Prevalence and Molecular Characterization of Babesia ovis Infecting Sheep in Nigeria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Study Population
2.3. Sample Collection
2.4. Thin Blood Smear Technique
2.5. Packed Cell Volume Determination
2.6. DNA Extraction of Blood Samples
2.7. Primer Set Determination
2.8. PCR Conditions
2.9. Agarose Gel Electrophoresis
2.9.1. Sequencing
2.9.2. Nucleotide Sequence
2.9.3. Statistical Data Analysis
3. Results
3.1. Demographics Distribution of Different Variables of Sheep from This Study
3.2. Prevalence of Babesia Ovis Infection in Sheep Detected by Microscopy and PCR
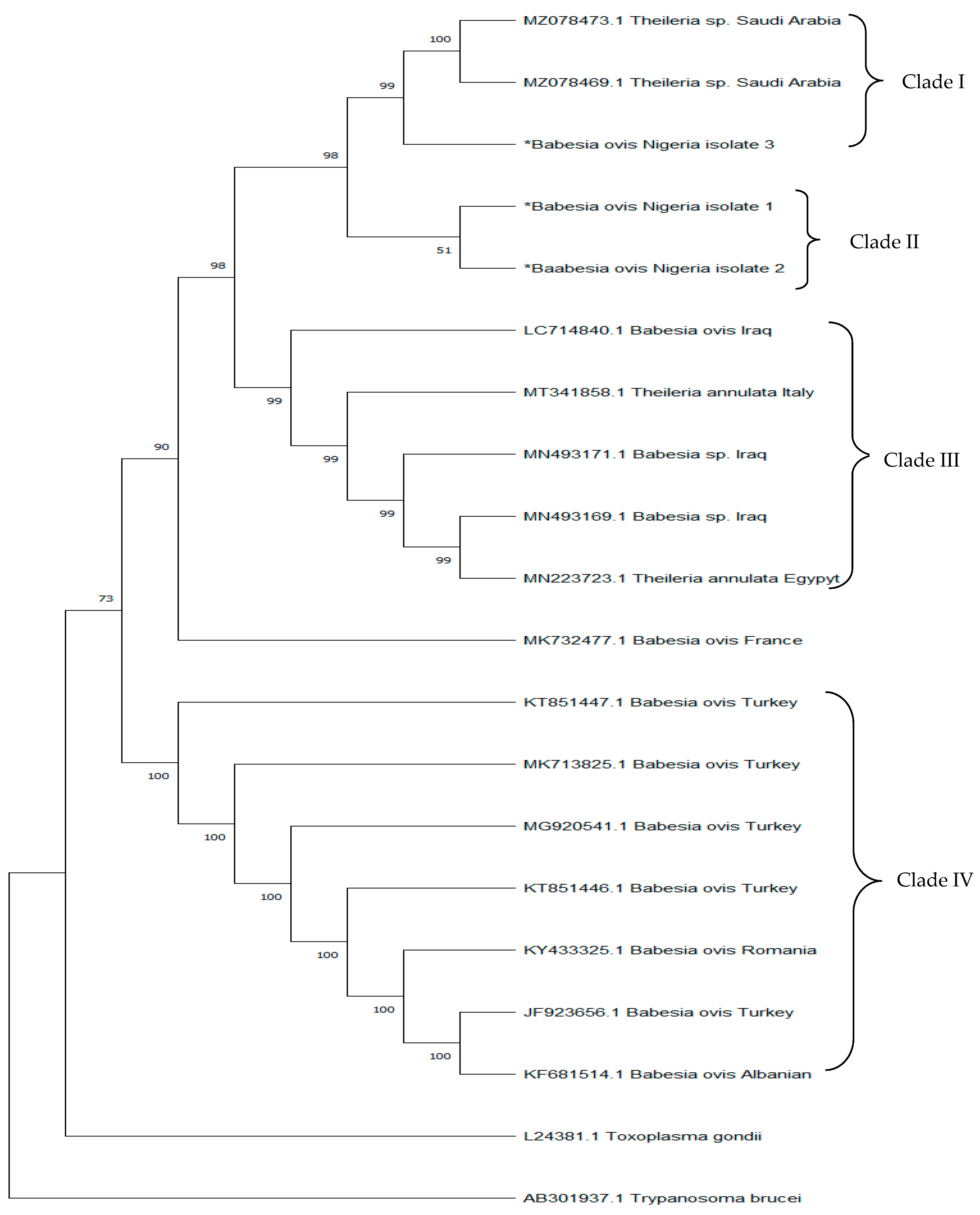
3.3. Sequence Similarities and Phylogenetic Analysis
4. Discussion
Limitation
5. Conclusions
6. Recommendation
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iangopathy, M.; Palavesam, A.; Amaresan, S.; Muthusamy, R. Economic Impact of Gastrointestinal Nematodes in Sheep on Meat Production. Int. J. Livest. Res. 2019, 9, 44–48. [Google Scholar]
- Kasozik, I.; Matovu, E.; Tayebwa, D.S.; Natuhwera, J.; Mugezi, I.; Muhero, M. Epidemiology of increasing hemoparasite burden in Ugandan cattle. Open J. Vetrinary Med. 2014, 4, 220–231. [Google Scholar] [CrossRef] [Green Version]
- Akande, F.A.; Takeet, M.I.; Makanju, O. Haemoparasites of cattle in Abeokuta, South-west, Nigeria. Sci. World J. 2010, 5, 19–21. [Google Scholar]
- Cornillot, E.; Hadj-Kaddour, K.; Dassouli, A.; Noel, B.; Ranwez, V.; Vacherie, B. Sequencing of the smallest Apicomplexan genome from the human pathogen Babesia microti. Nucleic Acids Res. 2012, 40, 9102–9114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, J.S.; Luo, J.; Schnittger, L.; Seitzer, U.; Jongejan, F.; Yin, H. Phylogenetic position of small ruminant infecting piroplasms. Ann. N. Y. Acad. Sci. 2006, 1081, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, L.; Rodriguez, A.E.; Florin-Christensen, M.; Morrison, D.A. Babesia: A world emerging. Infect. Genet. Evol. 2012, 12, 1788–1809. [Google Scholar] [CrossRef]
- Sevinc, F.; Sevinc, M.; Ekici, O.D.; Yıldız, R.; Isık, N.; Aydogdu, U. Babesia ovis infections: Detailed clinical and laboratory observations in the pre- and post-treatment periods of 97 field cases. Vet. Parasitol. 2013, 191, 35–43. [Google Scholar] [CrossRef]
- Sevinc, F.; Xuan, X. Major tick-borne parasitic diseases of animals: A frame of references in Turkey. Eurasian J. Vet. Sci. 2015, 31, 132–142. [Google Scholar] [CrossRef]
- Zahid, I.A.; Latif, M.; Baloch, K.B. Incidence and treatment of theileriasis and Babesiosis. Pak. Vet. J. 2005, 25, 137–140. [Google Scholar]
- Walker, A.R.; Bouattour, A.; Camicas, J.L.; Estrada-Peria, A.; Horak, I.; Latif, A.; Pegram, R.; Preston, P.M. Ticks of Domestic Animals in Africa: A Guide to Identification of Species; Bioscience Reports: Edinburgh, UK, 2003. [Google Scholar]
- Lopes, L.B.; Nicolino, R.; Capanema, R.O.; Oliveira, C.S.F.; Haddad, J.P.A.; Eckstein, C. Economic impacts of parasitic diseases in cattle. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Res. 2015, 10, 1–10. [Google Scholar] [CrossRef]
- Huruma, G.; Abdurhaman, M.; Gebre, S.; Deresa, B. Identification of tick species and their prevalence in and around Sebeta town, Ethiopia. J. Parasitol. Vector Biol. 2015, 7, 1–8. [Google Scholar]
- Marufu, M.C. Prevalence of Ticks and Tick-borne Diseases in Cattle on Communal Rangelands in the Highland Areas of the Eastern Cape Province, South Africa. Master’s Thesis, Department of Livestock and Pasture Science Faculty of Science and Agriculture, University of Fort Hare, Alice, South Africa, 2008; pp. 1–134. [Google Scholar]
- Yiwombe, K. An Investigation to Determine the Resistance of the Boophilus Tick (Blue Tick) to Amitraz in Selected Areas of Zimbabwe. Bachelor’s Thesis, Midlands State University, Gweru, Zimbabwe, 2013. [Google Scholar]
- Esmaeilnejad, B.; Tavassoli, M.; Asri-Rezaei, S.; Dalir-Naghadeh, B.; Mardani, K.; Jalilzadeh-Amin, G.; Golabi, M.; Arjmand, J. PCR-Based Detection of Babesia ovis in Rhipicephalus bursa and Small Ruminants. J. Parasitol. Res. 2014, 2014, 294704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onoja, I.I.; Malachy, P.; Mshelia, W.P.; Okaiyeto, S.O.; Danbirni, S.; Kwananshie, G. Prevalence of Babesiosisin cattle and goats at Zaria abbatoir. Niger. J. Vet. Adv. 2013, 3, 211–214. [Google Scholar]
- Fakhar, M.; Hajihasani, A.; Masoufi, S.; Alizadeh, H.; Shirzad, H.; Piri, F.; Pagheh, A.S. An epidemiological survey on bovjne and ovine Babesiosis in Kurdistan province, Western Iran. Trop. Anim. Health Prod. 2012, 44, 319–322. [Google Scholar] [CrossRef]
- Ijaz, M.; Rehman, A.; Ali, M.M.; Umar, M.; Khalid, S.; Mehmoo, K.; Hanif, A. Clinico-epidemiology and therapeutical trials on Babesiosis in sheep and goats in Lahore, Pakistan. J. Anim. Plant Sci. 2013, 23, 666–669. [Google Scholar]
- Urquhart, G.M.; Armour, J.; Duncan, J.L.; Dunn, A.M.; Jennings, F.W. Veterinary Parasitology, 2nd ed.; Blackwell Science Incorporated: Hoboken, NJ, USA, 1996; pp. 242–253. [Google Scholar]
- Radostits, O.M.; Gay, G.C.; Hinchiff, K.W.; Constable, P.O. Veterinary Medicine: A Text Book of the Diseases of Cattle, Sheep, Goat, Pigs and Horses, 10th ed.; Saunders Elsevier: London, UK, 2007; pp. 1110–1489, 1527–1530. [Google Scholar]
- Rahbari, S.; Nabian, S.; Khaki, Z.; Alidadi, N.; Ashrafihelan, J. Clinical hematologic and pathologic aspects of experimental ovine Babesiosis in Iran. Iran. J. Vet. Res. 2008, 9, 58–64. [Google Scholar]
- Takeet, M.I.; Akande, F.A.; Abakpa, S.A.V. Haemoprotozoan parasites of sheep in Abeokuta, Nigeria. Niger. J. Parasitol. 2009, 30, 142–146. [Google Scholar]
- Adejinmi, J.O.; Sadiq, N.A.; Fashanu, S.O.; Lasisi, O.T.; Ekundayo, S. Studies on the blood parasites of sheep in Ibadan, Nigeria. Afr. J. Biomed. Res. 2004, 7, 41–43. [Google Scholar] [CrossRef]
- Jatau, I.D.; Abdulganiyu, A.; Lawal, A.I.; Okubanjo, O.O.; Yusuf, K.H. Gastrointestinal and Haemoparasitism of sheep and goats at slaughtered Abattoir in Kano, Northern Nigeria. Sahel J. Vet. Sci. 2011, 9, 7–11. [Google Scholar]
- Samaila, A.B.; Musa, B.L. Prevalence of Haemoparasite of sheep and goats slaughtered in Bauchi Abattoir. Int. J. Appl. Biol. Res. 2012, 4, 128–133. [Google Scholar]
- Opara, M.N.; Santali, A.; Mohammed, B.R.; Jegede, O.C. Prevalence of Haemoparasites of small Ruminants in Lafia Nassarawa State. A Guinea Savannah zone in Nigeria. J. Vet. Adv. 2016, 6, 1251–1257. [Google Scholar]
- Egbe-Nwiyi, T.N.; Sheriff, G.A.; Paul, B.T. Prevalence of tick-borne haemoparasiticdiseases (TBHDS) and Hematological changes in sheep and goats in Maiduguri Abattoir. J. Vet. Med. Anim. Health 2018, 10, 28–33. [Google Scholar] [CrossRef] [Green Version]
- Takeet, M.I.; Oyewusi, A.J.; Abakpa, S.A.V.; Daramola, O.D.; Peters, S.O. Genetic diversity among Babesia rossi detected in naturally infected dog in Abeokuta, Nigeria, based 18S rRNA gene sequences. Acta Parasitol. 2017, 62, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Adams, K.M.G.; Paul, J.; Zaman, V. Medical and Veterinary Protozoology, an Illustrated Guide, Revised ed.; Churchill Livingstone: Edinburgh, UK; London, UK, 1977; pp. 32–49. [Google Scholar]
- Cheesbrough, M. Destrict Laboratory Practice in Tropical Countries; Cambridge University Press: Cambridge, UK, 2005; pp. 64–67. [Google Scholar]
- Nagore, D.; García-Sanmartín, J.; García-Pírez, A.L.; Juste, R.A. Hurtado Identification, genetic diversity and prevalence of Theileria and Babesia species in a sheep population from Nortern Spain. Int. J. Parasitol. 2004, 34, 1059–1067. [Google Scholar] [CrossRef]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy; W. H. Freeman & Co.: New York, NY, USA, 1973. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar]
- Tajima, F.; Nei, M. Estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol. 1984, 1, 269–285. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Shahzad, W.; Noor, H.; Ahmad, M.U.; Munir, R.; Saghar, M.S.; Hassan, M.M.; Ahmad, N.; Akbar, G.; Mehmood, F. Prevalence and Molecular Diagnosis of Babesia ovis and Theileria ovis in Lohi Sheep at Livestock Experiment Station (LES), Bahadurnagar, Okara, Pakistan. Iran. J. Parasitol. 2013, 8, 570–578. [Google Scholar]
- Naderi, A.; Nayebzadeh, H.; Gholami, S. Detection of Babesia infection among human, goats and sheep using microscopic and molecular methods in the city of Kuhdasht in Lorestan Province, West of Iran. J. Parasit. Dis. Off. Organ Indian Soc. Parasitol. 2017, 41, 837–842. [Google Scholar] [CrossRef]
- Dumanli, N.; Aktas, M.; Cetinkaya, B.; Cakmak, A.; Koroglu, E.; Saki, C.E. Prevalence and distribution of tropical theileriosis in eastern Turkey. Vet. Parasitol. 2005, 127, 9–15. [Google Scholar] [CrossRef]
- Iqbal, F.; Fatima, M.; Shahnawaz, S.; Naeem, M.; Shaikh, R.S.; Shaikh, A.S.; Aktas, M.; Ali, M. A Study on the determination of risk factors associated with babesiosis and prevalence of Babesia sp., by PCR amplification, in small ruminants from Southern Punjab (Pakistan). Parasite 2011, 18, 229–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onyiche, T.E.; Mofokeng, L.S.; Thekisoe, O.; MacLeod, E.T. Molecular survey for tick-borne pathogens and associated risk factors in sheep and goats in Kano Metropolis, Nigeria. Vet. Parasitol. Reg. Stud. Rep. 2022, 33, 100753. [Google Scholar] [CrossRef] [PubMed]
- Aktas, M.; Altay, K.; Dumanli, N. Determination of prevalence and risk factors for infection with Babesia ovis in small ruminants from Turkey by polymerase chain reaction. Parasitol. Res. 2007, 100, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Altay, K.; Aktas, M.; Dumanli, N. Detection of Babesia ovis by PCR in Rhipicephalus bursa collected from naturally infested sheep and goats. Res. Vet. Sci. 2008, 85, 116–119. [Google Scholar] [CrossRef]
- O’Connor, R.M.; Long, J.A.; Allred, D.R. Cytoadherence of Babesia bovis-infected erythrocytes to bovine brain capillary endothelial cells provides an in vitro model for sequestration. Infect. Immun. 1999, 67, 3921–3928. [Google Scholar] [CrossRef] [Green Version]
- Rjeibi, M.R.; Gharbi, M.; Mhadhbi, M.; Mabrouk, W.; Ayari, B.; Nasfi, I.; Jedidi, M.; Sassi, L.; Rekik, M.; Darghouth, M.A. Prevalence of piroplasms in small ruminants in North-West Tunisia and the first genetic characterisation of Babesia ovis in Africa. Parasite 2014, 21, 23. [Google Scholar] [CrossRef] [Green Version]
- Adua, M.M.; Idahor, K.O.; Panda, A.I.; Omeje, J.N. Prevalence of haemoparasites (Babesia species) in sheep under the traditional system of management in Lafia metropolis Nasarawa state, Nigeria. NSUK J. Sci. Technol. 2016, 6, 17–21. [Google Scholar]
- Carr, A.L.; Salgado, V.L. Ticks home in on body heat: A new understanding of Haller’s organ and repellent action. PLoS ONE 2019, 14, e0221659. [Google Scholar] [CrossRef] [Green Version]
- Bello, A.M.; Lawal, J.R.; Dauda, J.; Wakili, Y.; Mshellia, E.M.; Abubakar, M.I.; Biu, A.A. Prevalence of haemoparasite in balami sheep from Miduguri, Northeastern, Nigeria. Direct Res.J. Vet. Med. Anim. Sci. 2017, 2, 28–35. [Google Scholar]
- Ngole, I.U.; Ndamukong, K.J.; Mbuh, J.V. Internal parasites and haematological values in cattle slaughtered in Buea subdivision in Cameroun. Trop. Anim. Health Prod. 2003, 35, 409–413. [Google Scholar] [CrossRef]
- Masiga, D.K.; Okech, G.; Irungu, P.; Ouma, J.; Wekesa, S.; Ouma, B.; Guya, S.O.; Ndung’u, J.M. Growth and mortality in sheep and goats under high tsetse challenge in Kenya. Trop. Anim. Health Prod. 2002, 34, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Ademola, I.O.; Onyiche, T.E. Haemoparasites and haematological parameters of slaughtered Ruminants and pigs at Bodija Abattoir, Ibadan, Nigeria. Afr. J. Biomed. Res. 2013, 16, 101–105. [Google Scholar]
- Pedroni, M.J.; Sondgeroth, K.S.; Gallego-Lopez, G.M.; Echaide, I.; Lau, A.O. Comparative transcriptome analysis of geographically distinct virulent and attenuated Babesia bovis strains reveals similar gene expression changes through attenuation. BMC Genom. 2013, 14, 763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pradeep, R.K.; Nimisha, M.; Sruthi, M.K.; Vidya, P.; Amrutha, B.R.; Kurbet, P.S.; Kumar, K.G.A.; Varghese, A.; Deepa, C.K.; Dinesh, C.N. Molecular characterization of South Indian field isolates of bovine Babesia spp. and Anaplasma spp. Parasitol. Res. 2018, 118, 617–630. [Google Scholar] [CrossRef]
- Laha, R.; Mondal, B.; Biswas, S.K.; Chand, K.; Das, M.; Sarma, D.; Goswami, A.; Sen, A. Detection of Babesia bigemina infection in cattle from North-Eastern India by polymerase chain reaction and its genetic relatedness with other isolates. Trop. Anim. Health Prod. 2015, 47, 633–636. [Google Scholar] [CrossRef]
- Lack, J.B.; Reichard, M.V.; van den Bussche, R.A. Phylogeny and evolution of the piroplasmida as inferred from 18S rRNA sequences. Int. J. Parasitol. 2012, 42, 353–363. [Google Scholar] [CrossRef]
| Variables | Frequency (n)/(%) |
|---|---|
| Breeds | |
| West African Dwarf | 114 (57.6) |
| Balami | 10 (5.1) |
| Yankasa | 45 (22.7) |
| Uda | 29 (14.6) |
| Sex of Animal | |
| Male | 72 (36.4) |
| Female | 126 (63.6) |
| Age of Animal | |
| ≤1.5 | 70 (35.4) |
| >1.5–≤3 | 83 (41.9) |
| >3–≤4.5 | 27 (13.6) |
| >4.5 | 18 (9.1) |
| PCV Evaluation | |
| Anemic | 102 (51.5) |
| Normal | 89 (44.9) |
| Polycythemic | 7 (3.5) |
| Skin Coat Color | |
| White black | 145 (73.2) |
| Brown white | 13 (6.6) |
| White | 18 (9.1) |
| Black | 22 (11.1) |
| Body Condition Score Evaluation | |
| Emaciated | 8 (4.0) |
| Thin | 25 (12.6) |
| Average | 109 (55.1) |
| Fat | 42 (21.2) |
| Obese | 14 (7.1) |
| Location | |
| Abuja | 99 (50) |
| Abeokuta | 99 (50) |
| Total | 198 (100) |
| Variables | Number of Sampled Animals | Number of Infected Animals | Prevalence (%) | 95% CI | p-Value |
|---|---|---|---|---|---|
| Age of Animal (Years) | |||||
| ≤1.5 | 0.123 | ||||
| >1.5–≤3 | 70 | 19 | 27.1 | 16.45–37.62 | |
| >3–≤ 4.5 | 83 | 38 | 45.8 | 40.04–63.90 | |
| >4.5 | 27 | 10 | 37.0 | 6.77–23.75 | |
| 18 | 6 | 33.3 | 3.08–17.04 | ||
| Sex of Animal | |||||
| Male | 72 | 27 | 37.5 | 25.97–49.09 | 0.889 |
| Female | 126 | 46 | 36.5 | 50.91–74.03 | |
| Breeds | |||||
| WAD | 114 | 45 | 39.5 | 49.52–72.79 | 0.792 |
| Balami | 10 | 4 | 40.0 | 1.51–13.44 | |
| Yankasa | 45 | 15 | 33.3 | 11.98–31.62 | |
| Uda | 29 | 9 | 31.0 | 5.80–22.12 | |
| Coat Colour | |||||
| White black | 145 | 52 | 35.9 | 59.45–81.23 | 0.831 |
| Brown white | 13 | 4 | 30.8 | 1.51–13.44 | |
| White | 18 | 8 | 44.4 | 4.85–20.46 | |
| Black | 22 | 9 | 40.9 | 5.80–22.12 | |
| PCV Evaluation | 102 | ||||
| Anemic | 89 | 36 | 35.3 | 37.40–61.28 | 0.510 |
| Normal | 7 | 33 | 37.1 | 33.52–57.30 | |
| Polycythemia | 4 | 57.1 | 1.51–13.44 | ||
| Body Condition Score Evaluation | |||||
| Emaciated | |||||
| Thin | 8 | 6 | 75.0 | 3.08–17.04 | 0.180 |
| Average | 25 | 11 | 44.0 | 7.77–25.36 | |
| Fat | 109 | 36 | 33.0 | 37.40–61.28 | |
| Obese | 42 | 15 | 35.7 | 11.98–31.62 | |
| 14 | 5 | 35.7 | 2.26–15.26 | ||
| Location | |||||
| Abeokuta | 99 | 38.4 | 40.04–63.90 | 0.659 | |
| Abuja | 99 | 38 | 35.4 | 36.10–59.96 | |
| 35 | |||||
| Total | 198 | 73 | 36.9% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adewumi, T.S.; Takeet, M.I.; Akande, F.A.; Sonibare, A.O.; Okpeku, M. Prevalence and Molecular Characterization of Babesia ovis Infecting Sheep in Nigeria. Sustainability 2022, 14, 16974. https://doi.org/10.3390/su142416974
Adewumi TS, Takeet MI, Akande FA, Sonibare AO, Okpeku M. Prevalence and Molecular Characterization of Babesia ovis Infecting Sheep in Nigeria. Sustainability. 2022; 14(24):16974. https://doi.org/10.3390/su142416974
Chicago/Turabian StyleAdewumi, Taiye Samson, Michael Irewole Takeet, Foluke Adedayo Akande, Adekayode Olarewaju Sonibare, and Moses Okpeku. 2022. "Prevalence and Molecular Characterization of Babesia ovis Infecting Sheep in Nigeria" Sustainability 14, no. 24: 16974. https://doi.org/10.3390/su142416974
APA StyleAdewumi, T. S., Takeet, M. I., Akande, F. A., Sonibare, A. O., & Okpeku, M. (2022). Prevalence and Molecular Characterization of Babesia ovis Infecting Sheep in Nigeria. Sustainability, 14(24), 16974. https://doi.org/10.3390/su142416974






