Enhancing Forest Canopy Height Retrieval: Insights from Integrated GEDI and Landsat Data Analysis
Abstract
:1. Introduction
2. Materials and Methods
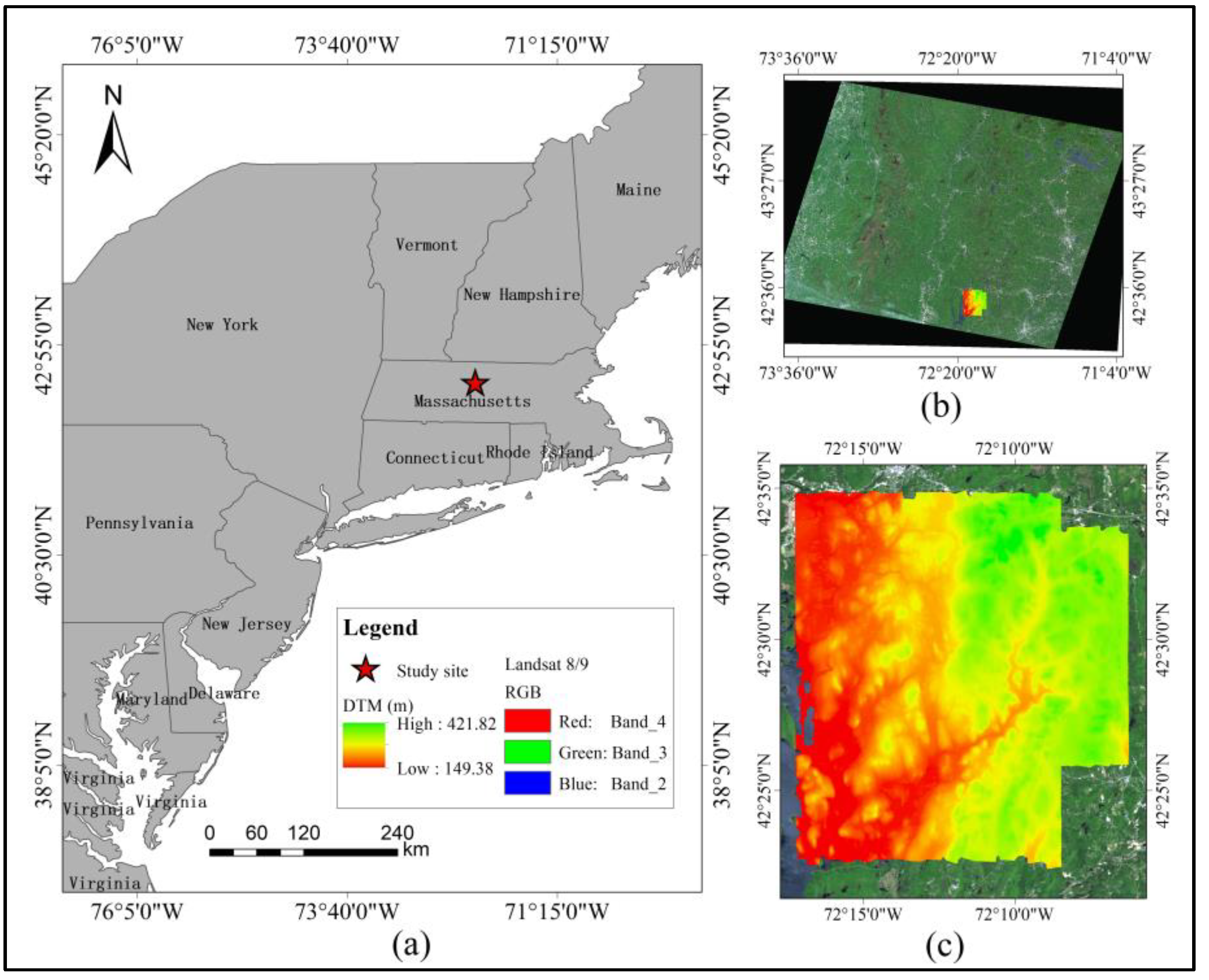
2.1. Research Area
2.2. Data Acquisition and Processing
2.2.1. ALS Data Processing
2.2.2. GEDI L2A Data Processing
2.2.3. Landsat 8 and 9 Data Processing
2.3. Forest Canopy Height Inversion Model Construction
2.3.1. BP Neural Network
2.3.2. Activation Function
2.3.3. Determination of the Number of Neurons in the Hidden Layer
2.3.4. Independent Variables Extraction
2.3.5. Importance Analysis of Independent Variables
2.3.6. Model Construction
2.4. Accuracy Verification Method
3. Results
3.1. Ground Elevation Inversion Using GEDI L2A Data
3.2. Forest Canopy Height Inversion Using GEDI L2A Data
3.3. BP Neural Network Model Inversion Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alexander, C.; Korstjens, A.H.; Hill, R.A. Influence of micro-topography and crown characteristics on tree height estimations in tropical forests based on lidar canopy height models. Int. J. Appl. Earth Obs. Geoinf. 2018, 65, 105–113. [Google Scholar] [CrossRef]
- Asner, G.P.; Mascaro, J. Mapping tropical forest carbon: Calibrating plot estimates to a simple lidar metric. Remote Sens. Environ. 2014, 140, 614–624. [Google Scholar] [CrossRef]
- Lucas, R.; Van De Kerchove, R.; Otero, V.; Lagomasino, D.; Fatoyinbo, L.; Omar, H.; Satyanarayana, B.; Dahdouh-Guebas, F. Structural characterisation of mangrove forests achieved through combining multiple sources of remote sensing data. Remote Sens. Environ. 2020, 237, 111543. [Google Scholar] [CrossRef]
- Tuominen, S.; Eerikäinen, K.; Schibalski, A.; Haakana, M.; Lehtonen, A. Mapping biomass variables with a multi-source forest inventory technique. Silva Fenn. 2010, 44, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Su, Y.; Xue, B.; Liu, J.; Zhao, X.; Fang, J.; Guo, Q. Mapping global forest aboveground biomass with spaceborne lidar, optical imagery, and forest inventory data. Remote Sens. 2016, 8, 565. [Google Scholar] [CrossRef] [Green Version]
- Lefsky, M.A.; Cohen, W.B.; Parker, G.G.; Harding, D.J. Lidar remote sensing for ecosystem studies: Lidar, an emerging remote sensing technology that directly measures the three-dimensional distribution of plant canopies, can accurately estimate vegetation structural attributes and should be of particular interest to forest, landscape, and global ecologists. BioScience 2002, 52, 19–30. [Google Scholar] [CrossRef]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Koch, B. A framework for mapping tree species combining hyperspectral and lidar data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Jin, S.; Su, Y.; Gao, S.; Hu, T.; Liu, J.; Guo, Q. The transferability of random forest in canopy height estimation from multi-source remote sensing data. Remote Sens. 2018, 10, 1183. [Google Scholar] [CrossRef] [Green Version]
- Shao, Z.; Zhang, L.; Wang, L. Stacked sparse autoencoder modeling using the synergy of airborne lidar and satellite optical and sar data to map forest above-ground biomass. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 5569–5582. [Google Scholar] [CrossRef]
- Klosterman, S.; Melaas, E.; Wang, J.A.; Martinez, A.; Frederick, S.; O’Keefe, J.; Orwig, D.A.; Wang, Z.; Sun, Q.; Schaaf, C.; et al. Fine-scale perspectives on landscape phenology from unmanned aerial vehicle (uav) photography. Agric. For. Meteorol. 2018, 248, 397–407. [Google Scholar] [CrossRef]
- Lu, B.; He, Y. Species classification using unmanned aerial vehicle (uav)-acquired high spatial resolution imagery in a heterogeneous grassland. ISPRS J. Photogramm. Remote Sens. 2017, 128, 73–85. [Google Scholar] [CrossRef]
- Kayitakire, F.; Hamel, C.; Defourny, P. Retrieving forest structure variables based on image texture analysis and ikonos-2 imagery. Remote Sens. Environ. 2006, 102, 390–401. [Google Scholar] [CrossRef]
- Irons, J.R.; Dwyer, J.L.; Barsi, J.A. The next landsat satellite: The landsat data continuity mission. Remote Sens. Environ. 2012, 122, 11–21. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Wang, C.; Nie, S.; Pan, F.; Xi, X.; Hu, Z. Mapping forest height using photon-counting lidar data and landsat 8 oli data: A case study in virginia and north carolina, USA. Ecol. Indic. 2020, 114, 106287. [Google Scholar] [CrossRef]
- Wang, Z.; Schaaf, C.B.; Lewis, P.; Knyazikhin, Y.; Schull, M.A.; Strahler, A.H.; Yao, T.; Myneni, R.B.; Chopping, M.J.; Blair, B.J. Retrieval of canopy height using moderate-resolution imaging spectroradiometer (modis) data. Remote Sens. Environ. 2011, 115, 1595–1601. [Google Scholar] [CrossRef] [Green Version]
- Gupta, P.; Christopher, S.A.; Wang, J.; Gehrig, R.; Lee, Y.; Kumar, N. Satellite remote sensing of particulate matter and air quality assessment over global cities. Atmos. Environ. 2006, 40, 5880–5892. [Google Scholar] [CrossRef]
- Smith, L.C. Satellite remote sensing of river inundation area, stage, and discharge: A review. Hydrol. Process. 1997, 11, 1427–1439. [Google Scholar] [CrossRef]
- Kumar, P.; Krishna, A.P. Insar-based tree height estimation of hilly forest using multitemporal radarsat-1 and sentinel-1 sar data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 5147–5152. [Google Scholar] [CrossRef]
- Pourshamsi, M.; Xia, J.; Yokoya, N.; Garcia, M.; Lavalle, M.; Pottier, E.; Balzter, H. Tropical forest canopy height estimation from combined polarimetric sar and lidar using machine-learning. ISPRS J. Photogramm. Remote Sens. 2021, 172, 79–94. [Google Scholar] [CrossRef]
- Niculescu, S.; Lardeux, C.; Grigoras, I.; Hanganu, J.; David, L. Synergy between lidar, radarsat-2, and spot-5 images for the detection and mapping of wetland vegetation in the danube delta. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 3651–3666. [Google Scholar] [CrossRef] [Green Version]
- Mallet, C.; Bretar, F. Full-waveform topographic lidar: State-of-the-art. ISPRS J. Photogramm. Remote Sens. 2009, 64, 1–16. [Google Scholar] [CrossRef]
- White, J.C.; Wulder, M.A.; Vastaranta, M.; Coops, N.C.; Pitt, D.; Woods, M. The utility of image-based point clouds for forest inventory: A comparison with airborne laser scanning. Forests 2013, 4, 518–536. [Google Scholar] [CrossRef] [Green Version]
- Wilkes, P.; Jones, S.D.; Suarez, L.; Mellor, A.; Woodgate, W.; Soto-Berelov, M.; Haywood, A.; Skidmore, A.K. Mapping forest canopy height across large areas by upscaling als estimates with freely available satellite data. Remote Sens. 2015, 7, 12563–12587. [Google Scholar] [CrossRef] [Green Version]
- Ben-Arie, J.R.; Hay, G.J.; Powers, R.P.; Castilla, G.; St-Onge, B. Development of a pit filling algorithm for lidar canopy height models. Comput. Geosci. 2009, 35, 1940–1949. [Google Scholar] [CrossRef]
- Disney, M.I.; Kalogirou, V.; Lewis, P.; Prieto-Blanco, A.; Hancock, S.; Pfeifer, M. Simulating the impact of discrete-return lidar system and survey characteristics over young conifer and broadleaf forests. Remote Sens. Environ. 2010, 114, 1546–1560. [Google Scholar] [CrossRef]
- Ma, Q.; Su, Y.; Guo, Q. Comparison of canopy cover estimations from airborne lidar, aerial imagery, and satellite imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 4225–4236. [Google Scholar] [CrossRef]
- Verrelst, J.; Rivera, J.P.; Veroustraete, F.; Muñoz-Marí, J.; Clevers, J.G.P.W.; Camps-Valls, G.; Moreno, J. Experimental sentinel-2 lai estimation using parametric, non-parametric and physical retrieval methods—A comparison. ISPRS J. Photogramm. Remote Sens. 2015, 108, 260–272. [Google Scholar] [CrossRef]
- Li, G.; Xie, Z.; Jiang, X.; Lu, D.; Chen, E. Integration of ziyuan-3 multispectral and stereo data for modeling aboveground biomass of larch plantations in north China. Remote Sens. 2019, 11, 2328. [Google Scholar] [CrossRef] [Green Version]
- Strahler, A.H. Vegetation canopy reflectance modeling—Recent developments and remote sensing perspectives. Remote Sens. Rev. 1997, 15, 179–194. [Google Scholar] [CrossRef]
- Stojanova, D.; Panov, P.; Gjorgjioski, V.; Kobler, A.; Džeroski, S. Estimating vegetation height and canopy cover from remotely sensed data with machine learning. Ecol. Inform. 2010, 5, 256–266. [Google Scholar] [CrossRef]
- Massman, W. A comparative study of some mathematical models of the mean wind structure and aerodynamic drag of plant canopies. Bound. Layer Meteorol. 1987, 40, 179–197. [Google Scholar] [CrossRef]
- Pittman, J.J.; Arnall, D.B.; Interrante, S.M.; Moffet, C.A.; Butler, T.J. Estimation of biomass and canopy height in bermudagrass, alfalfa, and wheat using ultrasonic, laser, and spectral sensors. Sensors 2015, 15, 2920–2943. [Google Scholar] [CrossRef] [Green Version]
- Verstraete, M.M.; Pinty, B.; Myneni, R.B. Potential and limitations of information extraction on the terrestrial biosphere from satellite remote sensing. Remote Sens. Environ. 1996, 58, 201–214. [Google Scholar] [CrossRef]
- Pourshamsi, M.; Garcia, M.; Lavalle, M.; Balzter, H. A machine-learning approach to polinsar and lidar data fusion for improved tropical forest canopy height estimation using nasa afrisar campaign data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 3453–3463. [Google Scholar] [CrossRef] [Green Version]
- Han, T.; Jiang, D.; Zhao, Q.; Wang, L.; Yin, K. Comparison of random forest, artificial neural networks and support vector machine for intelligent diagnosis of rotating machinery. Trans. Inst. Meas. Control 2018, 40, 2681–2693. [Google Scholar] [CrossRef]
- Rodriguez-Galiano, V.; Sanchez-Castillo, M.; Chica-Olmo, M.; Chica-Rivas, M. Machine learning predictive models for mineral prospectivity: An evaluation of neural networks, random forest, regression trees and support vector machines. Ore Geol. Rev. 2015, 71, 804–818. [Google Scholar] [CrossRef]
- Otchere, D.A.; Arbi Ganat, T.O.; Gholami, R.; Ridha, S. Application of supervised machine learning paradigms in the prediction of petroleum reservoir properties: Comparative analysis of ann and svm models. J. Pet. Sci. Eng. 2021, 200, 108182. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, cnn architectures, challenges, applications, future directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Han, L.; Li, L. Harmonizing surface reflectance between landsat-7 etm+, landsat-8 oli, and sentinel-2 msi over China. Environ. Sci. Pollut. Res. 2022, 29, 70882–70898. [Google Scholar] [CrossRef] [PubMed]
- Scholl, V.M.; Cattau, M.E.; Joseph, M.B.; Balch, J.K. Integrating national ecological observatory network (neon) airborne remote sensing and in-situ data for optimal tree species classification. Remote Sens. 2020, 12, 1414. [Google Scholar] [CrossRef]
- Liu, A.; Cheng, X.; Chen, Z. Performance evaluation of gedi and icesat-2 laser altimeter data for terrain and canopy height retrievals. Remote Sens. Environ. 2021, 264, 112571. [Google Scholar] [CrossRef]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote sensing technologies for enhancing forest inventories: A review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef] [Green Version]
- Potapov, P.; Li, X.; Hernandez-Serna, A.; Tyukavina, A.; Hansen, M.C.; Kommareddy, A.; Pickens, A.; Turubanova, S.; Tang, H.; Silva, C.E.; et al. Mapping global forest canopy height through integration of gedi and landsat data. Remote Sens. Environ. 2021, 253, 112165. [Google Scholar] [CrossRef]
- Dubayah, R.; Blair, J.B.; Goetz, S.; Fatoyinbo, L.; Hansen, M.; Healey, S.; Hofton, M.; Hurtt, G.; Kellner, J.; Luthcke, S.; et al. The global ecosystem dynamics investigation: High-resolution laser ranging of the earth’s forests and topography. Sci. Remote Sens. 2020, 1, 100002. [Google Scholar] [CrossRef]
- Adam, M.; Urbazaev, M.; Dubois, C.; Schmullius, C. Accuracy assessment of gedi terrain elevation and canopy height estimates in european temperate forests: Influence of environmental and acquisition parameters. Remote Sens. 2020, 12, 3948. [Google Scholar] [CrossRef]
- Urbazaev, M.; Hess, L.L.; Hancock, S.; Sato, L.Y.; Ometto, J.P.; Thiel, C.; Dubois, C.; Heckel, K.; Urban, M.; Adam, M.; et al. Assessment of terrain elevation estimates from icesat-2 and gedi spaceborne lidar missions across different land cover and forest types. Sci. Remote Sens. 2022, 6, 100067. [Google Scholar] [CrossRef]
- Masek, J.G.; Wulder, M.A.; Markham, B.; McCorkel, J.; Crawford, C.J.; Storey, J.; Jenstrom, D.T. Landsat 9: Empowering open science and applications through continuity. Remote Sens. Environ. 2020, 248, 111968. [Google Scholar] [CrossRef]
- Watanabe, F.S.Y.; Alcântara, E.; Rodrigues, T.W.P.; Imai, N.N.; Barbosa, C.C.F.; Rotta, L.H.d.S. Estimation of chlorophyll-a concentration and the trophic state of the barra bonita hydroelectric reservoir using oli/landsat-8 images. Int. J. Environ. Res. Public Health 2015, 12, 10391–10417. [Google Scholar] [CrossRef] [Green Version]
- Goh, A.T.C. Back-propagation neural networks for modeling complex systems. Artif. Intell. Eng. 1995, 9, 143–151. [Google Scholar] [CrossRef]
- Zhang, G.; Xia, B.; Wang, J. Intelligent state of charge estimation of lithium-ion batteries based on l-m optimized back-propagation neural network. J. Energy Storage 2021, 44, 103442. [Google Scholar] [CrossRef]
- Poorani, S.; Balasubramanie, P. Seizure detection based on eeg signals using asymmetrical back propagation neural network method. Circuits Syst. Signal Process. 2021, 40, 4614–4632. [Google Scholar] [CrossRef]
- Hochreiter, S. The vanishing gradient problem during learning recurrent neural nets and problem solutions. Int. J. Uncertain. Fuzziness Knowl. Based Syst. 1998, 6, 107–116. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.-H.; Kim, Y.-T.; Lee, S.-R. Shallow landslide susceptibility models based on artificial neural networks considering the factor selection method and various non-linear activation functions. Remote Sens. 2020, 12, 1194. [Google Scholar] [CrossRef] [Green Version]
- Varshney, M.; Singh, P. Optimizing nonlinear activation function for convolutional neural networks. Signal Image Video Process. 2021, 15, 1323–1330. [Google Scholar] [CrossRef]
- Chen, J.-C.; Wang, Y.-M. Comparing activation functions in modeling shoreline variation using multilayer perceptron neural network. Water 2020, 12, 1281. [Google Scholar] [CrossRef]
- Lederer, J. Activation functions in artificial neural networks: A systematic overview. arXiv 2021, arXiv:2101.09957. [Google Scholar] [CrossRef]
- Dubey, S.R.; Singh, S.K.; Chaudhuri, B.B. Activation functions in deep learning: A comprehensive survey and benchmark. Neurocomputing 2022, 503, 92–108. [Google Scholar] [CrossRef]
- Tu, J.V. Advantages and disadvantages of using artificial neural networks versus logistic regression for predicting medical outcomes. J. Clin. Epidemiol. 1996, 49, 1225–1231. [Google Scholar] [CrossRef]
- Mastromichalakis, S. Alrelu: A different approach on leaky relu activation function to improve neural networks performance. arXiv 2020, arXiv:2012.07564. [Google Scholar] [CrossRef]
- Alhassan, A.M.; Zainon, W.M.N.W. Brain tumor classification in magnetic resonance image using hard swish-based relu activation function-convolutional neural network. Neural Comput. Appl. 2021, 33, 9075–9087. [Google Scholar] [CrossRef]
- Misra, D. Mish: A self regularized non-monotonic activation function. arXiv 2019, arXiv:1908.08681. [Google Scholar] [CrossRef]
- Göçken, M.; Özçalıcı, M.; Boru, A.; Dosdoğru, A.T. Integrating metaheuristics and artificial neural networks for improved stock price prediction. Expert Syst. Appl. 2016, 44, 320–331. [Google Scholar] [CrossRef]
- Xiao, Y.; Wu, J.; Lin, Z.; Zhao, X. A deep learning-based multi-model ensemble method for cancer prediction. Comput. Methods Programs Biomed. 2018, 153, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhao, W.; Zhang, Y.; Zio, E. Remaining useful life prediction using multi-scale deep convolutional neural network. Appl. Soft Comput. 2020, 89, 106113. [Google Scholar] [CrossRef]
- Zhu, W.-D.; Qian, C.-Y.; He, N.-Y.; Kong, Y.-X.; Zou, Z.-Y.; Li, Y.-W. Research on chlorophyll-a concentration retrieval based on bp neural network model—Case study of dianshan lake, China. Sustainability 2022, 14, 8894. [Google Scholar] [CrossRef]
- Ota, T.; Ahmed, O.S.; Franklin, S.E.; Wulder, M.A.; Kajisa, T.; Mizoue, N.; Yoshida, S.; Takao, G.; Hirata, Y.; Furuya, N.; et al. Estimation of airborne lidar-derived tropical forest canopy height using landsat time series in cambodia. Remote Sens. 2014, 6, 10750–10772. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.-J.; Lee, C.-W. Forest canopy height estimation using multiplatform remote sensing dataset. J. Sens. 2018, 2018, 1593129. [Google Scholar] [CrossRef] [Green Version]
- Peng, D.; Zhang, H.; Liu, L.; Huang, W.; Huete, A.R.; Zhang, X.; Wang, F.; Yu, L.; Xie, Q.; Wang, C.; et al. Estimating the aboveground biomass for planted forests based on stand age and environmental variables. Remote Sens. 2019, 11, 2270. [Google Scholar] [CrossRef] [Green Version]
- Bye, I.J.; North, P.R.J.; Los, S.O.; Kljun, N.; Rosette, J.A.B.; Hopkinson, C.; Chasmer, L.; Mahoney, C. Estimating forest canopy parameters from satellite waveform lidar by inversion of the flight three-dimensional radiative transfer model. Remote Sens. Environ. 2017, 188, 177–189. [Google Scholar] [CrossRef] [Green Version]
- Koetz, B.; Morsdorf, F.; Sun, G.; Ranson, K.J.; Itten, K.; Allgower, B. Inversion of a lidar waveform model for forest biophysical parameter estimation. IEEE Geosci. Remote Sens. Lett. 2006, 3, 49–53. [Google Scholar] [CrossRef]
- Staben, G.; Lucieer, A.; Scarth, P. Modelling lidar derived tree canopy height from landsat tm, etm+ and oli satellite imagery—A machine learning approach. Int. J. Appl. Earth Obs. Geoinf. 2018, 73, 666–681. [Google Scholar] [CrossRef]
- Campbell, M.J.; Dennison, P.E.; Kerr, K.L.; Brewer, S.C.; Anderegg, W.R.L. Scaled biomass estimation in woodland ecosystems: Testing the individual and combined capacities of satellite multispectral and lidar data. Remote Sens. Environ. 2021, 262, 112511. [Google Scholar] [CrossRef]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the modis vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS; NASA Special Publication; NASA: Washington, DC, USA, 1974; Volume 351, p. 309. [Google Scholar]
- Huete, A.R. A soil-adjusted vegetation index (savi). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Lymburner, L.; Beggs, P.J.; Jacobson, C.R. Estimation of canopy-average surface-specific leaf area using landsat tm data. Photogramm. Eng. Remote Sens. 2000, 66, 183–192. [Google Scholar]
- Jordan, C.F. Derivation of leaf-area index from quality of light on the forest floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Viña, A. Remote Detection of Biophysical Properties of Plant Canopies. 2002. Available online: http://calmaps.unl.edu/snrscoq/snrs_colloquium_2002_andres_vina.ppt (accessed on 16 May 2003).
- Richardson, A.J.; Wiegand, C. Distinguishing vegetation from soil background information. Photogramm. Eng. Remote Sens. 1977, 43, 1541–1552. [Google Scholar]
- Huete, A.R.; Liu, H.Q. An error and sensitivity analysis of the atmospheric- and soil-correcting variants of the ndvi for the modis-eos. IEEE Trans. Geosci. Remote Sens. 1994, 32, 897–905. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Wylie, B.; Yang, L.; Homer, C.; Zylstra, G. Derivation of a tasselled cap transformation based on landsat 7 at-satellite reflectance. Int. J. Remote Sens. 2002, 23, 1741–1748. [Google Scholar] [CrossRef]
- Guliyev, N.J.; Ismailov, V.E. A single hidden layer feedforward network with only one neuron in the hidden layer can approximate any univariate function. Neural Comput. 2016, 28, 1289–1304. [Google Scholar] [CrossRef] [Green Version]
- Chugh, S.; Ghosh, S.; Gulistan, A.; Rahman, B.M.A. Machine learning regression approach to the nanophotonic waveguide analyses. J. Light. Technol. 2019, 37, 6080–6089. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.; Ma, G.; Su, S.-F.; Huang, C.; Boswell, M.K.; Xue, P. A multi-layer perceptron approach for accelerated wave forecasting in lake michigan. Ocean Eng. 2020, 211, 107526. [Google Scholar] [CrossRef]
- Baressi Šegota, S.; Anđelić, N.; Kudláček, J.; Čep, R. Artificial neural network for predicting values of residuary resistance per unit weight of displacement. Pomor. Zb. 2019, 57, 9–22. [Google Scholar]
- Dubayah, R.; Hofton, M.; Blair, J.; Armston, J.; Tang, H.; Luthcke, S. Gedi l2a Elevation and Height Metrics Data Global Footprint Level v001; NASA EOSDIS Land Processes DAAC: Sioux Falls, SD, USA, 2020. [Google Scholar] [CrossRef]
- Wang, C.; Elmore, A.J.; Numata, I.; Cochrane, M.A.; Shaogang, L.; Huang, J.; Zhao, Y.; Li, Y. Factors affecting relative height and ground elevation estimations of gedi among forest types across the conterminous USA. GISci. Remote Sens. 2022, 59, 975–999. [Google Scholar] [CrossRef]
- Dorado-Roda, I.; Pascual, A.; Godinho, S.; Silva, C.A.; Botequim, B.; Rodríguez-Gonzálvez, P.; González-Ferreiro, E.; Guerra-Hernández, J. Assessing the accuracy of gedi data for canopy height and aboveground biomass estimates in mediterranean forests. Remote Sens. 2021, 13, 2279. [Google Scholar] [CrossRef]
- Wang, C.; Elmore, A.J.; Numata, I.; Cochrane, M.A.; Lei, S.; Hakkenberg, C.R.; Li, Y.; Zhao, Y.; Tian, Y. A framework for improving wall-to-wall canopy height mapping by integrating gedi lidar. Remote Sens. 2022, 14, 3618. [Google Scholar] [CrossRef]
- Guo, Z.-H.; Wu, J.; Lu, H.-Y.; Wang, J.-Z. A case study on a hybrid wind speed forecasting method using bp neural network. Knowl. Based Syst. 2011, 24, 1048–1056. [Google Scholar] [CrossRef]
- Li, G.; Shi, J. On comparing three artificial neural networks for wind speed forecasting. Appl. Energy 2010, 87, 2313–2320. [Google Scholar] [CrossRef]
- Yu, W.; Li, B.; Jia, H.; Zhang, M.; Wang, D. Application of multi-objective genetic algorithm to optimize energy efficiency and thermal comfort in building design. Energy Build. 2015, 88, 135–143. [Google Scholar] [CrossRef]
- Guresen, E.; Kayakutlu, G.; Daim, T.U. Using artificial neural network models in stock market index prediction. Expert Syst. Appl. 2011, 38, 10389–10397. [Google Scholar] [CrossRef]
- Lee, D.; Derrible, S.; Pereira, F.C. Comparison of four types of artificial neural network and a multinomial logit model for travel mode choice modeling. Transp. Res. Rec. 2018, 2672, 101–112. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Huang, Y.; Ma, G. A beetle antennae search improved bp neural network model for predicting multi-factor-based gas explosion pressures. J. Loss Prev. Process Ind. 2020, 65, 104117. [Google Scholar] [CrossRef]







| Parameter Name | Parameter Size |
|---|---|
| Track height | 400 km |
| Coverage | 51.6° N–51.6° S |
| Repetition rate | 242 Hz |
| Pulse width | 15 ns |
| Wavelength | 1064 nm |
| Footprint | 25 m |
| Geolocation error | 8 m |
| Along-track distances | 60 m |
| Across-track distances | 600 m |
| Study Area | Acquisition Date | File Size (GB) |
|---|---|---|
| HARV | 4 July 2022 | 2.05 |
| 8 July 2022 | 2.35 | |
| 14 July 2022 | 2.49 | |
| 3 August 2022 | 2.37 | |
| 9 August 2022 | 2.22 | |
| 26 August 2022 | 2.20 | |
| 2 September 2022 | 2.70 | |
| 6 September 2022 | 1.92 | |
| 23 September 2022 | 2.37 | |
| 27 September 2022 | 2.28 |
| Algorithm Setting Group | Smoothing Width (Noise) | Smoothing Width (Signal) | Waveform Signal Start Threshold | Waveform Signal End Threshold |
|---|---|---|---|---|
| a1 | 6.5σ | 6.5σ | 3σ | 6σ |
| a2 | 6.5σ | 3.5σ | 3σ | 3σ |
| a3 | 6.5σ | 3.5σ | 3σ | 6σ |
| a4 | 6.5σ | 6.5σ | 6σ | 6σ |
| a5 | 6.5σ | 3.5σ | 3σ | 2σ |
| a6 | 6.5σ | 3.5σ | 3σ | 4σ |
| Band Name | Band Range (μm) | Spatial Resolution (m) |
|---|---|---|
| Band 1 Coastal | 0.43–0.45 | 30 |
| Band 2 Blue | 0.45–0.51 | 30 |
| Band 3 Green | 0.53–0.59 | 30 |
| Band 4 Red | 0.64–0.67 | 30 |
| Band 5 NIR | 0.85–0.88 | 30 |
| Band 6 SWIR 1 | 1.57–1.65 | 30 |
| Band 7 SWIR 2 | 2.11–2.29 | 30 |
| Band 8 PAN | 0.50–0.68 | 15 |
| Band 9 Cirrus | 1.36–1.38 | 30 |
| Sensor | Path/Row | Study Area | Acquisition Date |
|---|---|---|---|
| OLI | 013/030 | HARV | 6 August 2022 |
| OLI-2 | 013/030 | 14 August 2022 |
| Independent Variable | Variable Information | References |
|---|---|---|
| rh25, rh50, rh60, rh75, rh85, rh90, rh95, rh100 | Relative height metrics at (25, 50, 60, 75, 85, 90, 95, 100)% | – |
| EVI | [73] | |
| NDVI | [74] | |
| SAVI | [75] | |
| SLAVI | [76] | |
| RVI | [77] | |
| VI3 | [78] | |
| PVI | [79] | |
| SARVI | [80] | |
| DVI | [81] | |
| ARVI | [80] | |
| TCG | Greenness of TCT | [82] |
| TCB | Brightness of TCT | [82] |
| TCW | Wetness of TCT | [82] |
| Model | Independent Variables | Number of Independent Variables |
|---|---|---|
| GEDI | rh75, rh60, rh85, rh95, rh100, rh25, rh90 | 7 |
| OLI/OLI-2 | SLAVI, TCB, VI3, EVI, ARVI, TCG, TCW, DVI, PVI | 9 |
| GEDI and OLI/OLI-2 | rh75, rh60, rh85, rh95, VI3, SLAVI, rh90, rh100, EVI, rh25, rh50, DVI, TCW | 13 |
| Data Group | Number of Neurons | ||
|---|---|---|---|
| Input Layer | Hidden Layer | Output Layer | |
| GEDI | 7 | 9 | 1 |
| OLI | 9 | 8 | 1 |
| OLI-2 | 9 | 8 | 1 |
| GEDI and OLI | 13 | 11 | 1 |
| GEDI and OLI-2 | 13 | 11 | 1 |
| Relative Height | Evaluation Indicator | |||
|---|---|---|---|---|
| R-Squared | MAE (m) | RMSE (m) | rRMSE | |
| rh90 | 0.30 | 3.66 | 5.50 | 24.04% |
| rh91 | 0.32 | 3.58 | 5.43 | 23.72% |
| rh92 | 0.33 | 3.51 | 5.36 | 23.42% |
| rh93 | 0.34 | 3.45 | 5.29 | 23.12% |
| rh94 | 0.35 | 3.41 | 5.24 | 22.89% |
| rh95 | 0.35 | 3.41 | 5.22 | 22.81% |
| rh96 | 0.34 | 3.44 | 5.24 | 22.88% |
| rh97 | 0.32 | 3.54 | 5.29 | 23.10% |
| rh98 | 0.28 | 3.71 | 5.42 | 23.68% |
| rh99 | 0.22 | 4.07 | 5.69 | 24.85% |
| rh100 | 0.04 | 4.95 | 6.38 | 27.87% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, W.; Yang, F.; Qiu, Z.; He, N.; Zhu, X.; Li, Y.; Xu, Y.; Lu, Z. Enhancing Forest Canopy Height Retrieval: Insights from Integrated GEDI and Landsat Data Analysis. Sustainability 2023, 15, 10434. https://doi.org/10.3390/su151310434
Zhu W, Yang F, Qiu Z, He N, Zhu X, Li Y, Xu Y, Lu Z. Enhancing Forest Canopy Height Retrieval: Insights from Integrated GEDI and Landsat Data Analysis. Sustainability. 2023; 15(13):10434. https://doi.org/10.3390/su151310434
Chicago/Turabian StyleZhu, Weidong, Fei Yang, Zhenge Qiu, Naiying He, Xiaolong Zhu, Yaqin Li, Yuelin Xu, and Zhigang Lu. 2023. "Enhancing Forest Canopy Height Retrieval: Insights from Integrated GEDI and Landsat Data Analysis" Sustainability 15, no. 13: 10434. https://doi.org/10.3390/su151310434
APA StyleZhu, W., Yang, F., Qiu, Z., He, N., Zhu, X., Li, Y., Xu, Y., & Lu, Z. (2023). Enhancing Forest Canopy Height Retrieval: Insights from Integrated GEDI and Landsat Data Analysis. Sustainability, 15(13), 10434. https://doi.org/10.3390/su151310434






