The Effect of the Artificial Reef on the Structure and Function of Sediment Bacterial Community
Abstract
:1. Introduction
2. Materials and Methods
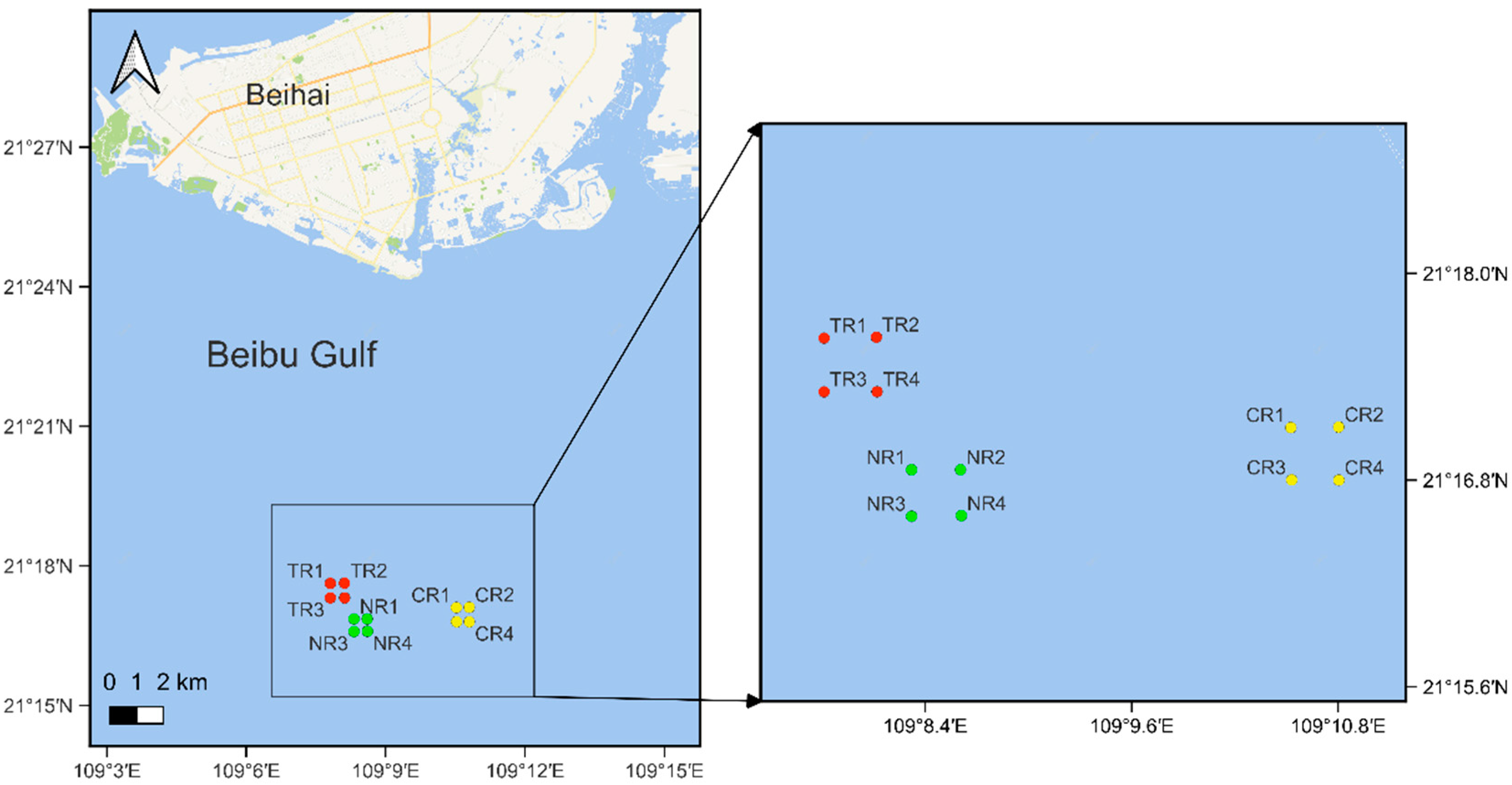
2.1. Study Area and Samples Collection
2.2. Physical and Chemical Parameters
2.3. DNA Extraction and PCR Amplification
2.4. Illumina NovaSeq Sequencing
2.5. Bioinformatics Analysis
3. Results
3.1. Changes in Physicochemical Characteristics of Overlying Water and Sediment
3.2. Sediment Bacterial Community Composition

3.3. Relationships between Sediment Microbial Communities and Environmental Factors
3.4. Bacterial Relationships within the Sediment Microbial Communities
3.5. Sediment Microbial Functional Annotation
4. Discussion
4.1. Changes in Physicochemical Characteristics of Overlying Water and Sediment
4.2. Sediment Bacterial Community Composition
4.3. Relationships between Sediment Microbial Communities and Environmental Factors
4.4. Bacterial Relationships within the Sediment Microbial Communities
4.5. Sediment Microbial Functional Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qin, M.; Yue, C.; Du, Y. Evolution of China’s marine ranching policy based on the perspective of policy tools. Mar. Policy 2020, 117, 103941. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, L. Evolution of marine ranching policies in China: Review, performance and prospects. Sci. Total Environ. 2020, 737, 139782. [Google Scholar] [CrossRef]
- Qin, M.; Wang, X.; Du, Y.; Wan, X. Influencing factors of spatial variation of national marine ranching in China. Ocean Coast. Manag. 2021, 199, 105407. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, T.; Cheng, T.C.E.; Wang, N. Evolution of blue carbon trading of China’s marine ranching under the blue carbon special subsidy mechanism. Ocean Coast. Manag. 2022, 222, 106123. [Google Scholar] [CrossRef]
- Tan, Y.; Lou, S. Research and development of a large-scale modern recreational fishery marine ranch System. Ocean Eng. 2021, 233, 108610. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, L.; Cong, W.; Jiang, Z.; Liang, Z. Comparative analysis of the ecological succession of microbial communities on two artificial reef materials. Microorganisms 2021, 9, 120. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.; Zhu, L.; Xie, W.; Zhang, J.; Wang, J.; Jiang, Z.; Liang, Z. Research on the influence of cut-opening factors on flow field effect of artificial reef. Ocean Eng. 2022, 249, 110890. [Google Scholar] [CrossRef]
- Jiang, Z.; Liang, Z.; Zhu, L.; Liu, Y. Numerical simulation of effect of guide plate on flow field of artificial reef. Ocean Eng. 2016, 116, 236–241. [Google Scholar] [CrossRef]
- Neumann, C.; Faria, E.F.; Dos Santos, A.C.P. Concrete leaching of a hydroelectric powerhouse due to 40 years of exposure to river water. Constr. Build. Mater. 2021, 302, 124253. [Google Scholar] [CrossRef]
- Yi, Y.; Zhu, D.; Guo, S.; Zhang, Z.; Shi, C. A review on the deterioration and approaches to enhance the durability of concrete in the marine environment. Cem. Concr. Compos. 2020, 113, 103695. [Google Scholar] [CrossRef]
- Zhang, D.; Li, M.; Yang, Y.; Yu, H.; Xiao, F.; Mao, C.; Huang, J.; Yu, Y.; Wang, Y.; Wu, B.; et al. Nitrite and nitrate reduction drive sediment microbial nitrogen cycling in a eutrophic lake. Water Res. 2022, 220, 118637. [Google Scholar] [CrossRef] [PubMed]
- Pierangeli, G.M.F.; Domingues, M.R.; Choueri, R.B.; Hanisch, W.S.; Gregoracci, G.B.; Benassi, R.F. Spatial variation and environmental parameters affecting the abundant and rare communities of bacteria and archaea in the sediments of tropical urban reservoirs. Microb. Ecol. 2022, 83, 2047. [Google Scholar] [CrossRef]
- Dong, X.; Zhang, C.; Li, W.; Weng, S.; Song, W.; Li, J.; Wang, Y. Functional diversity of microbial communities in inactive seafloor sulfide deposits. FEMS Microbiol. Ecol. 2021, 97, b108. [Google Scholar] [CrossRef]
- Roberto, A.A.; Van Gray, J.B.; Leff, L.G. Sediment bacteria in an urban stream: Spatiotemporal patterns in community composition. Water Res. 2018, 134, 353–369. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Han, T.; Huang, H.; Kuang, Z.; Qi, Z. The extent and pattern of mariculture impacts on spatial and seasonal variations of sediment bacterial communities among three coastal waters. Front. Mar. Sci. 2022, 9, 782456. [Google Scholar] [CrossRef]
- Fang, G.; Yu, H.; Sheng, H.; Tang, Y.; Liang, Z. Comparative analysis of microbial communities between water and sediment in Laoshan Bay marine ranching with varied aquaculture activities. Mar. Pollut. Bull. 2021, 173, 112990. [Google Scholar] [CrossRef]
- Vila-Costa, M.; Cerro-Gálvez, E.; Martínez-Varela, A.; Casas, G.; Dachs, J. Anthropogenic dissolved organic carbon and marine microbiomes. ISME J. 2020, 14, 2646–2648. [Google Scholar] [CrossRef]
- Kumar Parida, P.; Behera, B.K.; Dehury, B.; Rout, A.K.; Sarkar, D.J.; Rai, A.; Das, B.K.; Mohapatra, T. Community structure and function of microbiomes in polluted stretches of river Yamuna in New Delhi, India, using shotgun metagenomics. Environ. Sci. Pollut. R. 2022, 29, 1–15. [Google Scholar] [CrossRef]
- Li, N.; Dong, K.; Jiang, G.; Tang, J.; Xu, Q.; Li, X.; Kang, Z.; Zou, S.; Chen, X.; Adams, J.M.; et al. Stochastic processes dominate marine free-living Vibrio community assembly in a subtropical gulf. FEMS Microbiol. Ecol. 2020, 96, a198. [Google Scholar] [CrossRef]
- Lao, Q.; Su, Q.; Liu, G.; Shen, Y.; Chen, F.; Lei, X.; Qing, S.; Wei, C.; Zhang, C.; Gao, J. Spatial distribution of and historical changes in heavy metals in the surface seawater and sediments of the Beibu Gulf, China. Mar. Pollut. Bull. 2019, 146, 427–434. [Google Scholar] [CrossRef]
- Yao, Y.; Wang, C. Variations in Summer Marine Heatwaves in the South China Sea. J. Geophys. Res. Ocean. 2021, 126, e2021J–e17792J. [Google Scholar] [CrossRef]
- Hampel, J.J.; Moseley, R.D.; Hamdan, L.J. Microbiomes respond predictably to built habitats on the seafloor. Mol. Ecol. 2022. [Google Scholar] [CrossRef]
- Zhou, Z.; Meng, H.; Gu, W.; Li, J.; Deng, M.; Gu, J. High-throughput sequencing reveals the main drivers of niche-differentiation of bacterial community in the surface sediments of the northern South China sea. Mar. Environ. Res. 2022, 178, 105641. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Huang, T.; Chen, S.; Yang, X.; Lv, K.; Sekar, R. Abundance and diversity of bacteria in oxygen minimum drinking water reservoir sediments studied by quantitative PCR and pyrosequencing. Microb. Ecol. 2015, 69, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Li, J.; Yin, L.; Luo, X.; Ahmad, M.; Fang, B.; Li, S.; Deng, Q.; Wang, P.; Li, W. Habitat-dependent prokaryotic microbial community, potential keystone species, and network complexity in a subtropical estuary. Environ. Res. 2022, 212, 113376. [Google Scholar] [CrossRef]
- Chen, Q.; Chen, P. Changes in the heavy metals and petroleum hydrocarbon contents in seawater and surface sediment in the year following artificial reef construction in the Pearl River Estuary, China. Environ. Sci. Pollut. Res. 2020, 27, 6009–6021. [Google Scholar] [CrossRef]
- Oh, S.; Shin, W.S.; Kim, H.T. Effects of pH, dissolved organic matter, and salinity on ibuprofen sorption on sediment. Environ. Sci. Pollut. Res. Int. 2016, 23, 22882–22889. [Google Scholar] [CrossRef] [Green Version]
- Rajeev, M.; Sushmitha, T.J.; Toleti, S.R.; Pandian, S.K. Sediment-associated bacterial community and predictive functionalities are influenced by choice of 16S ribosomal RNA hypervariable region(s): An amplicon-based diversity study. Genomics 2020, 112, 4968–4979. [Google Scholar] [CrossRef]
- Hamdan, H.Z.; Salam, D.A. Microbial community evolution during the aerobic biodegradation of petroleum hydrocarbons in marine sediment microcosms: Effect of biostimulation and seasonal variations. Environ. Pollut. 2020, 265, 114858. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Lastauskien, E.; Valskys, V.; Stankeviiūt, J.; Kalcien, V.; Armalyt, J. The impact of intensive fish farming on pond sediment microbiome and antibiotic resistance gene composition. Front. Vet. Sci. 2021, 8, 673756. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef] [Green Version]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. GigaScience 2013, 2, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Wagner, H.H. Vegan Community Ecology Package Version 2.5. 2020. Available online: https://cran.r-project.org/web/packages/BiodiversityR/index.html (accessed on 7 November 2020).
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; Mcdonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Jiang, J.; Hu, X.; Hu, J.; Ren, C.; Zhou, S. The response of microbial community structure and sediment properties to anthropogenic activities in Caohai wetland sediments. Ecotox. Environ. Safe. 2021, 211, 111936. [Google Scholar] [CrossRef]
- Zhou, X.; Zhao, X.; Zhang, S.; Lin, J. Marine ranching construction and management in east china sea: Programs for sustainable fishery and aquaculture. Water-Sui. 2019, 11, 1237. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yao, L.; Chen, P.; Yu, J.; Wu, Q.E. Environmental influence on the spatiotemporal variability of fishing grounds in the Beibu Gulf, South China Sea. J. Mar. Sci. Eng. 2020, 8, 957. [Google Scholar] [CrossRef]
- Liu, S.; Zhou, X.; Zeng, C.; Frankstone, T.; Cao, L. Characterizing the development of Sea ranching in China. Rev. Fish Biol. Fisher. 2022, 32, 783–803. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, W. Maximum sustainable yield estimation of enhancement species with the characteristics of movement inside and outside marine ranching. J. Oceanol. Limnol. 2021, 39, 2380–2387. [Google Scholar] [CrossRef]
- Yanfeng, W.; Qiwei, H.U.; Jing, Y.U.; Pimao, C.; Liming, S. Effect assessment of fishery resources proliferation in Zhelin Bay marine ranching in eastern Guangdong. South China Fish. Sci. 2019, 15, 12. [Google Scholar] [CrossRef]
- Li, J.; Yin, Z.; Yang, J.; Chen, L.; Xu, M.; Zhang, Y.; Wu, Z.; Tian, T. Analysis of spring community structure and evaluation of ecological niche in tangshan marine ranching, China. Sustainability 2022, 14, 6999. [Google Scholar] [CrossRef]
- Uthaman, S.; Vishwakarma, V.; George, R.P.; Ramachandran, D.; Kumari, K.; Preetha, R.; Premila, M.; Rajaraman, R.; Mudali, U.K.; Amarendra, G. Enhancement of strength and durability of fly ash concrete in seawater environments: Synergistic effect of nanoparticles. Constr. Build. Mater. 2018, 187, 448–459. [Google Scholar] [CrossRef]
- Xu, Q.; Ji, T.; Yang, Z.; Ye, Y. Preliminary investigation of artificial reef concrete with sulphoaluminate cement, marine sand and sea water. Constr. Build. Mater. 2019, 211, 837–846. [Google Scholar] [CrossRef]
- Song, M.; Wang, J.; Nie, Z.; Wang, L.; Wang, J.; Zhang, J.; Wang, Y.; Guo, Z.; Jiang, Z.; Liang, Z. Evaluation of artificial reef habitats as reconstruction or enhancement tools of benthic fish communities in northern Yellow Sea. Mar. Pollut. Bull. 2022, 182, 113968. [Google Scholar] [CrossRef]
- Wang, L.; Liang, Z.; Guo, Z.; Cong, W.; Song, M.; Wang, Y.; Jiang, Z. Response mechanism of microbial community to seasonal hypoxia in marine ranching. Sci. Total Environ. 2022, 811, 152387. [Google Scholar] [CrossRef]
- White, C.A.; Nichols, P.D.; Ross, D.J.; Dempster, T. Dispersal and assimilation of an aquaculture waste subsidy in a low productivity coastal environment. Mar. Pollut. Bull. 2017, 120, 309–321. [Google Scholar] [CrossRef]
- Zlatkovic, S. Some metabolic, diversity and toxicity aspects of bacterial communities life in aquatic sediments. MedCrave Online 2017, 5, 156. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Sun, J.; Fang, E.; Guo, B.; Dai, Y.; Gao, Y.; Wang, H.; Zhang, X.; Xu, X.; Yu, Y.; et al. Impact of artificial reefs on sediment bacterial structure and function in Bohai Bay. Can. J. Microbiol. 2018, 65, 191–200. [Google Scholar] [CrossRef]
- Eraqi, W.A.; Elrakaiby, M.T.; Megahed, S.A.; Yousef, N.H.; Elshahed, M.S.; Yassin, A.S. Spatiotemporal analysis of the water and sediment Nile microbial community along an urban metropolis. Microb. Ecol. 2021, 82, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shi, Q.; Qu, J.; He, M.; Liu, Q. A pollution risk assessment and source analysis of heavy metals in sediments: A case study of Lake Gehu, China. Chinese J. Anal. Chem. 2022, 50, 100077. [Google Scholar] [CrossRef]
- Kalkan, S. Heavy metal resistance of marine bacteria on the sediments of the Black Sea. Mar. Pollut. Bull. 2022, 179, 113652. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Wan, J.; Wang, X.; Peng, C.; Wang, G.; Liang, W.; Zhang, W. Mixed bacteria-loaded biochar for the immobilization of arsenic, lead, and cadmium in a polluted soil system: Effects and mechanisms. Sci. Total Environ. 2022, 811, 152112. [Google Scholar] [CrossRef] [PubMed]
- Thanh-Nho, N.; Marchand, C.; Strady, E.; Vinh, T.; Nhu-Trang, T. Metals geochemistry and ecological risk assessment in a tropical mangrove (Can Gio, Vietnam). Chemosphere 2019, 219, 365–382. [Google Scholar] [CrossRef]
- Liu, E.; Yang, Y.; Xie, Z.; Wang, J.; Chen, M. Influence of sulfate reduction on arsenic migration and transformation in groundwater environment. Water 2022, 14, 942. [Google Scholar] [CrossRef]
- Zhuang, M.; Sanganyado, E.; Li, P.; Liu, W. Distribution of microbial communities in metal-contaminated nearshore sediment from Eastern Guangdong, China. Environ. Pollut. 2019, 250, 482–492. [Google Scholar] [CrossRef]
- Vipindas, P.V.; Mujeeb, R.K.M.; Jabir, T.; Thasneem, T.R.; Mohamed Hatha, A.A. Diversity of sediment bacterial communities in the South Eastern Arabian Sea. Reg. Stud. Mar. Sci. 2020, 35, 101153. [Google Scholar] [CrossRef]
- Qi, Q.; Hu, C.; Lin, J.; Wang, X.; Tang, C.; Dai, Z.; Xu, J. Contamination with multiple heavy metals decreases microbial diversity and favors generalists as the keystones in microbial occurrence networks. Environ. Pollut. 2022, 306, 119406. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, Y.; Sun, W.; Xie, S. Biodegradation of nonylphenol by two alphaproteobacterial strains in liquid culture and sediment microcosm. Int. Biodeter. Biodegr. 2014, 92, 1–5. [Google Scholar] [CrossRef]
- Kunihiro, T.; Takasu, H.; Miyazaki, T.; Uramoto, Y.; Kinoshita, K.; Yodnarasri, S.; Hama, D.; Wada, M.; Kogure, K.; Ohwada, K.; et al. Increase in Alphaproteobacteria in association with a polychaete, Capitella sp. I, in the organically enriched sediment. ISME J. 2011, 5, 1818–1831. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, D.; Sun, W.; Sun, X.; Yan, G.; Gao, W.; Lin, H. Characterizing sediment bacterial community and identifying the biological indicators in a seawater-freshwater transition zone during the wet and dry seasons. Environ. Sci. Pollut. R. 2022, 29, 41219–41230. [Google Scholar] [CrossRef]
- Jung, J.; Park, W. Acinetobacter species as model microorganisms in environmental microbiology: Current state and perspectives. Appl. Microbiol. Biot. 2015, 99, 2533–2548. [Google Scholar] [CrossRef]
- Bonthond, G.; Merselis, D.G.; Dougan, K.E.; Graff, T.; Todd, W.; Fourqurean, J.W.; Rodriguez-Lanetty, M. Inter-domain microbial diversity within the coral holobiont Siderastrea siderea from two depth habitats. PeerJ 2018, 6, e4323. [Google Scholar] [CrossRef] [Green Version]
- Montagna, M.; Sassera, D.; Epis, S.; Bazzocchi, C.; Vannini, C.; Lo, N.; Sacchi, L.; Fukatsu, T.; Petroni, G.; Bandi, C. “Candidatus Midichloriaceae” fam. nov. (Rickettsiales), an Ecologically Widespread Clade of Intracellular Alphaproteobacteria. Appl. Environ. Microb. 2013, 79, 3241–3248. [Google Scholar] [CrossRef] [Green Version]
- Poudyal, S.; Pulpipat, T.; Wang, P.; Chen, S. Comparison of the pathogenicity of Francisella orientalis in Nile tilapia (Oreochromis niloticus), Asian seabass (Lates calcarifer) and largemouth bass (Micropterus salmoides) through experimental intraperitoneal infection. J. Fish Dis. 2020, 43, 1097–1106. [Google Scholar] [CrossRef]
- Duodu, S.; Larsson, P.; Sjödin, A.; Forsman, M.; Colquhoun, D.J. The distribution of francisella-like bacteria associated with coastal waters in Norway. Microb. Ecol. 2012, 64, 370–377. [Google Scholar] [CrossRef]
- Berrada, Z.L.; Telford, S.R. Diversity of Francisella Species in Environmental Samples from Martha’s Vineyard, Massachusetts. Microb. Ecol. 2010, 59, 277–283. [Google Scholar] [CrossRef] [Green Version]
- Seshadri, R.; Paulsen, I.T.; Eisen, J.A.; Read, T.D.; Nelson, K.E.; Nelson, W.C.; Ward, N.L.; Tettelin, H.; Davidsen, T.M.; Beanan, M.J.; et al. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. USA 2003, 100, 5455–5460. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Tian, J.; Gatesoupe, F.; Li, W.; Zou, H.; Yang, B.; Wang, G. Intestinal microbiota of gibel carp (Carassius auratus gibelio) and its origin as revealed by 454 pyrosequencing. World J. Microbiol. Biotechnol. 2013, 29, 1585–1595. [Google Scholar] [CrossRef]
- Peng, W.; Li, X.; Lin, M.; Fan, W. Microbiological analysis of cadmium-contaminated sediments during biostabilization with indigenous sulfate-reducing bacteria. J. Soil. Sediment. 2020, 20, 584–593. [Google Scholar] [CrossRef]
- Boeraş, I.; Burcea, A.; Coman, C.; Bănăduc, D.; Curtean-Bănăduc, A. Bacterial microbiomes in the sediments of lotic systems ecologic drivers and role: A case study from the Mures River, Transylvania, Romania. Water-Sui. 2021, 13, 3518. [Google Scholar] [CrossRef]
- Huang, W.; Chen, X.; Wang, K.; Chen, J.; Zheng, B.; Jiang, X. Comparison among the microbial communities in the lake, lake wetland, and estuary sediments of a plain river network. MicrobiologyOpen 2019, 8, e644. [Google Scholar] [CrossRef]
- Sikorski, J.; Möhle, M.; Wackernagel, W. Identification of complex composition, strong strain diversity and directional selection in local Pseudomonas stutzeri populations from marine sediment and soils. Environ. Microbiol. 2002, 4, 465–476. [Google Scholar] [CrossRef]
- Chen, Q.; Li, Y.; Liu, M.; Zhu, B.; Mu, J.; Chen, Z. Removal of Pb and Hg from marine intertidal sediment by using rhamnolipid biosurfactant produced by a Pseudomonas aeruginosa strain. Environ. Technol. Innov. 2021, 22, 101456. [Google Scholar] [CrossRef]
- Park, Y.; Ko, J.; Yun, S.; Lee, E.Y.; Kim, S.; Kang, S.; Lee, B.; Kim, S. Enhancement of bioremediation by Ralstonia sp. HM-1 in sediment polluted by Cd and Zn. Bioresour. Technol. 2008, 99, 7458–7463. [Google Scholar] [CrossRef]
- Urbanczyk, H.; Ast, J.C.; Dunlap, P.V. Phylogeny, genomics, and symbiosis of Photobacterium. FEMS Microbiol. Rev. 2011, 35, 324–342. [Google Scholar] [CrossRef]
- Kim, M.; Cha, I.; Lee, K.; Lee, E.; Park, S. Genomics reveals the metabolic potential and functions in the redistribution of dissolved organic matter in marine environments of the genus Thalassotalea. Microorganisms 2020, 8, 1412. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, W.; Liu, Y.; Cai, M.; Luo, Z.; Li, M. Reveals microbial diversity and metabolic potentials of seawater and surface. Front. Microbiol. 2018, 9, 2402. [Google Scholar] [CrossRef] [Green Version]
- Mikhailov, I.S.; Zakharova, Y.R.; Bukin, Y.S.; Galachyants, Y.P.; Petrova, D.P.; Sakirko, M.V.; Likhoshway, Y.V. Co-occurrence networks among bacteria and microbial eukaryotes of lake Baikal during a spring phytoplankton bloom. Microb. Ecol. 2019, 77, 96–109. [Google Scholar] [CrossRef]
- Kurm, V.; van der Putten, W.H.; Weidner, S.; Geisen, S.; Snoek, B.L.; Bakx, T.; Hol, W.H.G. Competition and predation as possible causes of bacterial rarity. Environ. Microbiol. 2019, 21, 1356–1368. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; He, R.; Wang, W.; Zhao, D.; Zeng, J.; Huang, R.; Duan, M.; Yu, Z. Composition and co-occurrence patterns of Phragmites australis rhizosphere bacterial community. Aquat. Ecol. 2021, 55, 695–710. [Google Scholar] [CrossRef]
- Yuan, Q.; Wang, P.; Wang, X.; Hu, B.; Tao, L. Phytoremediation of cadmium-contaminated sediment using Hydrilla verticillata and Elodea canadensis harbor two same keystone rhizobacteria Pedosphaeraceae and Parasegetibacter. Chemosphere 2022, 286, 131648. [Google Scholar] [CrossRef] [PubMed]
- Kett, G.F.; Culloty, S.C.; Jansen, M.A.K.; Lynch, S.A. Development of a sensitive polymerase chain reaction (PCR) and digoxigenin (DIG)-labeled in situ hybridization (ISH) for the detection of Vibrio bacteria in the Pacific oyster Crassostrea gigas. Aquac. Rep. 2022, 22, 100961. [Google Scholar] [CrossRef]
- Möller, L.; Kreikemeyer, B.; Luo, Z.; Jost, G.; Labrenz, M. Impact of coastal aquaculture operation systems in Hainan island (China) on the relative abundance and community structure of Vibrio in adjacent coastal systems. Estuar. Coast. Shelf Sci. 2020, 233, 106542. [Google Scholar] [CrossRef]
- Jung, S.W.; Kang, J.; Park, J.S.; Joo, H.M.; Suh, S.; Kang, D.; Lee, T.; Kim, H. Dynamic bacterial community response to Akashiwo sanguinea (Dinophyceae) bloom in indoor marine microcosms. Sci. Rep. 2021, 11, 6983. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, M.; Jiafeng, H.; Guo, X.; Zhang, Y.; Liu, D.; Wu, R.; He, H.; Wang, J. Diversity of the microbial community and cultivable protease-producing bacteria in the sediments of the Bohai Sea, Yellow Sea and South China Sea. PLoS ONE 2019, 14, e215328. [Google Scholar] [CrossRef] [Green Version]
- Liang, S.; Deng, J.; Jiang, Y.; Wu, S.; Zhou, Y.; Zhu, W. Functional distribution of bacterial community under different land use patterns based on FaProTax function prediction. Pol. J. Environ. Stud. 2020, 29, 1–17. [Google Scholar] [CrossRef]
- Lin, G.; Lin, X. Bait input altered microbial community structure and increased greenhouse gases production in coastal wetland sediment. Water Res. 2022, 218, 118520. [Google Scholar] [CrossRef]
- Lee, Y.; Choi, Y. Complete genome sequence and analysis of three kinds of β-agarase of Cellulophaga lytica DAU203 isolated from marine sediment. Mar. Genom. 2017, 35, 43–46. [Google Scholar] [CrossRef]
- Shi, P.; Wang, H.; Feng, M.; Cheng, H.; Yang, Q.; Yan, Y.; Xu, J.; Zhang, M. The coupling response between different bacterial metabolic functions in water and sediment improve the ability to mitigate climate change. Water 2022, 14, 1203. [Google Scholar] [CrossRef]
- Nielsen, L.P.; Risgaard-Petersen, N. Rethinking sediment biogeochemistry after the discovery of electric currents. Annu. Rev. Mar. Sci. 2015, 7, 425–442. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Qin, C.; Ma, H.; Xi, S.; Zuo, T.; Pan, W.; Li, C. Response of protist community dynamics and co-occurrence patterns to the construction of artificial reefs: A case study in Daya Bay, China. Sci. Total Environ. 2020, 742, 140575. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tong, F.; Chen, G.; Feng, X.; Liu, Y.; Chen, P. The Effect of the Artificial Reef on the Structure and Function of Sediment Bacterial Community. Sustainability 2022, 14, 14728. https://doi.org/10.3390/su142214728
Tong F, Chen G, Feng X, Liu Y, Chen P. The Effect of the Artificial Reef on the Structure and Function of Sediment Bacterial Community. Sustainability. 2022; 14(22):14728. https://doi.org/10.3390/su142214728
Chicago/Turabian StyleTong, Fei, Guobao Chen, Xue Feng, Yan Liu, and Pimao Chen. 2022. "The Effect of the Artificial Reef on the Structure and Function of Sediment Bacterial Community" Sustainability 14, no. 22: 14728. https://doi.org/10.3390/su142214728





