Cadmium Accumulation in Plants: Insights from Phylogenetic Variation into the Evolution and Functions of Membrane Transporters
Abstract
:1. Introduction
2. Phylogenetic Variations in Cd Accumulation in Plants
3. Physiological Processes Mediating Cadmium Accumulation in Plants
3.1. Cd Uptake by Roots
3.2. Root-to-Shoot Cd Translocation
3.3. Intracellular Cd Sequestration
3.4. Cd Accumulation in Shoots and Grains
4. Evolution of Cadmium Transporters and Their Functions in Cadmium Accumulation
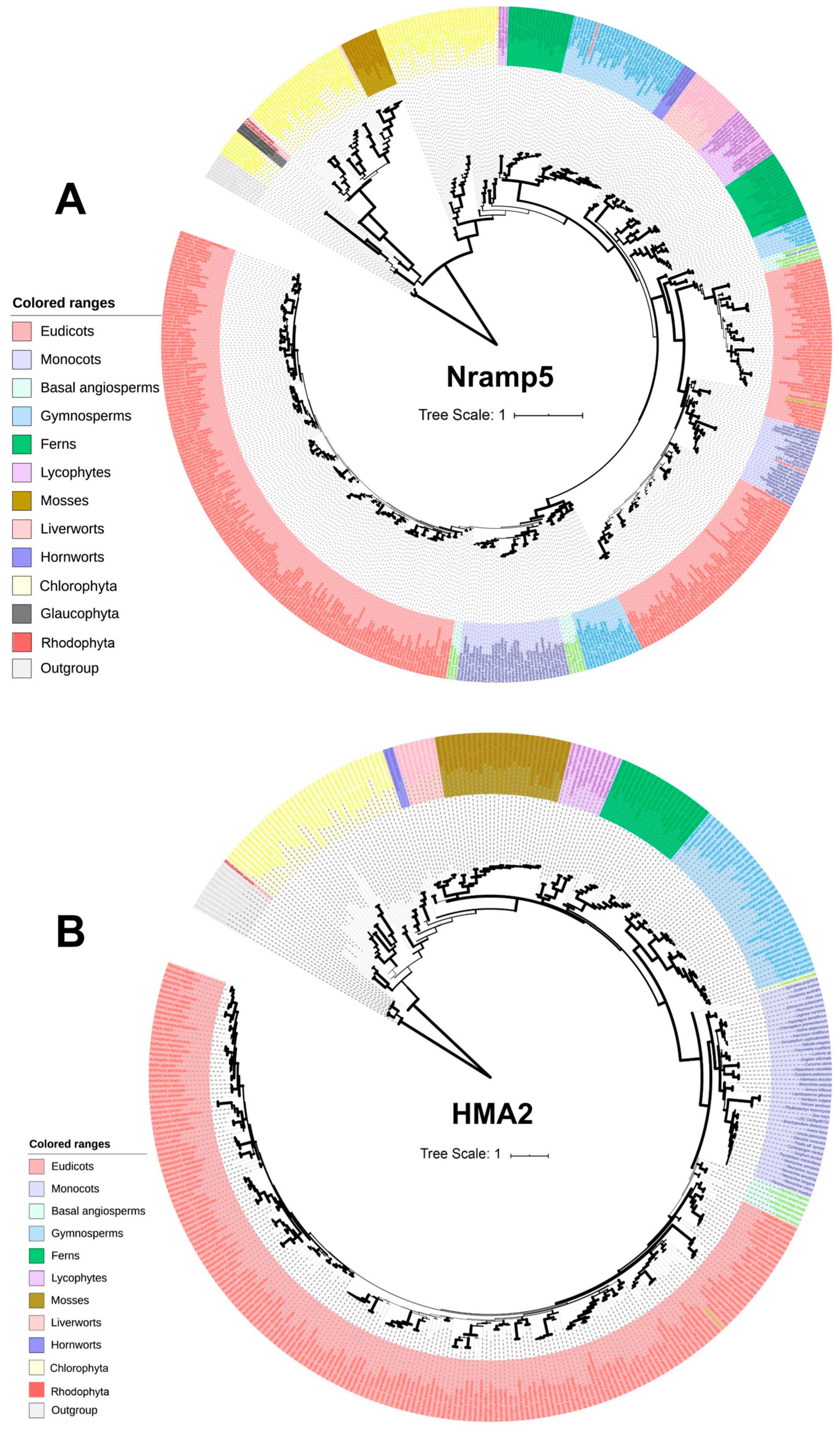
4.1. Evolution of Cd Transporters
4.2. Function of Cd Transporters
4.2.1. NRAMPs
4.2.2. HMAs
4.2.3. ZIPs
4.2.4. CDFs
4.2.5. OPTs
4.2.6. ABCCs
4.2.7. CAXs
4.2.8. Other Transporters Involved in the Uptake and Intervascular Transfer of Cd
| Family | Gene Symbol | Expression Organ and Localization | Plant Species | Possible Properties | References |
|---|---|---|---|---|---|
| NRAMPs | AtNRAMP3/4 | root, leaf (tonoplast) | Arabidopsis thaliana | Cd, Fe, Mn | [103,154] |
| NcNRAMP1 | root, shoot (PM, tonoplast) | Noccaea caerulescens | Cd | [110] | |
| TcNRAMP3/4 | root, shoot (tonoplast) | Thlaspi caerulescens | Cd, Fe, Mn | [155] | |
| OsNRAMP1 | root, shoot (PM) | Oryza sativa | Cd | [104] | |
| OsNRAMP5 | root (PM) | Oryza sativa | Cd, Fe, Mn | [156,157] | |
| HvNRAMP5 | root (PM) | Hordeum vulgare | Cd, Mn | [109] | |
| HMAs | AtHMA1 | root, shoot (chloroplast envelope) | Arabidopsis thaliana | Cd, Zn, Cu | [158,159] |
| AtHMA3 | root, collar, leaf (tonoplast) | Arabidopsis thaliana | Cd, Zn, Pb, Co | [160,161,162] | |
| AtHMA2/4 | root, stem, leaf (PM) | Arabidopsis thaliana | Cd, Zn | [163,164,165,166] | |
| OsHMA2 | root (PM) | Oryza sativa | Cd, Zn | [91,117] | |
| OsHMA3 | root (tonoplast) | Oryza sativa | Cd, Zn | [118,167,168] | |
| OsHMA9 | vascular bundle and anther (PM) | Oryza sativa | Cd, Cu, Zn, Pb | [169] | |
| TaHMA2 | root, shoot (PM) | Triticum aestivum | Cd, Zn | [170] | |
| GmHMA3 | root (ER) | Glycine max | Cd, Zn | [171] | |
| TcHMA3 | root, shoot (tonoplast) | Thlaspi caerulescens | Cd | [172] | |
| SaHMA3 | root, shoot (tonoplast) | Sedum alfredii | Cd | [173] | |
| SpHMA3 | root, shoot (tonoplast) | Sedum plumbizincicola | Cd, Zn | [115] | |
| ABCCs | AtABCC1/2 | root, shoot (tonoplast) | Arabidopsis thaliana | Cd-PC, Hg-PC, As(III)-PC | [82,174] |
| AtABCC3 | root, shoot (tonoplast) | Arabidopsis thaliana | Cd-PC | [143] | |
| AtPDR8 | root, shoot (PM) | Arabidopsis thaliana | Cd | [144] | |
| OsABCG36 | root, shoot (PM) | Arabidopsis thaliana | Cd | [146] | |
| CDFs | OsMTP1 | root, leaf (tonoplast) | Oryza sativa | Cd, Ni, Fe | [132] |
| TgMTP1 | root, leaf (tonoplast) | Thlaspi goesingense | Cd, Zn, Co, Ni | [133,175] | |
| CitMTP1 | root, leaf (tonoplast) | Citrus sinensis | Cd, Zn, Mn, Cu | [129] | |
| CsMTP1/4 | root, hypocotyl, cotyledon, petiole, leaf (tonoplast) | Cucumis sativus | Cd, Mn, Zn | [134] | |
| OPTs | ZmYS1 | leaf blade and sheath, crown, seminal root (PM) | Zea mays | Cu, Ni, Cd, Fe, Zn, Mn | [66] |
| OsYSL2 | shoot phloem (PM) | Oryza sativa | Fe(II)-NA, Mn-NA, Cd-NA | [138,176] | |
| AtOPT3 | root, shoot (PM) | Arabidopsis thaliana | Cd, Zn, Fe | [177] | |
| SnYSL3 | root, shoot (PM) | Sedum nigrum | Fe(II)-NA, Mn-NA, Cd-NA | [137] | |
| ZIPs | OsZIP1 | root, shoot (ER, PM) | Oryza sativa | Cd, Zn, Cu | [178] |
| TcIRT1 | root (PM) | Thlaspi caerulescens | Cd, Zn, Fe(II, III); Mn | [179] | |
| TcZNT1 | root, shoot (PM) | Thlaspi caerulescens | Cd, Zn | [126,127] | |
| CAXs | AtCAX2/4 | root (tonoplast) | Arabidopsis thaliana | Cd, Zn, Mn | [81,148] |
| AhCAX1 | root, shoot (tonoplast) | Arabidopsis halleri | Cd | [150] | |
| SaCAX2 | root, shoot (tonoplast) | Sedum alfredii | Cd | [151] | |
| Others | OsLCT1 | leaf, node, phloem parenchyma (PM) | Oryza sativa | Cd | [90,180] |
| OsLCD | root, shoot (cytoplasm, nucleus) | Oryza sativa | Cd | [181] | |
| OsCd1 | root (PM) | Oryza sativa | Cd | [70] | |
| CAL1 | root, leaf sheath, internode (CW) | Oryza sativa | Cd | [152] |
5. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rai, P.K.; Lee, S.S.; Zhang, M.; Tsang, Y.F.; Kim, K.H. Heavy metals in food crops: Health risks, fate, mechanisms, and management. Environ. Int. 2019, 125, 365–385. [Google Scholar] [CrossRef] [PubMed]
- Clemens, S. Safer food through plant science: Reducing toxic element accumulation in crops. J. Exp. Bot. 2019, 70, 5537–5557. [Google Scholar] [CrossRef] [PubMed]
- Gallego, S.M.; Pena, L.B.; Barcia, R.A.; Azpilicueta, C.E.; Iannone, M.F.; Rosales, E.P.; Zawoznik, M.S.; Groppa, M.D.; Benavides, M.P. Unravelling cadmium toxicity and tolerance in plants: Insight into regulatory mechanisms. Environ. Exp. Bot. 2012, 83, 33–46. [Google Scholar] [CrossRef]
- Ismael, M.A.; Elyamine, A.M.; Moussa, M.G.; Cai, M.M.; Zhao, X.H.; Hu, C.X. Cadmium in plants: Uptake, toxicity, and its interactions with selenium fertilizers. Metallomics 2019, 11, 255–277. [Google Scholar] [CrossRef]
- Khan, A.R.; Ullah, I.; Khan, A.L.; Park, G.S.; Waqas, M.; Hong, S.J.; Jung, B.K.; Kwak, Y.; Lee, I.J.; Shin, J.H. Improvement in phytoremediation potential of Solanum nigrum under cadmium contamination through endophytic-assisted Serratia sp. RSC-14 inoculation. Environ. Sci. Pollut. Res. 2015, 22, 14032–14042. [Google Scholar] [CrossRef]
- Clemens, S.; Ma, J.F. Toxic heavy metal and metalloid accumulation in crop plants and foods. Annu. Rev. Plant Biol. 2016, 67, 489–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahid, M.; Dumat, C.; Khalid, S.; Schreck, E.; Xiong, T.; Niazi, N.K. Foliar heavy metal uptake, toxicity and detoxification in plants: A comparison of foliar and root metal uptake. J. Hazard. Mater. 2017, 325, 36–58. [Google Scholar] [CrossRef] [Green Version]
- Shi, G.L.; Li, D.J.; Wang, Y.F.; Liu, C.H.; Hu, Z.B.; Lou, L.Q.; Rengel, Z.; Cai, Q.S. Accumulation and distribution of arsenic and cadmium in winter wheat (Triticum aestivum L.) at different developmental stages. Sci. Total Environ. 2019, 667, 532–539. [Google Scholar] [CrossRef]
- Fasani, E.; Manara, A.; Martini, F.; Furini, A.; DalCorso, G. The potential of genetic engineering of plants for the remediation of soils contaminated with heavy metals. Plant Cell Environ. 2018, 41, 1201–1232. [Google Scholar] [CrossRef]
- Cappa, J.J.; Pilon-Smits, E.A.H. Evolutionary aspects of elemental hyperaccumulation. Planta 2014, 239, 267–275. [Google Scholar] [CrossRef] [Green Version]
- Zeng, F.; Mao, Y.; Cheng, W.; Wu, F.; Zhang, G. Genotypic and environmental variation in chromium, cadmium and lead concentrations in rice. Environ. Pollut. 2008, 153, 309–314. [Google Scholar] [CrossRef]
- Wu, D.; Sato, K.; Ma, J.F. Genome-wide association mapping of cadmium accumulation in different organs of barley. New Phytol. 2015, 208, 817–829. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wan, H.; Qian, J.; Guo, J.; Sun, C.; Wen, J.; Yi, B.; Ma, C.; Tu, J.; Song, L.; et al. Genome-wide association study of cadmium accumulation at the seedling stage in rapeseed (Brassica napus L.). Front. Plant Sci. 2018, 9, 375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klimmek, S.; Stan, H.J.; Wilke, A.; Bunke, G.; Buchholz, R. Comparative analysis of the biosorption of cadmium, lead, nickel, and zinc by algae. Environ. Sci. Technol. 2001, 35, 4283–4288. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekharaiah, P.S.; Sanyal, D.; Dasgupta, S.; Banik, A. Cadmium biosorption and biomass production by two freshwater microalgae Scenedesmus acutus and Chlorella pyrenoidosa: An integrated approach. Chemosphere 2021, 269, 128755. [Google Scholar]
- Park, E.K.; Lee, S.E. Cadmium uptake by non-viable biomass from a marine brown alga Ecklonia radiata turn. Biotechnol. Bioprocess Eng. 2002, 7, 221–224. [Google Scholar] [CrossRef]
- Mazur, L.P.; Cechinel, M.A.P.; de Souza, S.M.A.G.U.; Boaventura, R.A.R.; Vilar, V.J.P. Brown marine macroalgae as natural cation exchangers for toxic metal removal from industrial wastewaters: A review. J. Environ. Manag. 2018, 223, 215–253. [Google Scholar] [CrossRef]
- Vigneault, B.; Campbell, P.G.C. Uptake of cadmium by freshwater green algae: Effects of pH and aquatic humic substances. J. Phycol. 2005, 41, 55–61. [Google Scholar] [CrossRef]
- Monteiro, C.M.; Castro, P.M.L.; Malcata, F.X. Use of the microalga Scenedesmus obliquus to remove cadmium cations from aqueous solutions. World J. Microb. Biot. 2009, 25, 1573–1578. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.H.; Zhang, J.; Liu, Y.; Jia, Y.; Jiao, Y.N.; Xu, B.; Chen, Z.D. Diversity, phylogeny, and adaptation of bryophytes: Insights from genomic and transcriptomic data. J. Exp. Bot. 2022, 73, 4306–4322. [Google Scholar] [CrossRef]
- Shaw, A.J.; Szövényi, P.; Shaw, B. Bryophyte diversity and evolution: Windows into the early evolution of land plants. Am. J. Bot. 2011, 98, 352–369. [Google Scholar] [CrossRef] [PubMed]
- Stanković, J.D.; Sabovljević, A.D.; Sabovljević, M.S. Bryophytes and heavy metals: A review. Acta Bot. Croat. 2018, 77, 109–118. [Google Scholar] [CrossRef]
- Mahapatra, B.; Dhal, N.K.; Dash, A.K.; Panda, B.P.; Panigrahi, K.C.S.; Pradhan, A. Perspective of mitigating atmospheric heavy metal pollution: Using mosses as biomonitoring and indicator organism. Environ. Sci. Pollut. Res. 2019, 26, 29620–29638. [Google Scholar] [CrossRef]
- Vukojević, V.; Sabovljević, M.; Jovanović, S. Mosses accumulate heavy metals from the substrata of coal ash. Arch. Biol. Sci. 2005, 57, 101–106. [Google Scholar] [CrossRef]
- Macedo-Miranda, G.; Avila-Pérez, P.; Gil-Vargas, P.; Zarazúa, G.; Sánchez-Meza, J.C.; Zepeda-Gómez, C.; Tejeda, S. Accumulation of heavy metals in mosses: A biomonitoring study. Springerplus 2016, 5, 715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pipíška, M.; Horník, M.; Remenárová, L.; Augustín, J.; Lesný, J. Biosorption of Cadmium, Cobalt and Zinc by Moss Rhytidiadelphus squarrosus in the Single and Binary Component Systems. Acta Chim. Slov. 2010, 57, 163–172. [Google Scholar]
- Pteridophyte Phylogeny Group, I. A community-derived classification for extant lycophytes and ferns. J. Syst. Evol. 2016, 54, 563–603. [Google Scholar] [CrossRef]
- Mehltreter, K. Phenology and habitat specifity of tropical ferns. In Biology and Evolution of Ferns and Lycophytes; Ranker, T.A., Haufler, C.H., Eds.; Cambridge University Press: Cambridge, UK, 2008; pp. 201–221. [Google Scholar]
- Meharg, A.A. Variation in arsenic accumulation—Hyperaccumulation in ferns and their allies. New Phytol. 2003, 157, 25–31. [Google Scholar] [CrossRef]
- Schmitt, M.; Mehltreter, K.; Sundue, M.; Testo, W.; Watanabe, T.; Jansen, S. The evolution of aluminum accumulation in ferns and lycophytes. Am. J. Bot. 2017, 104, 573–583. [Google Scholar] [CrossRef] [Green Version]
- Arora, A.; Saxena, S.; Sharma, D.K. Tolerance and phytoaccumulation of chromium by three Azolla species. World J. Microbiol. Biotechnol. 2006, 22, 97–100. [Google Scholar] [CrossRef]
- De La Torre, A.R.; Li, Z.; Van de Peer, Y.; Ingvarsson, P.K. Contrasting rates of molecular evolution and patterns of selection among gymnosperms and flowering plants. Mol. Biol. Evol. 2017, 34, 1363–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magallón, S.; Hilu, K.W.; Quandt, D. Land plant evolutionary timeline: Gene effects are secondary to fossil constraints in relaxed clock estimation of age and substitution rates. Am. J. Bot. 2013, 100, 556–573. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.Q.; Ran, J.H. Evolution and biogeography of gymnosperms. Mol. Phylogenet. Evol. 2014, 75, 24–40. [Google Scholar] [CrossRef]
- Kim, C.G.; Bell, J.N.B.; Power, S.A. Effects of soil cadmium on Pinus sylvestris L. seedlings. Plant Soil 2003, 257, 443–449. [Google Scholar] [CrossRef]
- Sousa, N.R.; Ramos, M.A.; Marques, A.P.G.C.; Castro, P.M.L. A genotype dependent-response to cadmium contamination in soil is displayed by Pinus pinaster in symbiosis with different mycorrhizal fungi. Appl. Soil Ecol. 2014, 76, 7–13. [Google Scholar] [CrossRef]
- Hashemi, S.A.; Farajpour, G. Investigation of cadmium pollution in the spruce saplings near the metal production factory. Toxicol. Ind. Health 2016, 32, 323–327. [Google Scholar] [CrossRef]
- Österås, A.H.; Greger, M. Interactions between calcium and copper or cadmium in Norway spruce. Biol. Plant. 2006, 50, 647–652. [Google Scholar] [CrossRef]
- Guo, B.; Liu, C.; Ding, N.; Fu, Q.; Lin, Y.; Li, H.; Li, N. Silicon alleviates cadmium toxicity in two cypress varieties by strengthening the exodermis tissues and stimulating phenolic exudation of roots. J. Plant Growth Regul. 2016, 35, 420–429. [Google Scholar] [CrossRef]
- Zeng, P.; Guo, Z.; Xiao, X.; Cao, X.; Peng, C. Response to cadmium and phytostabilization potential of Platycladus orientalis in contaminated soil. Int. J. Phytoremediat. 2018, 20, 1337–1345. [Google Scholar] [CrossRef] [PubMed]
- Bonet, A.; Lelu-Walter, M.A.; Faugeron, C.; Gloaguen, V.; Saladin, G. Physiological responses of the hybrid larch (Larix × eurolepis Henry) to cadmium exposure and distribution of cadmium in plantlets. Environ. Sci. Pollut. Res. 2016, 23, 8617–8626. [Google Scholar] [CrossRef] [PubMed]
- Moudouma, C.F.M.; Riou, C.; Gloaguen, V.; Saladin, G. Hybrid larch (larix x eurolepis henry): A good candidate for cadmium phytoremediation? Environ. Sci. Pollut. Res. 2013, 20, 1889–1894. [Google Scholar] [CrossRef] [PubMed]
- Field, T.S.; Arens, N.C. The ecophysiology of early angiosperms. Plant Cell Environ. 2007, 30, 291–309. [Google Scholar] [CrossRef]
- The Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Field, T.S.; Arens, N.C. Form, function and environments of the early angiosperms: Merging extant phylogeny and ecophysiology with fossils. New Phytol. 2005, 166, 383–408. [Google Scholar] [CrossRef]
- Broadley, M.R.; Willey, N.J.; Wilkins, J.C.; Baker, A.J.M.; Mead, A.; White, P.J. Phylogenetic variation in heavy metal accumulation in angiosperms. New Phytol. 2001, 152, 9–27. [Google Scholar] [CrossRef]
- Reeves, R.D.; Baker, A.J.M.; Jaffré, T.; Erskine, P.D.; Echevarria, G.; van der Ent, A. A global database for plants that hyperaccumulate metal and metalloid trace elements. New Phytol. 2017, 218, 407–411. [Google Scholar] [CrossRef] [Green Version]
- Qiu, R.L.; Tang, Y.T.; Zeng, X.W.; Thangavel, P.; Tang, L.; Gan, Y.Y.; Ying, R.R.; Wang, S.Z. Mechanisms of Cd hyperaccumulation and detoxification in heavy metal hyperaccumulators: How plants cope with Cd. Progress. Bot. 2012, 73, 127–159. [Google Scholar]
- Xu, W.M.; Xiang, P.; Liu, X.; Ma, L.Q. Closely-related species of hyperaccumulating plants and their ability in accumulation of A, Cd, Cu, Mn, Ni, Pb and Zn. Chemosphere 2020, 251, 126334. [Google Scholar] [CrossRef] [PubMed]
- Reeves, R.D.; van der Ent, A.; Baker, A.J.M. Global distribution and ecology of hyperaccumulator Plants. In Agromining: Farming for Metals; van der Ent, A., Echevarria, G., Baker, A.J.M., Morel, J.L., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 75–92. [Google Scholar]
- Verbruggen, N.; Juraniec, M.; Baliardini, C.; Meyer, C.L. Tolerance to cadmium in plants: The special case of hyperaccumulators. BioMetals 2013, 26, 633–638. [Google Scholar] [CrossRef]
- Tian, S.; Lu, L.; Labavitch, J.; Yang, X.; He, Z.; Hu, H.; Sarangi, R.; Newville, M.; Commisso, J.; Brown, P. Cellular sequestration of cadmium in the hyperaccumulator plant species Sedum alfredii. Plant Physiol. 2011, 157, 1914–1925. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Tian, S.; Yang, X.; Wang, X.; Brown, P.; Li, T.; He, Z. Enhanced root-to-shoot translocation of cadmium in the hyperaccumulating ecotype of Sedum alfredii. J. Exp. Bot. 2008, 59, 3203–3213. [Google Scholar] [CrossRef] [Green Version]
- Deng, D.M.; Deng, J.C.; Li, J.T.; Zhang, J.; Hu, M.; Lin, Z.; Liao, B. Accumulation of zinc, cadmium, and lead in four populations of Sedum alfredii growing on lead/zinc mine spoils. J. Integr. Plant Biol. 2008, 50, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Sobczyk, M.K.; Smith, J.A.C.; Pollard, A.J.; Filatov, D.A. Evolution of nickel hyperaccumulation and serpentine adaptation in the Alyssum serpyllifolium species complex. Heredity 2017, 118, 31–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynolds, R.J.B.; Cappa, J.J.; Pilon-Smits, E.A.H. Evolutionary aspects of plant selenium accumulation. In Selenium in Plants: Molecular, Physiological, Ecological and Evolutionary Aspects; Pilon-Smits, E.A.H., Winkel, L.H.E., Lin, Z.-Q., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 189–205. [Google Scholar]
- Hanikenne, M.; Nouet, C. Metal hyperaccumulation and hypertolerance: A model for plant evolutionary genomics. Curr. Opin. Plant Biol. 2011, 14, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Roux, C.; Castric, V.; Pauwels, M.; Wright, S.I.; Saumitou-Laprade, P.; Vekemans, X. Does speciation between Arabidopsis halleri and Arabidopsis lyrata coincide with major changes in a molecular target of adaptation? PLoS ONE 2011, 6, e26872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clemens, S. Evolution and function of phytochelatin synthases. J. Plant Physiol. 2006, 163, 319–332. [Google Scholar] [CrossRef]
- Zhao, F.J.; Hamon, R.E.; Enzo, L.; McLaughlin, M.J.; McGrath, S.P. Characteristics of cadmium uptake in two contrasting ecotypes of the hyperaccumulator Thlaspi caerulescens. J. Exp. Bot. 2002, 53, 535–543. [Google Scholar] [CrossRef] [Green Version]
- Meychik, N.; Nikolaeva, Y.; Kushunina, M. The significance of ion-exchange properties of plant root cell walls for nutrient and water uptake by plants. Plant Physiol. Biochem. 2021, 166, 140–147. [Google Scholar] [CrossRef]
- Haider, F.U.; Liqun, C.; Coulter, J.A.; Cheema, S.A.; Wu, J.; Zhang, R.; Wenjun, M.; Farooq, M. Cadmium toxicity in plants: Impacts and remediation strategies. Ecotoxicol. Environ. Saf. 2021, 211, 111887. [Google Scholar] [CrossRef]
- Verbruggen, N.; Hermans, C.; Schat, H. Molecular mechanisms of metal hyperaccumulation in plants. New Phytol. 2009, 181, 759–776. [Google Scholar] [CrossRef]
- Lux, A.; Martinka, M.; Vaculík, M.; White, P.J. Root responses to cadmium in the rhizosphere: A review. J. Exp. Bot. 2011, 62, 21–37. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, A.; Yamaji, N.; Yokosho, K.; Ma, J.F. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 2012, 24, 2155–2167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaaf, G.; Ludewig, U.; Erenoglu, B.E.; Mori, S.; Kitahara, T.; von Wirén, N. ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. J. Biol. Chem. 2004, 279, 9091–9096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.H.; Ouyang, Y.N.; Fan, Y.C.; Qiu, B.Y.; Zhang, G.P.; Zeng, F.R. The pathway of transmembrane cadmium influx via calcium-permeable channels and its spatial characteristics along rice root. J. Exp. Bot. 2018, 69, 5279–5291. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Liu, X.; Peijnenburg, W.J.G.M.; Zhao, J.; Chen, X.; Yu, J.; Wu, H. Pathways of cadmium fluxes in the root of the halophyte Suaeda salsa. Ecotoxicol. Environ. Saf. 2012, 75, 1–7. [Google Scholar] [CrossRef] [PubMed]
- White, P.J.; Broadley, M.R. Calcium in plants. Ann. Bot. 2003, 92, 487–511. [Google Scholar] [CrossRef]
- Yan, H.; Xu, W.; Xie, J.; Gao, Y.; Wu, L.; Sun, L.; Feng, L.; Chen, X.; Zhang, T.; Dai, C.; et al. Variation of a major facilitator superfamily gene contributes to differential cadmium accumulation between rice subspecies. Nat. Commun. 2019, 10, 2562. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.L.; Tian, S.K.; Yang, X.E.; Li, T.Q.; He, Z.L. Cadmium uptake and xylem loading are active processes in the hyperaccumulator Sedum alfredii. J. Plant Physiol. 2009, 166, 579–587. [Google Scholar] [CrossRef]
- Ueno, D.; Iwashita, T.; Zhao, F.J.; Ma, J.F. Characterization of Cd translocation and identification of the Cd form in xylem sap of the Cd-hyperaccumulator Arabidopsis halleri. Plant Cell Physiol. 2008, 49, 540–548. [Google Scholar] [CrossRef] [Green Version]
- Uraguchi, S.; Mori, S.; Kuramata, M.; Kawasaki, A.; Arao, T.; Ishikawa, S. Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J. Exp. Bot. 2009, 60, 2677–2688. [Google Scholar] [CrossRef] [Green Version]
- Tao, Q.; Jupa, R.; Luo, J.; Lux, A.; Kováč, J.; Wen, Y.; Zhou, Y.; Jan, J.; Liang, Y.; Li, T. The apoplasmic pathway via the root apex and lateral roots contributes to Cd hyperaccumulation in the hyperaccumulator Sedum alfredii. J. Exp. Bot. 2017, 68, 739–751. [Google Scholar]
- Guo, J.; Ye, D.; Zhang, X.; Huang, H.; Wang, Y.; Zheng, Z.; Li, T.; Yu, H. Characterization of cadmium accumulation in the cell walls of leaves in a low-cadmium rice line and strengthening by foliar silicon application. Chemosphere 2022, 287, 132374. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Luo, N.; Li, Y.W.; Cai, Q.Y.; Li, H.Y.; Mo, C.H.; Wong, M.H. Cadmium in rice: Transport mechanisms, influencing factors, and minimizing measures. Environ. Pollut. 2017, 224, 622–630. [Google Scholar] [PubMed]
- Wu, F.B.; Dong, J.; Qian, Q.Q.; Zhang, G.P. Subcellular distribution and chemical form of Cd and Cd-Zn interaction in different barley genotypes. Chemosphere 2005, 60, 1437–1446. [Google Scholar] [CrossRef] [PubMed]
- Qiu, R.L.; Thangavel, P.; Hu, P.J.; Senthilkumar, P.; Ying, R.R.; Tang, Y.T. Interaction of cadmium and zinc on accumulation and sub-cellular distribution in leaves of hyperaccumulator Potentilla griffithii. J. Hazard. Mater. 2011, 186, 1425–1430. [Google Scholar] [CrossRef]
- Tian, S.; Xie, R.; Wang, H.; Hu, Y.; Hou, D.; Liao, X.; Brown, P.H.; Yang, H.; Lin, X.; Labavitch, J.M.; et al. Uptake, sequestration and tolerance of cadmium at cellular levels in the hyperaccumulator plant species Sedum alfredii. J. Exp. Bot. 2017, 68, 2387–2398. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.S.; Dietz, K.J.; Mimura, T. Vacuolar compartmentalization as indispensable component of heavy metal detoxification in plants. Plant Cell Environ. 2016, 39, 1112–1126. [Google Scholar] [CrossRef] [Green Version]
- Korenkov, V.; Hirschi, K.; Crutchfield, J.D.; Wagner, G.J. Enhancing tonoplast Cd/H antiport activity increases Cd, Zn, and Mn tolerance, and impacts root/shoot Cd partitioning in Nicotiana tabacum L. Planta 2007, 226, 1379–1387. [Google Scholar] [CrossRef]
- Park, J.; Song, W.Y.; Ko, D.; Eom, Y.; Hansen, T.H.; Schiller, M.; Lee, T.G.; Martinoia, E.; Lee, Y. The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 2012, 69, 278–288. [Google Scholar] [CrossRef]
- Zhang, J.; Martinoia, E.; Lee, Y. Vacuolar transporters for cadmium and arsenic in plants and their applications in phytoremediation and crop development. Plant Cell Physiol. 2018, 59, 1317–1325. [Google Scholar] [CrossRef]
- Ueno, D.; Ma, J.F.; Iwashita, T.; Zhao, F.J.; McGrath, S.P. Identification of the form of Cd in the leaves of a superior Cd-accumulating ecotype of Thlaspi caerulescens using 113Cd-NMR. Planta 2005, 221, 928–936. [Google Scholar] [CrossRef] [PubMed]
- Clemens, S.; Palmgren, M.G.; Krämer, U. A long way ahead: Understanding and engineering plant metal accumulation. Trends Plant Sci. 2002, 7, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Cózatl, D.G.; Butko, E.; Springer, F.; Torpey, J.W.; Komives, E.A.; Kehr, J.; Schroeder, J.I. Identification of high levels of phytochelatins.; glutathione and cadmium in the phloem sap of Brassica napus. A role for thiol-peptides in the long-distance transport of cadmium and the effect of cadmium on iron translocation. Plant J. 2008, 54, 249–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.A.; Castro-Guerrero, N.; Mendoza-Cozatl, D.G. Moving toward a precise nutrition: Preferential loading of seeds with essential nutrients over non-essential toxic elements. Front. Plant Sci. 2014, 5, 51. [Google Scholar] [CrossRef] [Green Version]
- Yamaji, N.; Ma, J.F. The node, a hub for mineral nutrient distribution in graminaceous plants. Trends Plant Sci. 2014, 19, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Fujimaki, S.; Suzui, N.; Ishioka, N.S.; Kawachi, N.; Ito, S.; Chino, M.; Nakamura, S.I. Tracing cadmium from culture to spikelet: Noninvasive imaging and quantitative characterization of absorption, transport, and accumulation of cadmium in an intact rice plant. Plant Physiol. 2010, 152, 1796–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uraguchi, S.; Kamiya, T.; Sakamoto, T.; Kasai, K.; Sato, Y.; Nagamura, Y.; Yoshida, A.; Kyozuka, J.; Ishikawa, S.; Fujiwara, T. Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proc. Nat. Acad. Sci. USA 2011, 108, 20959–20964. [Google Scholar] [CrossRef]
- Yamaji, N.; Xia, J.; Mitani-Ueno, N.; Yokosho, K.; Ma, J.F. Preferential delivery of zinc to developing tissues in rice is mediated by P-type heavy metal ATPase OsHMA2. Plant Physiol. 2013, 162, 927–939. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.H.; Chen, G.; Dai, F.; Wang, Y.; Hills, A.; Ruan, Y.L.; Zhang, G.P.; Franks, P.J.; Nevo, E.; Blatt, M.R.; et al. Molecular evolution of grass stomata. Trends Plant Sci. 2017, 22, 124–139. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Zeng, J.; Xia, K.; Fan, T.; Li, Y.; Wang, Y.; Xu, X.; Zhang, M. Evolutionary expansion and functional diversification of oligopeptide transporter gene family in rice. Rice 2012, 5, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ullah, I.; Wang, Y.; Eide, D.J.; Dunwell, J.M. Evolution, and functional analysis of Natural Resistance-Associated Macrophage Proteins (NRAMPs) from Theobroma cacao and their role in cadmium accumulation. Sci. Rep. 2018, 8, 14412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanikenne, M.; Krämer, U.; Demoulin, V.; Baurain, D.A. A comparative inventory of metal transporters in the green alga Chlamydomonas reinhardtii and the red alga Cyanidioschizon merolae. Plant Physiol. 2005, 137, 428–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomolplitinant, K.M.; Saier, M.H., Jr. Evolution of the oligopeptide transporter family. J. Membr. Biol. 2011, 240, 89–110. [Google Scholar] [CrossRef] [Green Version]
- One Thousand Plant Transcriptomes Initiative. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richer, E.; Courville, P.; Bergevin, I.; Cellier, M.F.M. Horizontal gene transfer of “prototype” Nramp in Bacteria. J. Mol. Evol. 2003, 57, 363–376. [Google Scholar] [CrossRef]
- Amborella Genome Project. The Amborella genome and the evolution of flowering plants. Science 2013, 342, 1241089. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Lanquar, V.; Lelièvre, F.; Bolte, S.; Hamès, C.; Alcon, C.; Neumann, D.; Vansuyt, G.; Curie, C.; Schröder, A.; Krämer, U.; et al. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J. 2005, 24, 4041–4051. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, R.; Ishimaru, Y.; Nakanishi, H.; Nishizawa, N.K. Role of the iron transporter OsNRAMP1 in cadmium uptake and accumulation in rice. Plant Signal. Behav. 2011, 6, 1813–1816. [Google Scholar] [CrossRef] [Green Version]
- Tang, Z.; Cai, H.; Li, J.; Lv, Y.; Zhang, W.; Zhao, F.J. Allelic variation of NtNramp5 associated with cultivar variation in cadmium accumulation in tobacco. Plant Cell Physiol. 2017, 58, 1583–1593. [Google Scholar] [CrossRef] [Green Version]
- Peris-Peris, C.; Serra-Cardona, A.; Sánchez-Sanuy, F.; Campo, S.; Ariño, J.; San Segundo, B. Two NRAMP6 isoforms function as iron and manganese transporters and contribute to disease resistance in rice. Mol. Plant-Microbe Interact. 2017, 30, 385–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaji, N.; Sasaki, A.; Xia, J.X.; Yokosho, K.; Ma, J.F. A node-based switch for preferential distribution of manganese in rice. Nat. Commun. 2013, 4, 2442. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Yamaji, N.; Kasai, T.; Ma, J.F. Plasma membrane-localized transporter for aluminum in rice. Proc. Nat. Acad. Sci. USA 2010, 107, 18381–18385. [Google Scholar] [CrossRef]
- Wu, D.; Yamaji, N.; Yamane, M.; Kashino-Fujii, M.; Sato, K.; Ma, J.F. The HvNramp5 transporter mediates uptake of cadmium and manganese.; but not iron. Plant Physiol. 2016, 172, 1899–1910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milner, M.J.; Mitani-Ueno, N.; Yamaji, N.; Yokosho, K.; Craft, E.; Fei, Z.; Ebbs, S.; Clemencia Zambrano, M.; Ma, J.F.; Kochian, L.V. Root and shoot transcriptome analysis of two ecotypes of Noccaea caerulescens uncovers the role of NcNramp1 in Cd hyperaccumulation. Plant J. 2014, 78, 398–410. [Google Scholar] [CrossRef]
- Williams, L.E.; Mills, R.F. P1B-ATPases—An ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci. 2005, 10, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Mills, R.F.; Krijger, G.C.; Baccarini, P.J.; Hall, J.L.; Williams, L.E. Functional expression of AtHMA4, a P1B-type ATPase of the Zn/Co/Cd/Pb subclass. Plant J. 2003, 35, 164–176. [Google Scholar] [CrossRef]
- Craciun, A.R.; Meyer, C.L.; Chen, J.; Roosens, N.; De Groodt, R.; Hilson, P.; Verbruggen, N. Variation in HMA4 gene copy number and expression among Noccaea caerulescens populations presenting different levels of Cd tolerance and accumulation. J. Exp. Bot. 2012, 63, 4179–4189. [Google Scholar] [CrossRef] [Green Version]
- Hanikenne, M.; Talke, I.N.; Haydon, M.J.; Lanz, C.; Nolte, A.; Motte, P.; Kroymann, J.; Weigel, D.; Krämer, U. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature 2008, 453, 391–395. [Google Scholar] [CrossRef]
- Liu, H.; Zhao, H.; Wu, L.; Liu, A.; Zhao, F.; Xu, W. Heavy metal ATPase 3 (HMA3) confers cadmium hypertolerance on the cadmium/zinc hyperaccumulator Sedum plumbizincicola. New Phytol. 2017, 215, 687–698. [Google Scholar] [CrossRef] [Green Version]
- Satoh-Nagasawa, N.; Mori, M.; Nakazawa, N.; Kawamoto, T.; Nagato, Y.; Sakurai, K.; Takahashi, H.; Watanabe, A.; Akagi, H. Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol. 2012, 53, 213–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, R.; Ishimaru, Y.; Shimo, H.; Ogo, Y.; Senoura, T.; Nishizawa, N.K.; Nakanishi, H. The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant Cell Environ. 2012, 35, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Miyadate, H.; Adachi, S.; Hiraizumi, A.; Tezuka, K.; Nakazawa, N.; Kawamoto, T.; Katou, K.; Kodama, I.; Sakurai, K.; Takahashi, H.; et al. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 2011, 189, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Wang, P.; Yang, M.; Lian, X.; Tang, Z.; Huang, C.F.; Salt, D.E.; Zhao, F.J. A loss-of-function allele of OsHMA3 associated with high cadmium accumulation in shoots and grain of Japonica rice cultivars. Plant Cell Environ. 2016, 39, 1941–1954. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.; Zhang, L.; Tang, Z.; Huang, X.Y.; Ma, J.F.; Zhao, F.J. Producing cadmium-free Indica rice by overexpressing OsHMA3. Environ. Int. 2019, 126, 619–626. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, J.; Tang, Z.; Huang, X.Y.; Wang, X.; Salt, D.E.; Zhao, F.J. Variation in the BrHMA3 coding region controls natural variation in cadmium accumulation in Brassica rapa vegetables. J. Exp. Bot. 2019, 70, 5865–5878. [Google Scholar] [CrossRef] [Green Version]
- Colangelo, E.P.; Guerinot, M.L. Put the metal to the petal: Metal uptake and transport throughout plants. Curr. Opin. Plant. Biol. 2006, 9, 322–330. [Google Scholar] [CrossRef]
- Vert, G.; Grotz, N.; Dédaldéchamp, F.; Gaymard, F.; Guerinot, M.L.; Briat, J.F.; Curie, C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 2002, 14, 1223–1233. [Google Scholar] [CrossRef] [Green Version]
- Bughio, N.; Yamaguchi, H.; Nishizawa, N.K.; Nakanishi, H.; Mori, S. Cloning an iron-regulated metal transporter from rice. J. Exp. Bot. 2002, 53, 1677–1682. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.; Chen, L.; Li, X. Arabidopsis and rice showed a distinct pattern in ZIPs genes expression profile in response to Cd stress. Bot. Stud. 2018, 59, 22. [Google Scholar] [CrossRef]
- Pence, N.S.; Larsen, P.B.; Ebbs, S.D.; Letham, D.L.D.; Lasat, M.M.; Garvin, D.F.; Eide, D.; Kochian, L.V. The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc. Nat. Acad. Sci. USA 2000, 97, 4956–4960. [Google Scholar] [CrossRef]
- Lin, Y.F.; Hassan, Z.; Talukdar, S.; Schat, H.; Aarts, M.G.M. Expression of the ZNT1 zinc transporter from the metal hyperaccumulator Noccaea caerulescens confers enhanced zinc and cadmium tolerance and accumulation to Arabidopsis thaliana. PLoS ONE 2016, 11, e0149750. [Google Scholar] [CrossRef]
- Wang, F.H.; Qiao, K.; Shen, Y.H.; Wang, H.; Chai, T.Y. Characterization of the gene family encoding metal tolerance proteins in Triticum urartu: Phylogenetic, transcriptional, and functional analyses. Metallomics 2021, 13, mfab038. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.Z.; Tong, Y.H.; Zhou, X.; Ling, L.L.; Chun, C.P.; Cao, L.; Zeng, M.; Peng, L.Z. Genome-wide identification of sweet orange (Citrus sinensis) metal tolerance proteins and analysis of their expression patterns under zinc, manganese, copper, and cadmium toxicity. Gene 2017, 629, 1–8. [Google Scholar] [CrossRef]
- Peiter, E.; Montanini, B.; Gobert, A.; Pedas, P.; Husted, S.; Maathuis, F.J.M.; Blaudez, D.; Chalot, M.; Sanders, D. A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proc. Nat. Acad. Sci. USA 2007, 104, 8532–8537. [Google Scholar] [CrossRef]
- Gustin, J.L.; Zanis, M.J.; Salt, D.E. Structure and evolution of the plant cation diffusion facilitator family of ion transporters. BMC Evol. Biol. 2011, 11, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, L.; Yang, S.; Liu, B.; Zhang, M.; Wu, K. Molecular characterization of a rice metal tolerance protein, OsMTP1. Plant Cell Rep. 2012, 31, 67–79. [Google Scholar] [CrossRef]
- Kim, D.; Gustin, J.L.; Lahner, B.; Persans, M.W.; Baek, D.; Yun, D.J.; Salt, D.E. The plant CDF family member TgMTP1 from the Ni/Zn hyperaccumulator Thlaspi goesingense acts to enhance efflux of Zn at the plasma membrane when expressed in Saccharomyces cerevisiae. Plant J. 2004, 39, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Migocka, M.; Kosieradzka, A.; Papierniak, A.; Maciaszczyk-Dziubinska, E.; Posyniak, E.; Garbiec, A.; Filleur, S. Two metal-tolerance proteins, MTP1 and MTP4, are involved in Zn homeostasis and Cd sequestration in cucumber cells. J. Exp. Bot. 2015, 66, 1001–1015. [Google Scholar] [CrossRef] [Green Version]
- Negishi, T.; Nakanishi, H.; Yazaki, J.; Kishimoto, N.; Fujii, F.; Shimbo, K.; Yamamoto, K.; Sakata, K.; Sasaki, T.; Kikuchi, S.; et al. cDNA microarray analysis of gene expression during Fe-deficiency stress in barley suggests that polar transport of vesicles is implicated in phytosiderophore secretion in Fe-deficient barley roots. Plant J. 2002, 30, 83–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrissey, J.; Guerinot, M.L. Iron uptake and transport in plants: The good, the bad, and the ionome. Chem. Rev. 2009, 109, 4553–4567. [Google Scholar] [CrossRef] [Green Version]
- Feng, S.; Tan, J.; Zhang, Y.; Liang, S.; Xiang, S.; Wang, H.; Chai, T. Isolation and characterization of a novel cadmium-regulated Yellow Stripe-Like transporter (SnYSL3) in Solanum nigrum. Plant Cell Rep. 2017, 36, 281–296. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Masuda, H.; Bashir, K.; Inoue, H.; Tsukamoto, T.; Takahashi, M.; Nakanishi, H.; Aoki, N.; Hirose, T.; Ohsugi, R.; et al. Rice metal-nicotianamine transporter, OsYSL2.; is required for the long-distance transport of iron and manganese. Plant J. 2010, 62, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Q.; Wu, Z.X.; Yang, Y.; Teng, Q.X.; Li, Y.D.; Lei, Z.N.; Jani, K.A.; Kaushal, N.; Chen, Z.S. ATP-binding cassette (ABC) transporters in cancer: A review of recent updates. J. Evid. Based Med. 2021, 14, 232–256. [Google Scholar] [CrossRef]
- Do, T.H.T.; Martinoia, E.; Lee, Y.; Hwang, J.U. 2021 update on ATP-binding cassette (ABC) transporters: How they meet the needs of plants. Plant Physiol. 2021, 187, 1876–1892. [Google Scholar] [CrossRef]
- Zhang, X.; Rui, H.; Zhang, F.; Hu, Z.; Xia, Y.; Shen, Z. Overexpression of a functional Vicia sativa PCS1 homolog increases cadmium tolerance and phytochelatins synthesis in Arabidopsis. Front. Plant Sci. 2018, 9, 107. [Google Scholar] [CrossRef] [Green Version]
- Bovet, L.; Eggmann, T.; Meylan-Bettex, M.; Polier, J.; Kammer, P.; Marin, E.; Feller, U.; Martinoia, E. Transcript levels of AtMRPs after cadmium treatment: Induction of AtMRP3. Plant Cell Environ. 2003, 26, 371–381. [Google Scholar] [CrossRef]
- Brunetti, P.; Zanella, L.; De Paolis, A.; Di Litta, D.; Cecchetti, V.; Falasca, G.; Barbieri, M.; Altamura, M.M.; Costantino, P.; Cardarelli, M. Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 2015, 66, 3815–3829. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.Y.; Bovet, L.; Maeshima, M.; Martinoia, E.; Lee, Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 2007, 50, 207–218. [Google Scholar] [CrossRef]
- Kim, D.Y.; Bovet, L.; Kushnir, S.; Noh, E.W.; Martinoia, E.; Lee, Y. AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiol. 2006, 140, 922–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, S.; Lu, Y.; Zhang, X.; Yang, G.; Chao, D.; Wang, Z.; Shi, M.; Chen, J.; Chao, D.Y.; Li, R.; et al. The ABC transporter ABCG36 is required for cadmium tolerance in rice. J. Exp. Bot. 2019, 70, 5909–5918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pittman, J.K.; Sreevidya, C.S.; Shigaki, T.; Ueoka-Nakanishi, H.; Hirschi, K.D. Distinct N-terminal regulatory domains of Ca2+/H+ antiporters. Plant Physiol. 2002, 130, 1054–1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korenkov, V.; King, B.; Hirschi, K.; Wagner, G.J. Root-selective expression of AtCAX4 and AtCAX2 results in reduced lamina cadmium in field-grown Nicotiana tabacum L. Plant Biotechnol. J. 2009, 7, 219–226. [Google Scholar] [CrossRef]
- Wu, Q.; Shigaki, T.; Williams, K.A.; Han, J.S.; Kim, C.K.; Hirschi, K.D.; Park, S. Expression of an Arabidopsis Ca2+/H+ antiporter CAX1 variant in petunia enhances cadmium tolerance and accumulation. J. Plant Physiol. 2011, 168, 167–173. [Google Scholar] [CrossRef]
- Baliardini, C.; Meyer, C.L.; Salis, P.; Saumitou-Laprade, P.; Verbruggen, N. CATION EXCHANGER1 cosegregates with cadmium tolerance in the metal hyperaccumulator Arabidopsis halleri and plays a role in limiting oxidative stress in Arabidopsis spp. Plant Physiol. 2015, 169, 549–559. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Zhang, J.; Lu, L.L.; Zhu, Z.Q.; Yang, X.E. Functional analysis of CAX2-like transporters isolated from two ecotypes of Sedum alfredii. Biol. Plantarum. 2016, 60, 37–47. [Google Scholar] [CrossRef]
- Luo, J.S.; Huang, J.; Zeng, D.L.; Peng, J.S.; Zhang, G.B.; Ma, H.L.; Guan, Y.; Yi, H.Y.; Fu, Y.L.; Gong, J.M. A defensin-like protein drives cadmium efflux and allocation in rice. Nat. Commun. 2018, 9, 645. [Google Scholar] [CrossRef] [Green Version]
- Perfus-Barbeoch, L.; Leonhardt, N.; Vavasseur, A.; Forestier, C. Heavy metal toxicity: Cadmium permeates through calcium channels and disturbs the plant water status. Plant J. 2002, 32, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Thomine, S.; Wang, R.; Ward, J.M.; Crawford, N.M.; Schroeder, J.I. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc. Nat. Acad. Sci. USA 2000, 97, 4991–4996. [Google Scholar] [CrossRef] [PubMed]
- Oomen, R.J.; Wu, J.; Lelievre, F.; Blanchet, S.; Richaud, P.; Barbier-Brygoo, H.; Aarts, M.G.M.; Thomine, S. Functional characterization of NRAMP3 and NRAMP4 from the metal hyperaccumulator Thlaspi caerulescens. New Phytol. 2009, 181, 637–650. [Google Scholar]
- Ishimaru, Y.; Bashir, K.; Nakanishi, H.; Nishizawa, N.K. OsNRAMP5, a major player for constitutive iron and manganese uptake in rice. Plant Signal. Behav. 2012, 7, 763–766. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Zhang, Y.; Zhang, L.; Hu, J.; Zhang, X.; Lu, K.; Dong, H.; Wang, D.; Zhao, F.J.; Huang, C.F.; et al. OsNRAMP5 contributes to manganese translocation and distribution in rice shoots. J. Exp. Bot. 2014, 65, 4849–4861. [Google Scholar]
- Moreno, I.; Norambuena, L.; Maturana, D.; Toro, M.; Vergara, C.; Orellana, A.; Zurita-Silva, A.; Ordenes, V.R. AtHMA1 is a thapsigargin-sensitive Ca2+/heavy metal pump. J. Biol. Chem. 2008, 283, 9633–9641. [Google Scholar]
- Kim, Y.Y.; Choi, H.; Segami, S.; Cho, H.; Martinoia, E.; Maeshima, M.; Lee, Y. AtHMA1 contributes to the detoxification of excess Zn (II) in Arabidopsis. Plant J. 2009, 58, 737–753. [Google Scholar] [PubMed] [Green Version]
- Gravot, A.; Lieutaud, A.; Verret, F.; Auroy, P.; Vavasseur, A.; Richaud, P. AtHMA3, a plant P1B-ATPase, functions as a Cd/Pb transporter in yeast. FEBS Lett. 2004, 561, 22–28. [Google Scholar]
- Morel, M.; Crouzet, J.; Gravot, A.; Auroy, P.; Leonhardt, N.; Vavasseur, A.; Richaud, P. AtHMA3, a P-1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2009, 149, 894–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, D.Y.; Silva, A.; Baxter, I.; Huang, Y.S.; Nordborg, M.; Danku, J.; Lahner, B.; Yakubova, E.; Salt, D.E. Genome-wide association studies identify heavy metal ATPase3 as the primary determinant of natural variation in leaf cadmium in Arabidopsis thaliana. PLoS Genet. 2012, 8, e1002923. [Google Scholar]
- Hussain, D.; Haydon, M.J.; Wang, Y.; Wong, E.; Sherson, S.M.; Young, J.; Camakaris, J.; Harper, J.F.; Cobbett, C.C. P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 2004, 16, 1327–1339. [Google Scholar]
- Lekeux, G.; Crowet, J.M.; Nouet, C.; Joris, M.; Jadoul, A.; Bosman, B.; Carnol, M.; Motte, P.; Lins, L.; Galleni, M.; et al. Homology modeling and in vivo functional characterization of the zinc permeation pathway in a heavy metal P-type ATPase. J. Exp. Bot. 2019, 70, 329–341. [Google Scholar] [PubMed] [Green Version]
- Verret, F.; Gravot, A.; Auroy, P.; Leonhardt, N.; David, P.; Nussaume, L.; Vavasseur, A.; Richaud, P. Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett. 2004, 576, 306–312. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.K.E.; Jarvis, R.S.; Sherson, S.M.; Cobbertt, C.S. Functional analysis of the heavy metal binding domains of the Zn/Cd-transporting ATPase, HMA2, in Arabidopsis thaliana. New Phytol. 2009, 181, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Ueno, D.; Yamaji, N.; Kono, I.; Huang, C.F.; Ando, T.; Yano, M.; Ma, J.F. Gene limiting cadmium accumulation in rice. Proc. Nat. Acad. Sci. USA 2010, 107, 16500–16505. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Yamaji, N.; Ma, J.F. Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J. Exp. Bot. 2014, 65, 6013–6021. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Kim, Y.Y.; Lee, Y.; An, G. Rice P1B-type heavy-metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiol. 2007, 145, 831–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, J.; Wang, J.; Chai, T.; Zhang, Y.; Feng, S.; Li, Y.; Zhao, H.; Liu, H.; Chai, X. Functional analyses of Ta HMA 2, a P(1B)-type ATPase in wheat. Plant Biotechnol. J. 2013, 11, 420–431. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, K.F.; Poysa, V.; Shi, C.; Zhou, Y.H. A single point mutation in GmHMA3 affects cadmium (Cd) translocation and accumulation in soybean seeds. Mol. Plant 2012, 5, 1154–1156. [Google Scholar] [CrossRef] [Green Version]
- Ueno, D.; Milner, M.J.; Yamaji, N.; Yokosho, K.; Koyama, E.; Zambrano, M.C.; Kaskie, M.; Ebbs, S.D.; Ma, J.F. Elevated expression of TcHMA3 plays a key role in the extreme Cd tolerance in a Cd-hyperaccumulating ecotype of Thlaspi caerulescens. Plant J. 2011, 66, 852–862. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, M.; Shohag, M.J.; Tian, S.; Song, H.; Feng, Y.; Yang, X. Enhanced expression of SaHMA3 plays critical roles in Cd hyperaccumulation and hypertolerance in Cd hyperaccumulator Sedum alfredii Hance. Planta 2016, 243, 577–589. [Google Scholar] [CrossRef]
- Song, W.Y.; Park, J.; Mendoza-Cózatl, D.G.; Suter-Grotemeyer, M.; Shim, D.; Hörtensteiner, S.; Geisler, M.; Weder, B.; Rea, P.A.; Rentsch, D.; et al. Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Nat. Acad. Sci. USA 2010, 107, 21187–21192. [Google Scholar] [CrossRef]
- Persans, M.W.; Nieman, K.; Salt, D.E. Functional activity and role of cation-efflux family members in Ni hyperaccumulation in Thlaspi goesingense. Proc. Nat. Acad. Sci. USA 2001, 98, 9995–10000. [Google Scholar] [CrossRef] [PubMed]
- Koike, S.; Inoue, H.; Mizuno, D.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J. 2010, 39, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Cózatl, D.G.; Xie, Q.; Akmakjian, G.Z.; Jobe, T.O.; Patel, A.; Stacey, M.G.; Song, L.; Demoin, D.; Jurisson, S.; Stacey, G.; et al. OPT3 is a component of the iron-signaling network between leaves and roots and misregulation of OPT3 leads to an over-accumulation of cadmium in seeds. Mol. Plant 2014, 7, 1455–1469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.S.; Feng, S.J.; Zhang, B.Q.; Wang, M.Q.; Cao, H.W.; Rono, J.K.; Chen, X.; Yang, Z.M. OsZIP1 functions as a metal efflux transporter limiting excess zinc.; copper and cadmium accumulation in rice. BMC Plant Biol. 2019, 19, 283. [Google Scholar] [CrossRef] [Green Version]
- Lombi, E.; Tearall, K.L.; Howarth, J.R.; Zhao, F.J.; Hawkesford, M.J.; McGrath, S.P. Influence of iron status on cadmium and zinc uptake by different ecotypes of the hyperaccumulator Thlaspi caerulescens. Plant Physiol. 2002, 128, 1359–1367. [Google Scholar] [CrossRef] [Green Version]
- Uraguchi, S.; Kamiya, T.; Clemens, S.; Fujiwara, T. Characterization of OsLCT1, a cadmium transporter from indica rice (Oryza sativa). Physiol. Plant. 2014, 151, 339–347. [Google Scholar] [CrossRef]
- Shimo, H.; Ishimaru, Y.; An., G.; Yamakawa, T.; Nakanishi, H.; Nishizawa, N.K. Low cadmium (LCD), a novel gene related to cadmium tolerance and accumulation in rice. J. Exp. Bot. 2011, 62, 5727–5734. [Google Scholar] [CrossRef] [Green Version]
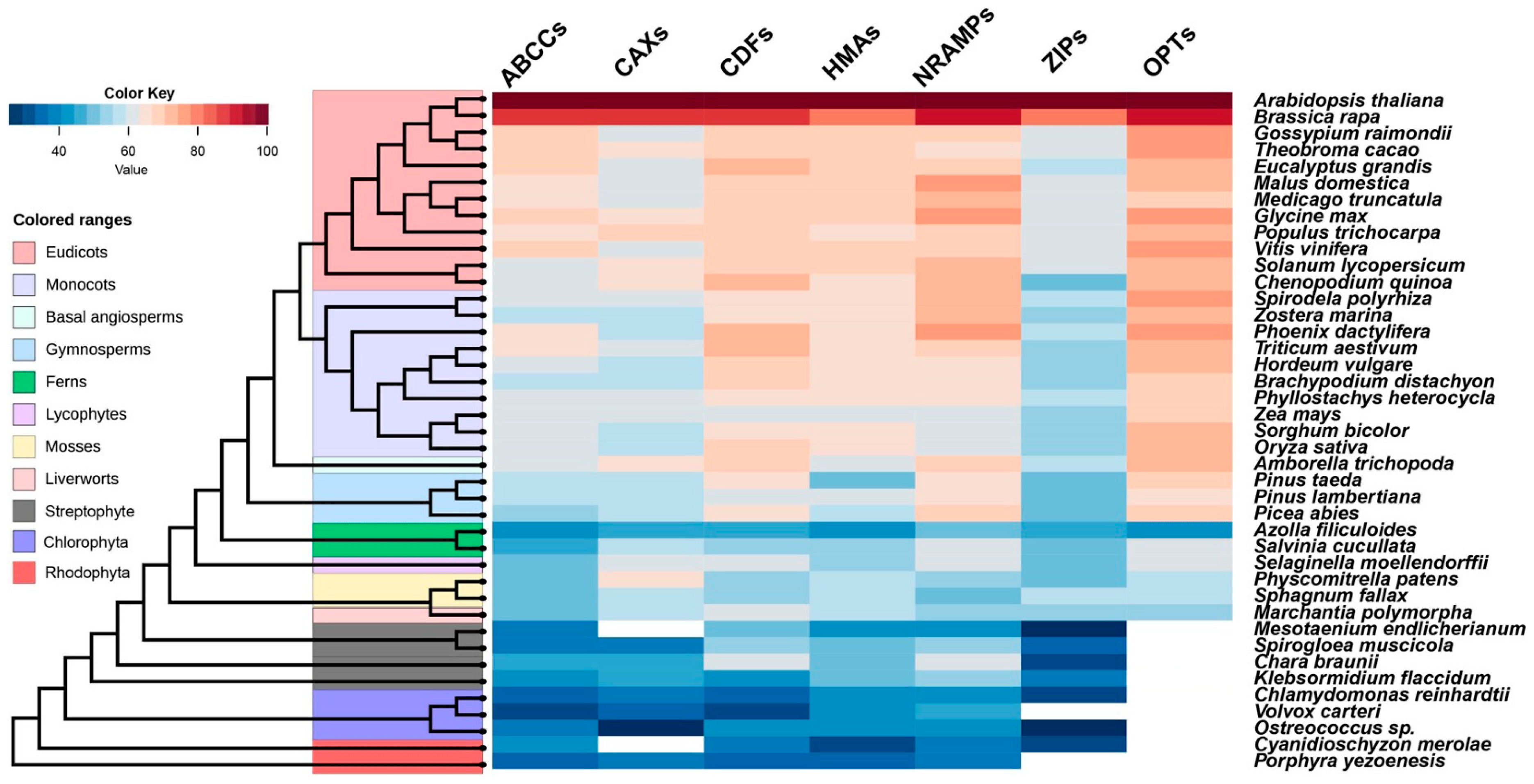
| Clade | Plant Species | NRAMPs | HMAs | ZIPs | CDFs | ABCCs | CAXs | OPTs |
|---|---|---|---|---|---|---|---|---|
| Eudicots | Arabidopsis thaliana | 6 | 8 | 15 | 12 | 15 | 11 | 9 |
| Brassica rapa | 9 | 15 | 28 | 18 | 17 | 12 | 11 | |
| Gossypium raimondii | 10 | 9 | 21 | 15 | 26 | 10 | 8 | |
| Theobroma cacao | 6 | 7 | 12 | 13 | 15 | 6 | 10 | |
| Eucalyptus grandis | 9 | 9 | 20 | 18 | 33 | 5 | 25 | |
| Malus domestica | 10 | 17 | 16 | 26 | 24 | 12 | 18 | |
| Medicago truncatula | 7 | 10 | 13 | 13 | 38 | 8 | 13 | |
| Glycine max | 13 | 18 | 19 | 23 | 37 | 17 | 13 | |
| Populus trichocarpa | 8 | 13 | 16 | 22 | 24 | 8 | 13 | |
| Vitis vinifera | 6 | 5 | 13 | 11 | 23 | 5 | 11 | |
| Solanum lycopersicum | 4 | 8 | 11 | 12 | 14 | 7 | 8 | |
| Chenopodium quinoa | 10 | 18 | 18 | 15 | 45 | 6 | 10 | |
| Monocots | Spirodela polyrhiza | 3 | 8 | 9 | 11 | 12 | 7 | 6 |
| Zostera marina | 5 | 7 | 13 | 9 | 10 | 6 | 4 | |
| Phoenix dactylifera | 10 | 21 | 14 | 19 | 32 | 16 | 11 | |
| Triticum aestivum | 18 | 26 | 25 | 19 | 53 | 15 | 22 | |
| Hordeum vulgare | 7 | 11 | 12 | 8 | 16 | 9 | 9 | |
| Brachypodium distachyon | 7 | 9 | 11 | 10 | 20 | 8 | 8 | |
| Phyllostachys heterocycla | 7 | 9 | 11 | 10 | 11 | 10 | 9 | |
| Zea mays | 8 | 12 | 10 | 11 | 12 | 11 | 8 | |
| Sorghum bicolor | 8 | 11 | 12 | 8 | 16 | 10 | 8 | |
| Oryza sativa | 7 | 8 | 11 | 9 | 16 | 7 | 9 | |
| Basal angiosperms | Amborella trichopoda | 3 | 7 | 8 | 8 | 14 | 4 | 8 |
| Gymnosperms | Pinus taeda | 13 | 8 | 11 | 8 | 11 | 11 | 34 |
| Pinus lambertiana | 9 | 11 | 13 | 10 | 10 | 7 | 11 | |
| Picea abies | 5 | 5 | 10 | 2 | 6 | 4 | 9 | |
| Ferns | Azolla filiculoides | 9 | 18 | 11 | 16 | 22 | 11 | 10 |
| Salvinia cucullata | 2 | 9 | 3 | 7 | 16 | 2 | 3 | |
| Lycophytes | Selaginella moellendorffii | 6 | 12 | 5 | 8 | 23 | 2 | 6 |
| Mosses | Physcomitrella patens | 6 | 18 | 7 | 12 | 15 | 6 | 2 |
| Sphagnum fallax | 6 | 8 | 5 | 9 | 16 | 4 | 11 | |
| Liverworts | Marchantia polymorpha | 5 | 6 | 5 | 5 | 15 | 3 | 5 |
| Streptophytes | Mesotaenium endlicherianum | 3 | 7 | 1 | 3 | 8 | 0 | 0 |
| Spirogloea muscicola | 9 | 19 | 3 | 17 | 11 | 3 | 0 | |
| Chara braunii | 1 | 3 | 2 | 2 | 2 | 2 | 0 | |
| Klebsormidium flaccidum | 3 | 6 | 2 | 5 | 3 | 1 | 0 | |
| Chlorophyta | Chlamydomonas reinhardtii | 2 | 4 | 2 | 4 | 4 | 3 | 0 |
| Volvox carteri | 2 | 4 | 0 | 1 | 4 | 3 | 0 | |
| Ostreococcus sp. | 1 | 5 | 1 | 3 | 2 | 2 | 0 | |
| Rhodophyta | Cyanidioschyzon merolae | 2 | 1 | 1 | 1 | 1 | 0 | 0 |
| Porphyra yezoensis | 1 | 3 | 0 | 2 | 2 | 1 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, Y.; Liu, H.; Chen, G.; Wu, X.; Zeng, F. Cadmium Accumulation in Plants: Insights from Phylogenetic Variation into the Evolution and Functions of Membrane Transporters. Sustainability 2023, 15, 12158. https://doi.org/10.3390/su151612158
Yi Y, Liu H, Chen G, Wu X, Zeng F. Cadmium Accumulation in Plants: Insights from Phylogenetic Variation into the Evolution and Functions of Membrane Transporters. Sustainability. 2023; 15(16):12158. https://doi.org/10.3390/su151612158
Chicago/Turabian StyleYi, Yun, Hongjiang Liu, Guang Chen, Xiaojian Wu, and Fanrong Zeng. 2023. "Cadmium Accumulation in Plants: Insights from Phylogenetic Variation into the Evolution and Functions of Membrane Transporters" Sustainability 15, no. 16: 12158. https://doi.org/10.3390/su151612158
APA StyleYi, Y., Liu, H., Chen, G., Wu, X., & Zeng, F. (2023). Cadmium Accumulation in Plants: Insights from Phylogenetic Variation into the Evolution and Functions of Membrane Transporters. Sustainability, 15(16), 12158. https://doi.org/10.3390/su151612158








