Forecasting the Number of Administrated Vaccine Doses
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chan, J.F.W.; Yuan, S.; Kok, K.H.; To, K.K.W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.Y.; Poon, R.W.S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Roda, W.C.; Varughese, M.B.; Han, D.; Li, M.Y. Why is it difficult to accurately predict the COVID-19 epidemic? Infect. Dis. Model. 2020, 5, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Z.; Dong, W.; Lu, Y.; Yang, P.; Wang, Q.; Jia, P. Real-time forecasting of hand-foot-and-mouth disease outbreaks using the integrating compartment model and assimilation filtering. Sci. Rep. 2019, 9, 2661. [Google Scholar] [CrossRef]
- Scarpino, S.V.; Petri, G. On the predictability of infectious disease outbreaks. Nat. Commun. 2019, 10, 898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, J.C. Mathematical models of SIR disease spread with combined non-sexual and sexual transmission routes. Infect. Dis. Model. 2017, 2, 35–55. [Google Scholar] [CrossRef] [PubMed]
- Werkman, M.; Green, D.M.; Murray, A.G.; Turnbull, J.F. The effectiveness of fallowing strategies in disease control in salmon aquaculture assessed with an SIS model. Prev. Vet. Med. 2011, 98, 64–73. [Google Scholar] [CrossRef] [Green Version]
- Fast, S.M.; Kim, L.; Cohn, E.L.; Mekaru, S.R.; Brownstein, J.S.; Markuzon, N. Predicting social response to infectious disease outbreaks from internet-based news streams. Ann. Oper. Res. 2018, 263, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Hong, K.J.; Do Shin, S.; Park, G.J.; Kim, S.; Hong, N. Forecasting respiratory infectious outbreaks using ED-based syndromic surveillance for febrile ED visits in a Metropolitan City. Am. J. Emerg. Med. 2019, 37, 183–188. [Google Scholar] [CrossRef] [Green Version]
- Rahimi, I.; Gandomi, A.H.; Asteris, P.G.; Chen, F. Analysis and Prediction of COVID-19 Using SIR, SEIQR, and Machine Learning Models: Australia, Italy, and UK Cases. Information 2021, 12, 109. [Google Scholar] [CrossRef]
- Çolak, A.B. Prediction of infection and death ratio of CoVID-19 virus in Turkey by using artificial neural network (ANN). Coronaviruses 2021, 2, 106–112. [Google Scholar] [CrossRef]
- Chimmula, V.K.R.; Zhang, L. Time series forecasting of COVID-19 transmission in Canada using LSTM networks. Chaos Solitons Fractals 2020, 135, 109864. [Google Scholar] [CrossRef]
- Da Silva, D.B.; Schmidt, D.; da Costa, C.A.; Da Rosa Righi, R.; Eskofier, B. DeepSigns: A predictive model based on Deep Learning for the early detection of patient health deterioration. Expert Syst. Appl. 2021, 165, 113905. [Google Scholar] [CrossRef]
- Çolak, A.B. An experimental study on the comparative analysis of the effect of the number of data on the error rates of artificial neural networks. Int. J. Energy Res. 2021, 45, 478–500. [Google Scholar] [CrossRef]
- Torres, J.F.; Hadjout, D.; Sebaa, A.; Martínez-Álvarez, F.; Troncoso, A. Deep learning for time series forecasting: A survey. Big Data 2021, 9, 3–21. [Google Scholar] [CrossRef]
- Shafiq, A.; Çolak, A.B.; Sindhu, T.N.; Lone, S.A.; Alsubie, A.; Jarad, F. Comparative Study of Artificial Neural Network versus Parametric Method in COVID-19 data Analysis. Results Phys. 2022, 38, 105613. [Google Scholar] [CrossRef]
- Alali, Y.; Harrou, F.; Sun, Y. A proficient approach to forecast COVID-19 spread via optimized dynamic machine learning models. Sci. Rep. 2022, 12, 2467. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, I.; Chen, F.; Gandomi, A.H. A review on COVID-19 forecasting models. Neural Comput. Appl. 2021, 1–11. [Google Scholar] [CrossRef]
- Kim, M.; Kang, J.; Kim, D.; Song, H.; Min, H.; Nam, Y.; Park, D.; Lee, J.G. Hi-covidnet: Deep learning approach to predict inbound COVID-19 patients and case study in South Korea. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Virtual Event, CA, USA, 6–10 July 2020; pp. 3466–3473. [Google Scholar]
- Miralles-Pechuán, L.; Jiménez, F.; Ponce, H.; Martínez-Villaseñor, L. A methodology based on deep q-learning/genetic algorithms for optimizing COVID-19 pandemic government actions. In Proceedings of the 29th ACM International Conference on Information & Knowledge Management, Virtual Event, Ireland, 19–23 October 2020; pp. 1135–1144. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M.; Furht, B. Deep Learning applications for COVID-19. J. Big Data 2021, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Farsani, R.M.; Pazouki, E. A transformer self-attention model for time series forecasting. J. Electr. Comput. Eng. Innov. 2021, 9, 1–10. [Google Scholar]
- La Gatta, V.; Moscato, V.; Postiglione, M.; Sperli, G. An epidemiological neural network exploiting dynamic graph structured data applied to the COVID-19 outbreak. IEEE Trans. Big Data 2021, 7, 45–55. [Google Scholar] [CrossRef]
- Cao, D.; Wang, Y.; Duan, J.; Zhang, C.; Zhu, X.; Huang, C.; Tong, Y.; Xu, B.; Bai, J.; Tong, J.; et al. Spectral temporal graph neural network for multivariate time-series forecasting. arXiv 2021, arXiv:2103.07719. [Google Scholar]
- Nytimes. Coronavirus (COVID-19) Data in the United States. 2021. Available online: https://github.com/nytimes/covid-19-data (accessed on 8 June 2022).
- Srk. Novel Corona Virus 2019 Dataset. 2021. Available online: https://www.kaggle.com/sudalairajkumar/novel-corona-virus-2019-dataset (accessed on 8 June 2022).
- Edouard, M. State-By-State Data on COVID-19 Vaccinations in the United States. 2021. Available online: https://ourworldindata.org/us-states-vaccinations (accessed on 8 June 2022).
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. arXiv 2017, arXiv:1706.03762. [Google Scholar]
- Bresson, X.; Laurent, T. Residual gated graph convnets. arXiv 2017, arXiv:1711.07553. [Google Scholar]
- Wang, Y.; Chen, Z.B. Dynamic graph Conv-LSTM model with dynamic positional encoding for the large-scale traveling salesman problem. Math. Biosci. Eng. 2022, 19, 9730–9748. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning (ICML), Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2016, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Hamilton, W.; Ying, Z.; Leskovec, J. Inductive representation learning on large graphs. Presented at Advances in Neural Information Processing Systems 30: Annual Conference on Neural Information Processing Systems 2017, Long Beach, CA, USA, 4–9 December 2017; pp. 1024–1034. [Google Scholar]
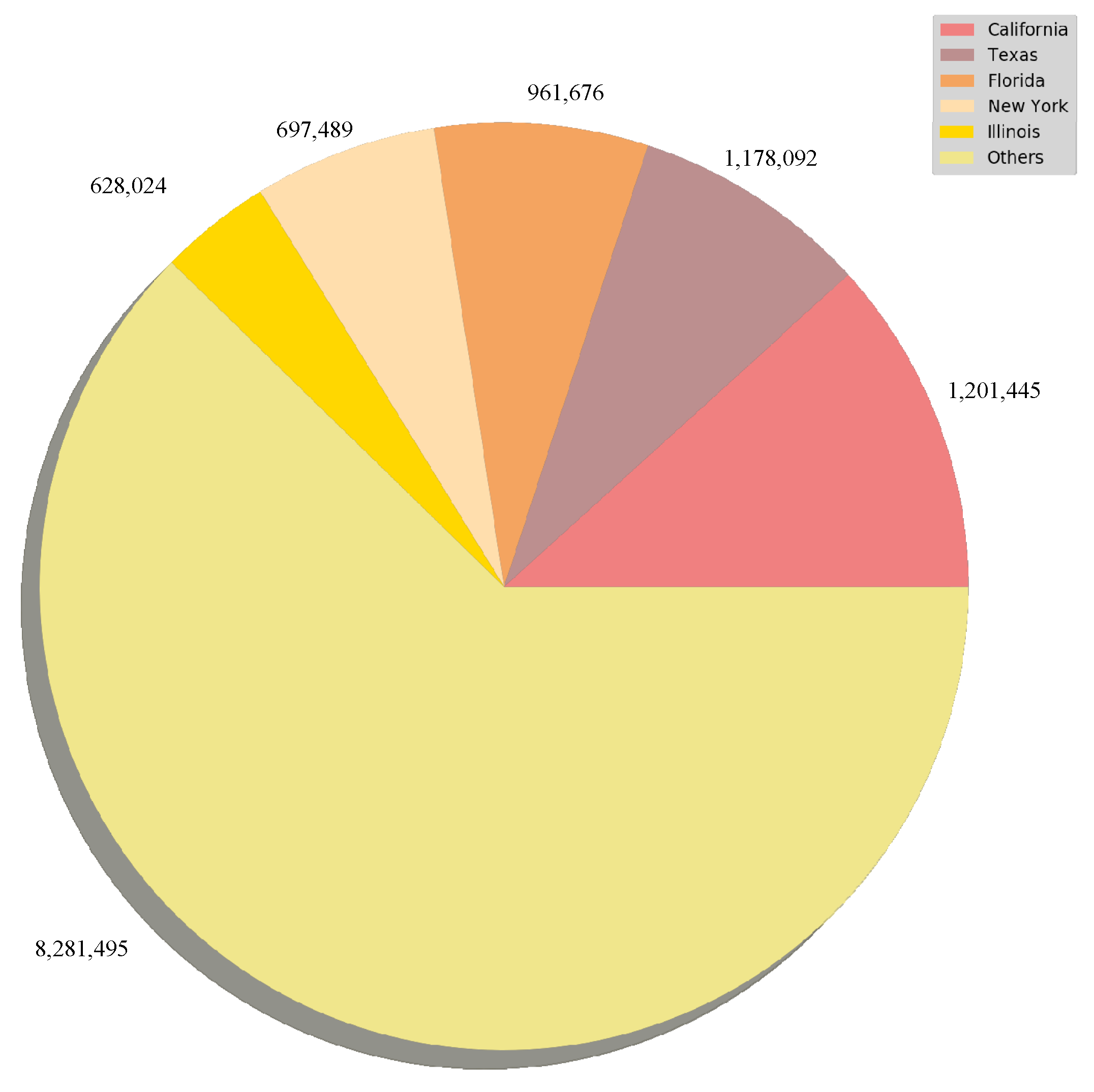







| Date | State | Cases | Deaths |
|---|---|---|---|
| 22 April 2021 | Texas | 2,868,207 | 49,984 |
| 22 April 2021 | Utah | 394,398 | 2178 |
| 22 April 2021 | Vermont | 22,325 | 243 |
| 22 April 2021 | Virgin Islands | 3068 | 27 |
| 22 April 2021 | Virginia | 650,981 | 10,653 |
| 22 April 2021 | Washington | 393,514 | 5472 |
| 22 April 2021 | West Virginia | 150,288 | 2808 |
| 22 April 2021 | Wisconsin | 654,681 | 7438 |
| 22 April 2021 | Wyoming | 57,613 | 705 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Wang, Y.; Ma, K. Integrating Transformer and GCN for COVID-19 Forecasting. Sustainability 2022, 14, 10393. https://doi.org/10.3390/su141610393
Li Y, Wang Y, Ma K. Integrating Transformer and GCN for COVID-19 Forecasting. Sustainability. 2022; 14(16):10393. https://doi.org/10.3390/su141610393
Chicago/Turabian StyleLi, Yulan, Yang Wang, and Kun Ma. 2022. "Integrating Transformer and GCN for COVID-19 Forecasting" Sustainability 14, no. 16: 10393. https://doi.org/10.3390/su141610393
APA StyleLi, Y., Wang, Y., & Ma, K. (2022). Integrating Transformer and GCN for COVID-19 Forecasting. Sustainability, 14(16), 10393. https://doi.org/10.3390/su141610393





