Evaluation of Reference Genes for Gene Expression Analysis in Eichhornia crassipes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials, Treatments, and Sampling
2.2. RNA Isolation and cDNA Synthesis
2.3. Candidate Reference Genes, Primer Design
2.4. qRT-PCR and Primer Specificity Test
2.5. Stability Analysis of Candidate Reference Genes and Determination of the Optimal Number of Reference Genes
3. Results
3.1. Evaluation of Primer Specificity
3.2. Expression Patterns of Candidate Reference Genes
3.3. Expression Stability of Candidate Reference Genes
3.3.1. GeNorm Analysis
3.3.2. NormFinder Analysis
3.3.3. BestKeeper Analysis
3.3.4. RefFinder Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 622, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Beaulieu, J.; Huggett, J.; Jaggi, R.; Kibenge, F.S.B.; Olsvik, P.A.; Penning, L.C.; Toegel, S. MIQE précis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol. Biol. 2010, 11, 74. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef]
- Thellin, O.; Zorzi, W.; Lakaye, B.; DeBorman, B.; Coumans, B. Housekeeping genes as internal standards: Use and limits. J. Biotechnol. 1999, 75, 291–295. [Google Scholar] [CrossRef]
- Bartlomiej, K.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar]
- Joseph, J.; Poolakkalody, N.J.; Shah, J. Plant reference genes for development and stress response studies. J. Biosci. 2018, 43, 173–187. [Google Scholar] [CrossRef]
- Gutierrez, L.; Mauriat, M.; Guénin, S.; Pelloux, J.; Lefebvre, J.; Van Wuytswinkel, O. The lack of a systematic validation of reference genes: A serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 2008, 6, 609–618. [Google Scholar] [CrossRef]
- Gao, K.; Khan, W.U.; Li, J.; Huang, S.; Yang, X.; Guo, T.; Guo, B.; Wu, R.; An, X. Identification and validation of reliable reference genes for gene expression studies in Koelreuteria paniculata. Genes 2022, 13, 714. [Google Scholar] [CrossRef]
- Penfound, W.T.; Earle, T.T. The biology of the water hyacinth. Ecol. Monogr. 1948, 18, 447–472. [Google Scholar] [CrossRef]
- Sharma, A.; Aggarwal, N.K.; Saini, A.; Yadav, A. Beyond biocontrol: Water hyacinth-opportunities and challenges. J. Environ. Sci. Technol. 2016, 9, 26–48. [Google Scholar] [CrossRef]
- Taylor, P.; Street, M.; Wt, L.; Aboul-Enein, A.M.; Al-Abd, A.M.; Shalaby, E.; Abul-Ela, F.; Nasr-Allah, A.A.; Mahmoud, A.M.; El-Shemy, H.A. Eichhornia crassipes (Mart) solms from water parasite to potential medicinal remedy. Plant Signal. Behav. 2011, 6, 834–836. [Google Scholar]
- Ben Bakrim, W.; Ezzariai, A.; Karouach, F.; Sobeh, M.; Kibret, M.; Hafidi, M.; Kouisni, L.; Yasri, A. Eichhornia crassipes (Mart.) Solms: A comprehensive review of its chemical composition, traditional use, and value-added products. Front. Pharmacol. 2022, 13, 842511. [Google Scholar] [CrossRef] [PubMed]
- Ayanda, O.I.; Ajayi, T.; Asuwaju, F.P. Eichhornia crassipes (Mart.) Solms: Uses, challenges, threats, and prospects. Sci. World J. 2020, 2020, 3452172. [Google Scholar] [CrossRef]
- Malik, A. Environmental challenge vis a vis opportunity: The case of water hyacinth. Environ. Int. 2007, 33, 122–138. [Google Scholar] [CrossRef]
- Villamagna, A.M.; Murphy, B.R. Ecological and socio-economic impacts of invasive water hyacinth (Eichhornia crassipes): A review. Freshw. Biol. 2010, 55, 282–298. [Google Scholar] [CrossRef]
- Mujere, N. Water Hyacinth: Characteristics, Problems, Control Options, and Beneficial Uses. In Impact of Water Pollution on Human Health and Environmental Sustainability; IGI Global: Hershey, PA, USA, 2015; pp. 343–361. ISBN 9781466695603. [Google Scholar]
- Gaurav, G.K.; Mehmood, T.; Cheng, L.; Klemeš, J.J.; Shrivastava, D.K. Water hyacinth as a biomass: A review. J. Clean. Prod. 2020, 277, 122214. [Google Scholar] [CrossRef]
- Ilo, O.P.; Simatele, M.D.; Nkomo, S.L.; Mkhize, N.M.; Prabhu, N.G. The benefits of water hyacinth (Eichhornia crassipes) for Southern Africa: A review. Sustainability 2020, 12, 9222. [Google Scholar] [CrossRef]
- Fu, M.; Jiang, L.; Yan, G. Identificatin of GS1a, GS1b and GS1c genes from Eichhornia crassipes and their transcript analysis in response to different nitrogen sources. Pak. J. Bot. 2018, 50, 2197–2204. [Google Scholar]
- Fu, M.; Jiang, L.; Li, Y.; Yan, G.; Zheng, L.; Peng, J. Identification of gene fragments related to nitrogen deficiency in Eichhornia crassipes (Pontederiaceae). Int. J. Trop. Biol. 2014, 62, 1637–1648. [Google Scholar] [CrossRef]
- Zhong, Y.; Lu, X.; Deng, Z.; Lu, Z.; Fu, M. A 1232 bp upstream sequence of glutamine synthetase 1b from Eichhornia crassipes is a root-preferential promoter sequence. BMC Plant Biol. 2021, 21, 66. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR Data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Poppe, B.; Van Roy, N.; Paepe, A. De Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 341. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D. MiRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Xu, J.; Audenaert, K.; Höfte, M.; De Vleesschauwer, D. Abscisic acid promotes susceptibility to the rice leaf blight pathogen Xanthomonas oryzae pv. oryzae by suppressing salicylic acid-mediated defenses. PLoS ONE 2013, 8, e67413. [Google Scholar]
- Ito, S.; Umehara, M.; Hanada, A.; Yamaguchi, S.; Asami, T. Effects of strigolactone-biosynthesis inhibitor TIS108 on Arabidopsis. Plant Signal. Behav. 2013, 8, e24193. [Google Scholar] [CrossRef]
- Xu, J. Global Switches and Fine-Tuning: The Multifaceted Role of Plant Hormones in Regulating Rice Immunity against the Leaf Blight Pathogen Xanthomonas oryzae pv. oryzae; Ghent University: Ghent, Belgium, 2014. [Google Scholar]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.-R. Eleven golden rules of quantitative RT-PCR. Plant Cell 2008, 20, 1736–1737. [Google Scholar] [CrossRef]
- Wan, H.; Zhao, Z.; Qian, C.; Sui, Y.; Malik, A.A.; Chen, J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 2010, 399, 257–261. [Google Scholar] [CrossRef]
- Wang, B.; Du, H.; Yao, Z.; Ren, C.; Ma, L. Validation of reference genes for accurate normalization of gene expression with quantitative real-time PCR in Haloxylon ammodendron under different abiotic stresses. Physiol. Mol. Biol. Plants 2018, 24, 455–463. [Google Scholar] [CrossRef]
- Liu, H.; Lu, Y.; Wang, X.; Wang, X.; Li, R.; Lu, C.; Lan, X.; Chen, Y. Selection and validation of reference genes for RT-qPCR analysis in tibetan medicinal plant Saussurea Laniceps callus under abiotic stresses and hormone treatments. Genes 2022, 13, 904. [Google Scholar] [CrossRef]
- Tu, C.; Xu, P.; Han, R.; Luo, J.; Xu, L. Defining suitable reference genes for qRT-PCR in Plagiodera versicolora (Coleoptera: Chrysomelidae) under different biotic or abiotic conditions. Agronomy 2022, 12, 1192. [Google Scholar] [CrossRef]
- Yang, J.; Han, F.; Yang, L.; Wang, J.; Jin, F.; Luo, A.; Zhao, F. Identification of reference genes for RT-qPCR analysis in Gleditsia microphylla under abiotic stress and hormone treatment. Genes 2022, 13, 1227. [Google Scholar] [CrossRef]
- Zhou, F.; Chen, Y.; Wu, H.; Yin, T. A selection of reliable reference genes for gene expression analysis in the female and male flowers of Salix suchowensis. Plants 2022, 11, 647. [Google Scholar] [CrossRef]
- Rodríguez-Parra, A.; Picazo-Aragonés, J.; Balao, F. Evaluation of reference genes in the polyploid complex. Plants 2022, 11, 518. [Google Scholar] [CrossRef]
- Su, X.; Lu, L.; Li, Y.; Zhen, C.; Hu, G.; Jiang, K.; Yan, Y.; Xu, Y.; Wang, G.; Shi, M.; et al. Reference gene selection for quantitative real-time PCR (qRT-PCR) expression analysis in Galium aparine L. PLoS ONE 2020, 15, e0226668. [Google Scholar] [CrossRef]
- Watson, M.A.; Carrier, J.C.; Cook, G.L. Effects oF exogenously supplied gibberellic acid (GA3) on paterns of water hyacinth development. Aquat. Bot. 1982, 13, 57–68. [Google Scholar] [CrossRef]
- Pieterse, A.H.; Aris, J.J.A.M.; Butter, M.E. Inhibition of float formation in water hyacinth by gibberellic acid. Nature 1976, 260, 423–424. [Google Scholar] [CrossRef]
- Reddy, K.R.; Agami, M.; Tucker, J.C. Influence of phosphorus on growth and nutrient storage by water hyacinth (Eichhornia crassipes (Mart.) Solms) plants. Aquat. Bot. 1990, 37, 355–365. [Google Scholar] [CrossRef]
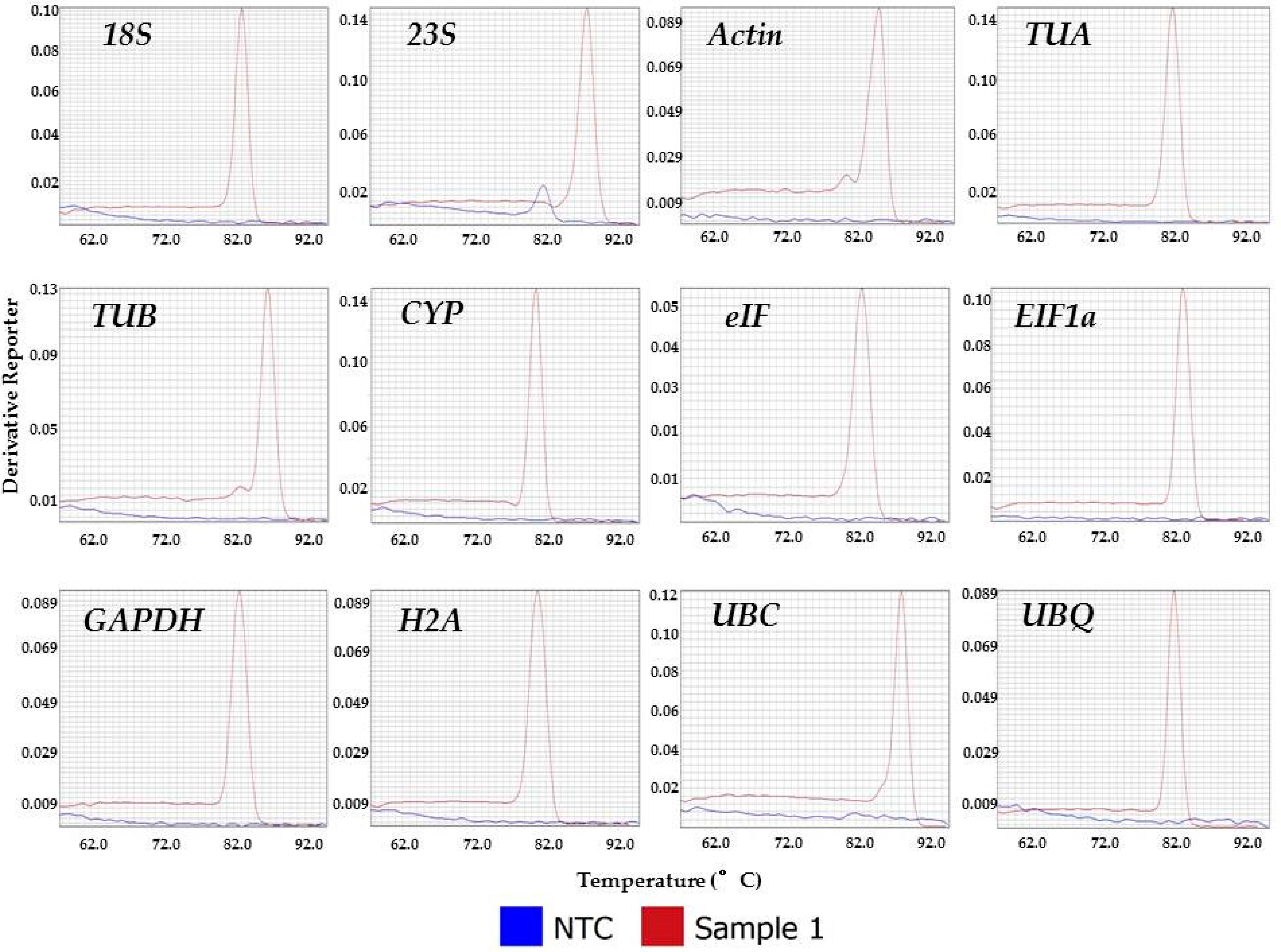
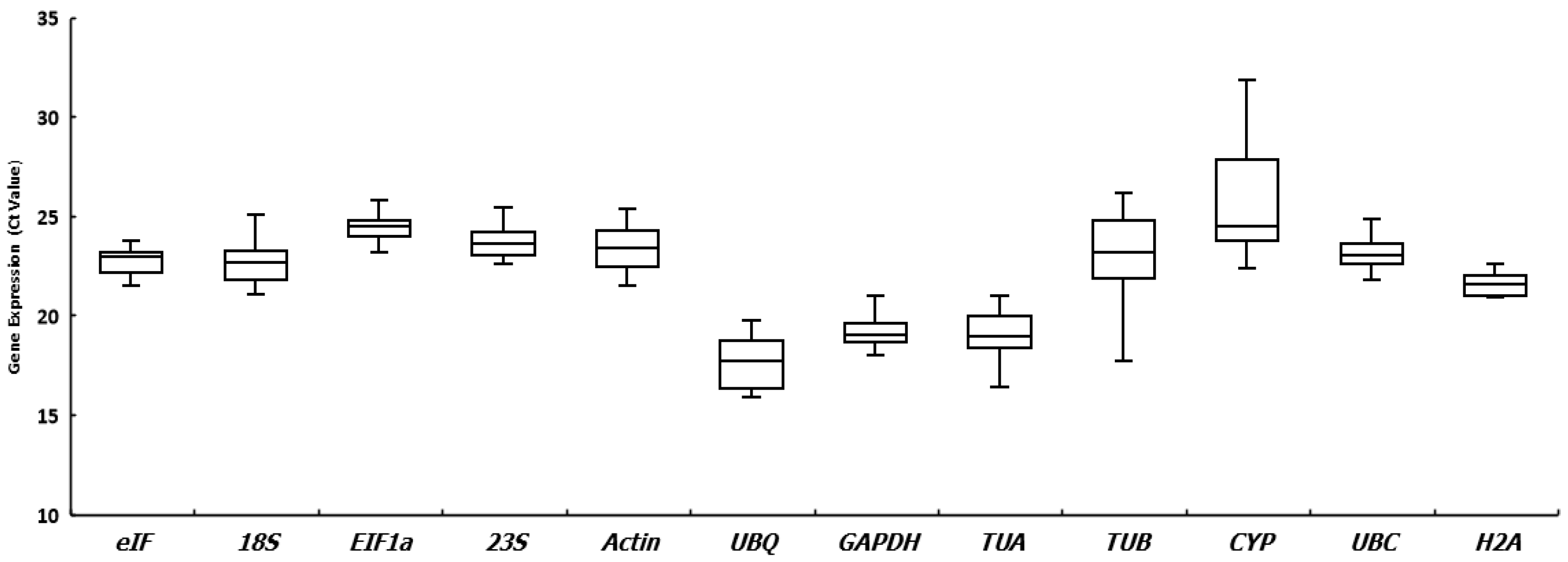

| Gene Name | Gene Product | Primer Sequence 5′–3′ | Annealing Temp (°C) | Amplicon Length (bp) | Tm(°C) |
|---|---|---|---|---|---|
| 18S | 18S ribosomal RNA | For: AGTGGCTATGCTATGCTGACAAGTC | 82.95 | 144 | 58.96 |
| Rev: TCATCTCCCTCTGGTCCATGTTCTC | 60.20 | ||||
| 23S | 23S ribosomal RNA | For: CACACCGTCTTGGAGGACATGC | 87.72 | 81 | 61.30 |
| Rev: AGGAGCGGAGAAGAACGAGGAC | 61.49 | ||||
| Actin | Actin | For: CTCATTGTGGACTGCGGCTTCTC | 84.70 | 85 | 60.98 |
| Rev: CGATACGACGGGCAGCATAGTTG | 60.69 | ||||
| TUA | Tubulin alpha | For: ACACAGCGATGAGAATGAGCAAGG | 81.68 | 128 | 59.72 |
| Rev: GGAGCAGAAGGAGCAGATGATGATG | 59.81 | ||||
| TUB | Tubulin beta | For: AGACCACCTCCCAGAACTTGCC | 86.55 | 150 | 62.23 |
| Rev: CTTGCCCTTGCCATTCTCTCTCAC | 60.62 | ||||
| CYP | Cyclophilin B | For: TCTCGCTAGGCCAGGTTTCATTATG | 80.61 | 127 | 58.79 |
| Rev: ATGCAATCCCAAGACTGTTCCTGAC | 59.41 | ||||
| ElF1a | Elongation factor 1a | For: AGATCCATAACCCAGTCCCTCAGTC | 83.43 | 150 | 60.07 |
| Rev: GGTGATCGAGTTGTCGCCTGTC | 60.94 | ||||
| GAPDH | Glycerol-3-phosphate dehydrogenase | For: CCGACCGTTGATGTCTCTGTTGTAG | 82.75 | 93 | 59.64 |
| Rev: CGATTCCTCCTTGATAGCAGCCTTG | 59.96 | ||||
| eIF | Eukayotic initiation factor | For: GTGGAGCCGATGCCGATAACTG | 82.25 | 81 | 61.15 |
| Rev: TGCTTGAATCCTCTTGCTTCTTCCC | 59.41 | ||||
| H2A | Histone | For: GCT TGGAAGGAGGACATGGATGC | 80.81 | 94 | 61.04 |
| Rev: TCTCTGGTGGCTGGATGAATGGG | 61.78 | ||||
| UBC | Ubiquitin conjugating enzym | For: AATCGTCCGCCTCCAACTCCTC | 88.40 | 115 | 62.09 |
| Rev: TCCGCTCCCAGATCCGTAAATCC | 61.41 | ||||
| UBQ | Polyubiquitin | For: ACACGAAACACACAACGGGACTC | 81.88 | 148 | 60.12 |
| Rev: GCACCTTGTAGACTCGTCATCATC C | 59.60 |
| Treatments | Rank | GeNorm | NormFinder | BestKeeper | RefFinder | ||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | ||
| Tissues | 1 | eIF/ElFIa | 0.688 | eIF | 0.37 | UBQ | 0.83047 | eIF | 0.344 |
| 2 | 23S | 0.808 | GAPDH | 0.43 | H2A | 0.948356 | ElFIa | 0.448 | |
| 3 | GAPDH | 0.91 | ElFIa | 0.56 | 18S | 1.107341 | 23S | 0.718 | |
| 4 | 18S | 0.966 | 23S | 0.57 | ElFIa | 1.155646 | GAPDH | 0.734 | |
| 5 | UBQ | 1.143 | 18S | 0.6 | Actin | 1.260154 | 18S | 0.877 | |
| 6 | H2A | 1.207 | UBC | 0.62 | GAPDH | 1.500039 | Actin | 1.009 | |
| 7 | UBC | 1.262 | TUA | 0.69 | eIF | 1.5658 | UBC | 1.207 | |
| 8 | TUA | 1.517 | H2A | 0.84 | UBC | 1.784104 | UBQ | 1.346 | |
| 9 | CYP | 1.692 | UBQ | 0.98 | 23S | 1.89264 | H2A | 1.439 | |
| 10 | Actin | 1.855 | Actin | 1.28 | CYP | 2.461265 | TUA | 1.883 | |
| 11 | TUB | 2.03 | CYP | 1.29 | TUA | 3.596017 | TUB | 2.346 | |
| 12 | TUB | 2.23 | TUB | 22.69811 | CYP | 2.508 | |||
| Hormones | 1 | eIF/Actin | 0.552 | eIF | 0.27 | UBQ | 0.889151 | Actin | 0.111 |
| 2 | UBC | 0.603 | Actin | 0.35 | CYP | 1.217228 | eIF | 0.276 | |
| 3 | GAPDH | 0.65 | UBC | 0.38 | UBC | 1.312575 | UBC | 0.304 | |
| 4 | ElFIa | 0.729 | UBQ | 0.4 | 18S | 1.399489 | GAPDH | 0.559 | |
| 5 | UBQ | 0.79 | ElFIa | 0.41 | eIF | 1.653703 | ElFIa | 0.706 | |
| 6 | 18S | 0.877 | 18S | 0.43 | ElFIa | 1.75747 | UBQ | 0.756 | |
| 7 | 23S | 0.939 | 23S | 0.49 | GAPDH | 1.950305 | 18S | 0.828 | |
| 8 | TUB | 1.122 | GAPDH | 0.5 | Actin | 2.06208 | 23S | 0.909 | |
| 9 | CYP | 1.283 | TUB | 0.77 | TUB | 2.49303 | TUB | 1.62 | |
| 10 | H2A | 1.402 | CYP | 0.89 | H2A | 2.838247 | CYP | 2.049 | |
| 11 | TUA | 1.528 | H2A | 0.94 | 23S | 4.371111 | TUA | 2.144 | |
| 12 | TUA | 0.95 | TUA | 6.043985 | H2A | 2.19 | |||
| P levels | 1 | TUA/H2A | 0.195 | UBQ | 0.14 | H2A | 1.151808 | TUA | 0.034 |
| 2 | UBQ | 0.256 | H2A | 0.24 | ElFIa | 1.266195 | H2A | 0.097 | |
| 3 | 23S | 0.293 | TUA | 0.25 | GAPDH | 1.340341 | UBQ | 0.131 | |
| 4 | ElFIa | 0.334 | 23S | 0.26 | UBQ | 1.965777 | 23S | 0.258 | |
| 5 | 18S | 0.377 | ElFIa | 0.34 | TUA | 2.086377 | ElFIa | 0.331 | |
| 6 | eIF | 0.404 | GAPDH | 0.38 | eIF | 2.112718 | GAPDH | 0.461 | |
| 7 | GAPDH | 0.442 | CYP | 0.41 | CYP | 2.120346 | eIF | 0.472 | |
| 8 | CYP | 0.494 | eIF | 0.41 | TUB | 2.289766 | 18S | 0.476 | |
| 9 | UBC | 0.686 | 18S | 0.49 | Actin | 2.34925 | Actin | 0.505 | |
| 10 | TUB | 0.901 | UBC | 0.62 | 23S | 2.473408 | CYP | 0.511 | |
| 11 | Actin | 1.187 | TUB | 0.84 | 18S | 2.53995 | TUB | 0.532 | |
| 12 | Actin | 1.1 | UBC | 3.807912 | UBC | 1.622 | |||
| Total | 1 | eIF/ElFIa | 0.738 | ElFIa | 0.26 | UBQ | 0.808993491 | eIF | 1.19 |
| 2 | GAPDH | 0.833 | eIF | 0.27 | 18S | 1.24039533 | ElFIa | 1.41 | |
| 3 | Actin | 0.924 | GAPDH | 0.27 | UBC | 1.435844982 | GPDH | 3.00 | |
| 4 | 18S | 0.968 | UBC | 0.32 | ElFIa | 1.468964977 | Actin | 4.86 | |
| 5 | 23S | 1.045 | 23S | 0.41 | Actin | 1.541997044 | 18S | 5.18 | |
| 6 | UBC | 1.115 | 18S | 0.54 | eIF | 1.61899665 | 23S | 5.18 | |
| 7 | UBQ | 1.244 | Actin | 0.55 | GAPDH | 1.653358981 | UBC | 6.44 | |
| 8 | H2A | 1.358 | TUA | 0.60 | CYP | 2.106868104 | UBQ | 8.00 | |
| 9 | TUA | 1.494 | UBQ | 0.93 | H2A | 2.639930593 | TUA | 9.24 | |
| 10 | TUB | 1.648 | H2A | 0.96 | 23S | 3.14792245 | H2A | 9.74 | |
| 11 | CYP | 1.844 | TUB | 1.00 | TUA | 4.785600612 | TUB | 11.00 | |
| 12 | CYP | 1.48 | TUB | 6.500722183 | CYP | 12.00 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Li, J.; Gao, T. Evaluation of Reference Genes for Gene Expression Analysis in Eichhornia crassipes. Sustainability 2022, 14, 11071. https://doi.org/10.3390/su141711071
Xu J, Li J, Gao T. Evaluation of Reference Genes for Gene Expression Analysis in Eichhornia crassipes. Sustainability. 2022; 14(17):11071. https://doi.org/10.3390/su141711071
Chicago/Turabian StyleXu, Jing, Jing Li, and Tianpeng Gao. 2022. "Evaluation of Reference Genes for Gene Expression Analysis in Eichhornia crassipes" Sustainability 14, no. 17: 11071. https://doi.org/10.3390/su141711071
APA StyleXu, J., Li, J., & Gao, T. (2022). Evaluation of Reference Genes for Gene Expression Analysis in Eichhornia crassipes. Sustainability, 14(17), 11071. https://doi.org/10.3390/su141711071







