Arsenic-Resistant Plant Growth Promoting Pseudoxanthomonas mexicana S254 and Stenotrophomonas maltophilia S255 Isolated from Agriculture Soil Contaminated by Industrial Effluent
Abstract
:1. Introduction
2. Materials and Methods
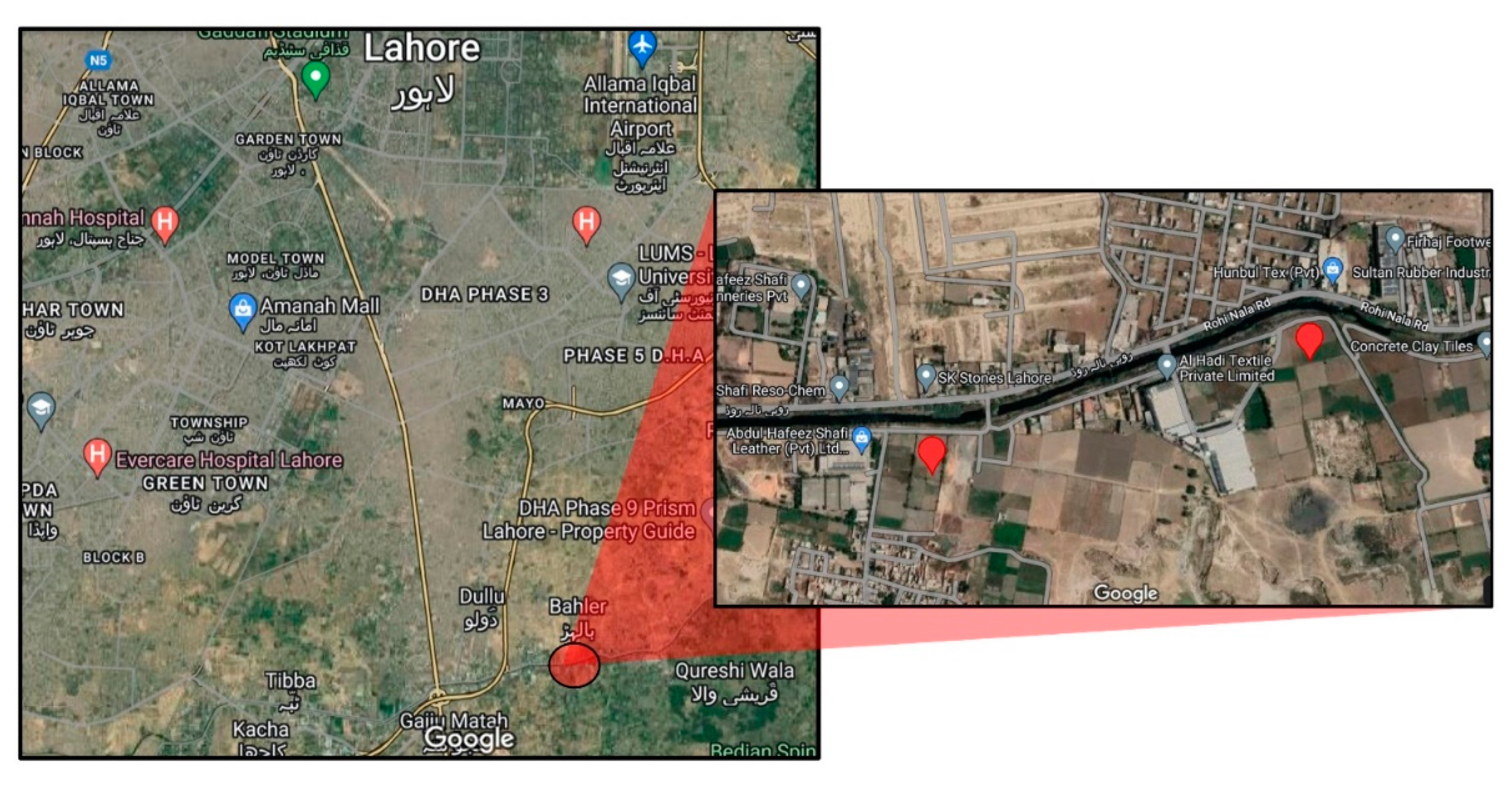
2.1. Soil Sampling
2.2. Isolation of As-Resistant Bacteria
2.3. Determination of As Resistance by the Isolates
2.4. Determination of Cross Element Resistance of the Isolates
2.5. Optimization of Culture Conditions
2.6. Qualitative Analysis of Bacterial Cultures for As Oxidation/Reduction
2.7. Estimation of Plant Growth-Promoting Activities
2.7.1. Phosphate Solubilization Activity
2.7.2. Hydrogen Cyanide (HCN) Production
2.7.3. Nitrogen Fixation
2.7.4. Auxin Estimation
2.8. 16S rRNA Gene Sequencing and Phylogenetic Analysis
2.9. Evaluation of Plant Growth in the Presence of the Bacterial Isolates
3. Statistical Analysis
4. Results
4.1. Isolation and Characterization of As-Resistant Bacteria
4.2. Phylogenetic Analysis of the Selected Isolates
4.3. Minimal Inhibitory Concentration (MIC) of the Isolates
4.4. Cross Element Resistance of the Isolates
4.5. Optimization for pH and Temperature
4.6. Qualitative Analysis of Bacterial Cultures for As(V) Reduction
4.7. Phosphate Solubilization, Hydrogen Cyanide (HCN) Production, Nitrogen Fixation, and Auxin Estimation
4.8. Effect of the Bacterial Isolates on Plant Growth
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bowell, R.J.; Alpers, C.N.; Jamieson, H.E.; Nordstrom, D.K.; Majzlan, J. The Environmental Geochemistry of Arsenic—An Overview. Rev. Mineral. Geochem. 2014, 79, 1–16. [Google Scholar] [CrossRef]
- Ventura-Lima, J.; Bogo, M.R.; Monserrat, J.M. Arsenic toxicity in mammals and aquatic animals: A comparative biochemical approach. Ecotoxicol. Environ. Saf. 2011, 74, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, J.; Lou, B.; Wu, R.; Wang, G.; Lu, C.; Wang, H.; Pi, J.; Xu, Y. The Role of Reactive Oxygen Species in Arsenic Toxicity. Biomolecules 2020, 10, 240. [Google Scholar] [CrossRef]
- Hughes, M.F.; Beck, B.D.; Chen, Y.; Lewis, A.S.; Thomas, D.J. Arsenic Exposure and Toxicology: A Historical Perspective. Toxicol. Sci. 2011, 123, 305–332. [Google Scholar] [CrossRef] [PubMed]
- Majeed, S.; Rashid, S.; Qadir, A.; Mackay, C.; Hayat, F. Spatial patterns of pollutants in water of metropolitan drain in Lahore, Pakistan, using multivariate statistical techniques. Environ. Monit. Assess. 2018, 190, 128. [Google Scholar] [CrossRef]
- Podgorski, J.E.; Eqani, S.A.M.A.S.; Khanam, T.; Ullah, R.; Shen, H.; Berg, M. Extensive arsenic contamination in high-pH unconfined aquifers in the Indus Valley. Sci. Adv. 2017, 3, e1700935. [Google Scholar] [CrossRef]
- Nawaz, A.R.; Anwar, U.; Ahmad, S. Assessing the Economic Impact of Tanneries’ Pollutants in Pakistan. J. Econ. Impact 2021, 3, 98–106. [Google Scholar] [CrossRef]
- Alka, S.; Shahir, S.; Ibrahim, N.; Ndejiko, M.J.; Vo, D.-V.N.; Manan, F.A. Arsenic removal technologies and future trends: A mini review. J. Clean. Prod. 2021, 278, 123805. [Google Scholar] [CrossRef]
- Saleem, H.; ul Ain Kokab, Q.; Rehman, Y. Arsenic respiration and detoxification by purple non-sulphur bacteria under anaerobic conditions. C. R. Biol. 2019, 342, 101–107. [Google Scholar] [CrossRef]
- Mohsin, H.; Asif, A.; Rehman, Y. Anoxic growth optimization for metal respiration and photobiological hydrogen production by arsenic-resistant Rhodopseudomonas and Rhodobacter species. J. Basic Microbiol. 2019, 59, 1208–1216. [Google Scholar] [CrossRef]
- Mohsin, H.; Shafique, M.; Rehman, Y. Genes and Biochemical Pathways Involved in Microbial Transformation of Arsenic. In Arsenic Toxicity: Challenges and Solutions; Kumar, N., Ed.; Springer: Singapore, 2021; pp. 391–413. [Google Scholar]
- Nookongbut, P.; Kantachote, D.; Krishnan, K.; Megharaj, M. Arsenic resistance genes of As-resistant purple nonsulfur bacteria isolated from As-contaminated sites for bioremediation application. J. Basic Microbiol. 2017, 57, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Shafique, M.; Jawaid, A.; Rehman, Y. Redox biotransformation of arsenic along with plant growth promotion by multi-metal resistance Pseudomonas sp. MX6. Comptes Rendus Biol. 2017, 340, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Qamar, N.; Rehman, Y.; Hasnain, S. Arsenic-resistant and plant growth-promoting Firmicutes and γ-Proteobacteria species from industrially polluted irrigation water and corresponding cropland. J. Appl. Microbiol. 2017, 123, 748–758. [Google Scholar] [CrossRef] [PubMed]
- Cavalca, L.; Zanchi, R.; Corsini, A.; Colombo, M.; Romagnoli, C.; Canzi, E.; Andreoni, V. Arsenic-resistant bacteria associated with roots of the wild Cirsium arvense (L.) plant from an arsenic polluted soil, and screening of potential plant growth-promoting characteristics. Syst. Appl. Microbiol. 2010, 33, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, M.K.; Yadav, P.; Shukla, A.; Srivastava, S. Utilizing the Potential of Microorganisms for Managing Arsenic Contamination: A Feasible and Sustainable Approach. Front. Environ. Sci. 2018, 6, 24. [Google Scholar] [CrossRef]
- Cappuccino, J.G.; Sherman, N. Microbiology: A Laboratory Manual; Pearson Benjamin Cummings: San Francisco, CA, USA, 2007. [Google Scholar]
- Saba; Rehman, Y.; Ahmed, M.; Sabri, A.N. Potential role of bacterial extracellular polymeric substances as biosorbent material for arsenic bioremediation. Bioremediat. J. 2019, 23, 72–81. [Google Scholar] [CrossRef]
- Salmassi, T.M.; Venkateswaren, K.; Satomi, M.; Newman, D.K.; Hering, J.G. Oxidation of arsenite by Agrobacterium albertimagni, AOL15, sp. nov. isolated from Hot Creek, California. Geomicrobiol. J. 2002, 19, 53–66. [Google Scholar] [CrossRef]
- Pikovskaya, R.I. Mobilization of phosphorus in soil connection with the vital activity of some microbial species. Microbiologiya 1948, 17, 362–370. [Google Scholar]
- Chung, H.; Park, M.; Madhaiyan, M.; Seshadri, S.; Song, J.; Cho, H.; Sa, T. Isolation and characterization of phosphate solubilizing bacteria from the rhizosphere of crop plants of Korea. Soil Biol. Biochem. 2005, 37, 1970–1974. [Google Scholar] [CrossRef]
- Batool, F.; Rehman, Y.; Hasnain, S. Phylloplane associated plant bacteria of commercially superior wheat varieties exhibit superior plant growth promoting abilities. Front. Life Sci. 2016, 9, 313–322. [Google Scholar] [CrossRef]
- Gordon, S.A.; Weber, R.P. Colorimetric estimation of indoleacetic acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernbom, N.; Ng, Y.Y.; Kjelleberg, S.; Harder, T.; Gram, L. Marine bacteria from Danish coastal waters show antifouling activity against the marine fouling bacterium Pseudoalteromonas sp. strain S91 and zoospores of the green alga Ulva australis independent of bacteriocidal activity. Appl. Environ. Microbiol. 2011, 77, 8557–8567. [Google Scholar] [CrossRef]
- Porsby, C.H.; Nielsen, K.F.; Gram, L. Phaeobacter and Ruegeria Species of the Roseobacter clade colonize separate niches in a Danish Turbot (Scophthalmus maximus)-rearing farm and antagonize Vibrio anguillarum under different growth conditions. Appl. Environ. Microbiol. 2008, 74, 7356–7364. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Khan, A.G. Relationships between chromium biomagnification ratio, accumulation factor, and mycorrhizae in plants growing on tannery effluent-polluted soil. Environ. Int. 2001, 26, 417–423. [Google Scholar] [CrossRef]
- Azizur Rahman, M.; Hasegawa, H.; Mahfuzur Rahman, M.; Nazrul Islam, M.; Majid Miah, M.A.; Tasmen, A. Effect of arsenic on photosynthesis, growth and yield of five widely cultivated rice (Oryza sativa L.) varieties in Bangladesh. Chemosphere 2007, 67, 1072–1079. [Google Scholar] [CrossRef]
- Clemens, S.; Ma, J.F. Toxic heavy metal and metalloid accumulation in crop plants and foods. Annu. Rev. Plant Biol. 2016, 67, 489–512. [Google Scholar] [CrossRef]
- Bachate, S.; Cavalca, L.; Andreoni, V. Arsenic resistant bacteria isolated from agricultural soils of Bangladesh and characterization of arsenate-reducing strains. J. Appl. Microbiol. 2009, 107, 145–156. [Google Scholar] [CrossRef]
- Sarkar, A.; Kazy, S.K.; Sar, P. Studies on arsenic transforming groundwater bacteria and their role in arsenic release from subsurface sediment. Environ. Sci. Pollut. Res. 2014, 21, 8645–8662. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, G.K.; Wang, H.; Zhang, Y.; Tian, Z.; Chai, W.; Lu, H. Class 1 In-Tn5393c array contributed to antibiotic resistance of non-pathogenic Pseudoxanthomonas mexicana isolated from a wastewater bioreactor treating streptomycin. Sci. Total Environ. 2022, 821, 153537. [Google Scholar] [CrossRef] [PubMed]
- An, S.-Q.; Berg, G. Stenotrophomonas maltophilia. Trends Microbiol. 2018, 26, 637–638. [Google Scholar] [CrossRef] [PubMed]
- Botes, E.; Van Heerden, E.; Litthauer, D. Hyper-resistance to arsenic in bacteria isolated from an antimony mine in South Africa. S. Afr. J. Sci. 2007, 103, 279–281. [Google Scholar]
- Xiong, W.; Yin, C.; Wang, Y.; Lin, S.; Deng, Z.; Liang, R. Characterization of an efficient estrogen-degrading bacterium Stenotrophomonas maltophilia SJTH1 in saline-, alkaline-, heavy metal-contained environments or solid soil and identification of four 17β-estradiol-oxidizing dehydrogenases. J. Hazard. Mater. 2020, 385, 121616. [Google Scholar] [CrossRef]
- Baldiris, R.; Acosta-Tapia, N.; Montes, A.; Hernández, J.; Vivas-Reyes, R. Reduction of Hexavalent Chromium and Detection of Chromate Reductase (ChrR) in Stenotrophomonas maltophilia. Molecules 2018, 23, 406. [Google Scholar] [CrossRef]
- Liaquat, F.; Munis, M.F.H.; Arif, S.; Haroon, U.; Shengquan, C.; Qunlu, L. Cd-tolerant SY-2 strain of Stenotrophomonas maltophilia: A potential PGPR, isolated from the Nanjing mining area in China. 3 Biotech 2020, 10, 519. [Google Scholar] [CrossRef]
- Gopi, K.; Jinal, H.N.; Prittesh, P.; Kartik, V.P.; Amaresan, N. Effect of copper-resistant Stenotrophomonas maltophilia on maize (Zea mays) growth, physiological properties, and copper accumulation: Potential for phytoremediation into biofortification. Int. J. Phytoremediat. 2020, 22, 662–668. [Google Scholar] [CrossRef]
- Kumar, S.; Bansal, K.; Patil, P.P.; Kaur, A.; Kaur, S.; Jaswal, V.; Gautam, V.; Patil, P.B. Genomic insights into evolution of extensive drug resistance in Stenotrophomonas maltophilia complex. Genomics 2020, 112, 4171–4178. [Google Scholar] [CrossRef]
- Aslam, F.; Yasmin, A.; Thomas, T. Essential Gene Clusters Identified in Stenotrophomonas MB339 for Multiple Metal/Antibiotic Resistance and Xenobiotic Degradation. Curr. Microbiol. 2018, 75, 1484–1492. [Google Scholar] [CrossRef]
- Raman, N.M.; Asokan, S.; Shobana Sundari, N.; Ramasamy, S. Bioremediation of chromium(VI) by Stenotrophomonas maltophilia isolated from tannery effluent. Int. J. Environ. Sci. Technol. 2018, 15, 207–216. [Google Scholar] [CrossRef]
- Patel, V.; Cheturvedula, S.; Madamwar, D. Phenanthrene degradation by Pseudoxanthomonas sp. DMVP2 isolated from hydrocarbon contaminated sediment of Amlakhadi canal, Gujarat, India. J. Hazard. Mater. 2012, 201–202, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gao, M.; Wei, J.; Ma, K.; Zhang, J.; Yang, Y.; Yu, S. Extracellular polymeric substances, microbial activity and microbial community of biofilm and suspended sludge at different divalent cadmium concentrations. Bioresour. Technol. 2016, 205, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Muehe, E.M.; Weigold, P.; Adaktylou, I.J.; Planer-Friedrich, B.; Kraemer, U.; Kappler, A.; Behrens, S.; Lovell, C.R. Rhizosphere Microbial Community Composition Affects Cadmium and Zinc Uptake by the Metal-Hyperaccumulating Plant Arabidopsis halleri. Appl. Environ. Microbiol. 2015, 81, 2173–2181. [Google Scholar] [CrossRef] [PubMed]
- Pathma, J.; Sakthivel, N. Molecular and functional characterization of bacteria isolated from straw and goat manure based vermicompost. Appl. Soil Ecol. 2013, 70, 33–47. [Google Scholar] [CrossRef]
- Lampis, S.; Santi, C.; Ciurli, A.; Andreolli, M.; Vallini, G. Promotion of arsenic phytoextraction efficiency in the fern Pteris vittata by the inoculation of As-resistant bacteria: A soil bioremediation perspective. Front. Plant Sci. 2015, 6, 80. [Google Scholar] [CrossRef]
- Liaqat, F.; Eltem, R. Identification and characterization of endophytic bacteria isolated from in vitro cultures of peach and pear rootstocks. 3 Biotech 2016, 6, 120. [Google Scholar] [CrossRef]
- Ulrich, K.; Kube, M.; Becker, R.; Schneck, V.; Ulrich, A. Genomic Analysis of the Endophytic Stenotrophomonas Strain 169 Reveals Features Related to Plant-Growth Promotion and Stress Tolerance. Front. Microbiol. 2021, 12, 1542. [Google Scholar] [CrossRef]




| Strains | Metals | |||||
|---|---|---|---|---|---|---|
| Co (1 mM) | Se (1 mM) | Zn (1 mM) | Ni (1 mM) | Cd (1 mM) | Cr (1 mM) | |
| S252 | + | + | + | + | − | + |
| S253 | + | + | + | + | + | + |
| S254 | ++ | +++ | +++ | +++ | − | + |
| S255 | +++ | +++ | +++ | +++ | +++ | +++ |
| S257 | + | + | + | + | − | + |
| S258 | + | + | + | + | − | + |
| S4E1 | + | + | + | + | − | + |
| S43 | + | + | + | + | − | + |
| S46 | + | + | + | + | − | + |
| S48 | + | + | + | + | − | + |
| Strains | Plant Growth-Promoting Activities | |||
|---|---|---|---|---|
| HCN Production | Phosphate Solubilization | Auxin Production | Nitrogen Fixation | |
| S252 | + | − | +++ | − |
| S253 | − | − | +++ | − |
| S254 | +++ | − | + | + |
| S255 | ++ | − | +++ | + |
| S257 | − | − | ++ | − |
| S258 | − | − | + | − |
| S4E1 | + | − | + | − |
| S43 | + | − | + | − |
| S46 | + | − | + | + |
| S48 | − | − | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huda, N.u.; Tanvir, R.; Badar, J.; Ali, I.; Rehman, Y. Arsenic-Resistant Plant Growth Promoting Pseudoxanthomonas mexicana S254 and Stenotrophomonas maltophilia S255 Isolated from Agriculture Soil Contaminated by Industrial Effluent. Sustainability 2022, 14, 10697. https://doi.org/10.3390/su141710697
Huda Nu, Tanvir R, Badar J, Ali I, Rehman Y. Arsenic-Resistant Plant Growth Promoting Pseudoxanthomonas mexicana S254 and Stenotrophomonas maltophilia S255 Isolated from Agriculture Soil Contaminated by Industrial Effluent. Sustainability. 2022; 14(17):10697. https://doi.org/10.3390/su141710697
Chicago/Turabian StyleHuda, Noor ul, Rabia Tanvir, Javaria Badar, Iftikhar Ali, and Yasir Rehman. 2022. "Arsenic-Resistant Plant Growth Promoting Pseudoxanthomonas mexicana S254 and Stenotrophomonas maltophilia S255 Isolated from Agriculture Soil Contaminated by Industrial Effluent" Sustainability 14, no. 17: 10697. https://doi.org/10.3390/su141710697
APA StyleHuda, N. u., Tanvir, R., Badar, J., Ali, I., & Rehman, Y. (2022). Arsenic-Resistant Plant Growth Promoting Pseudoxanthomonas mexicana S254 and Stenotrophomonas maltophilia S255 Isolated from Agriculture Soil Contaminated by Industrial Effluent. Sustainability, 14(17), 10697. https://doi.org/10.3390/su141710697







