Sources of Antibiotic Contamination in Wastewater and Approaches to Their Removal—An Overview
Abstract
:1. Introduction
2. Antibiotics in Water
2.1. Source
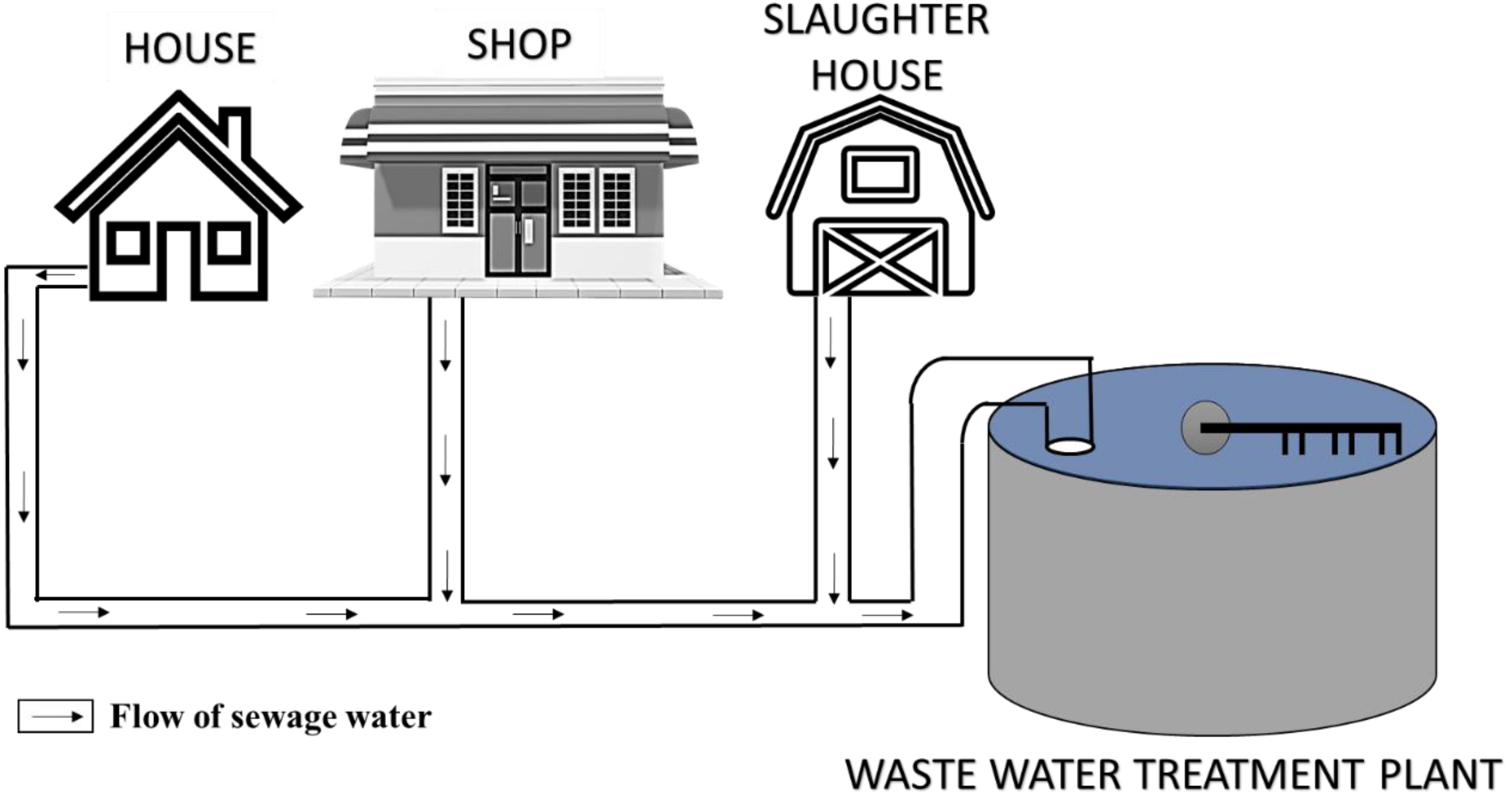
2.1.1. Domestic Wastewater
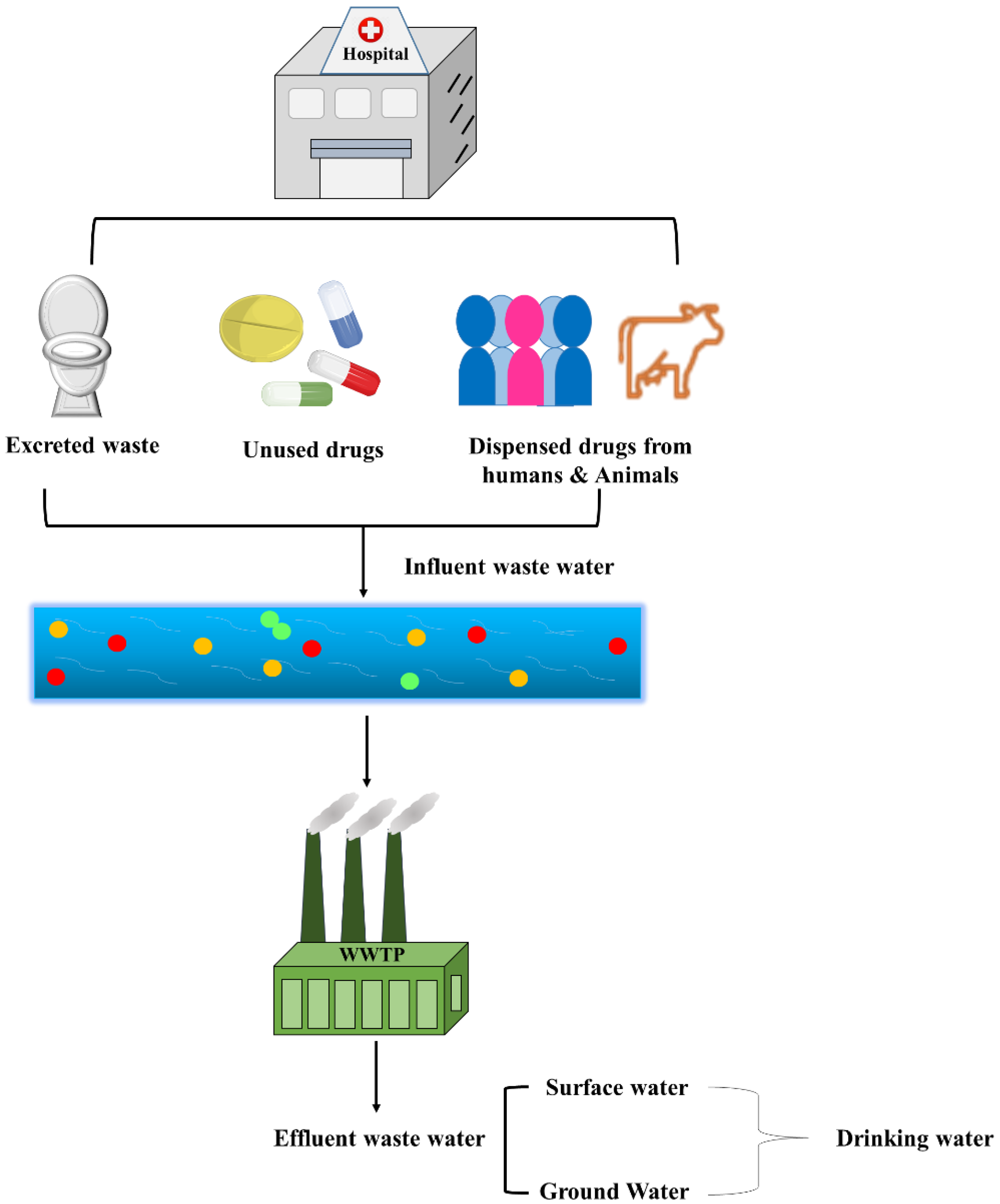
2.1.2. Effluent from Hospitals
2.1.3. Effluent from Slaughter Houses
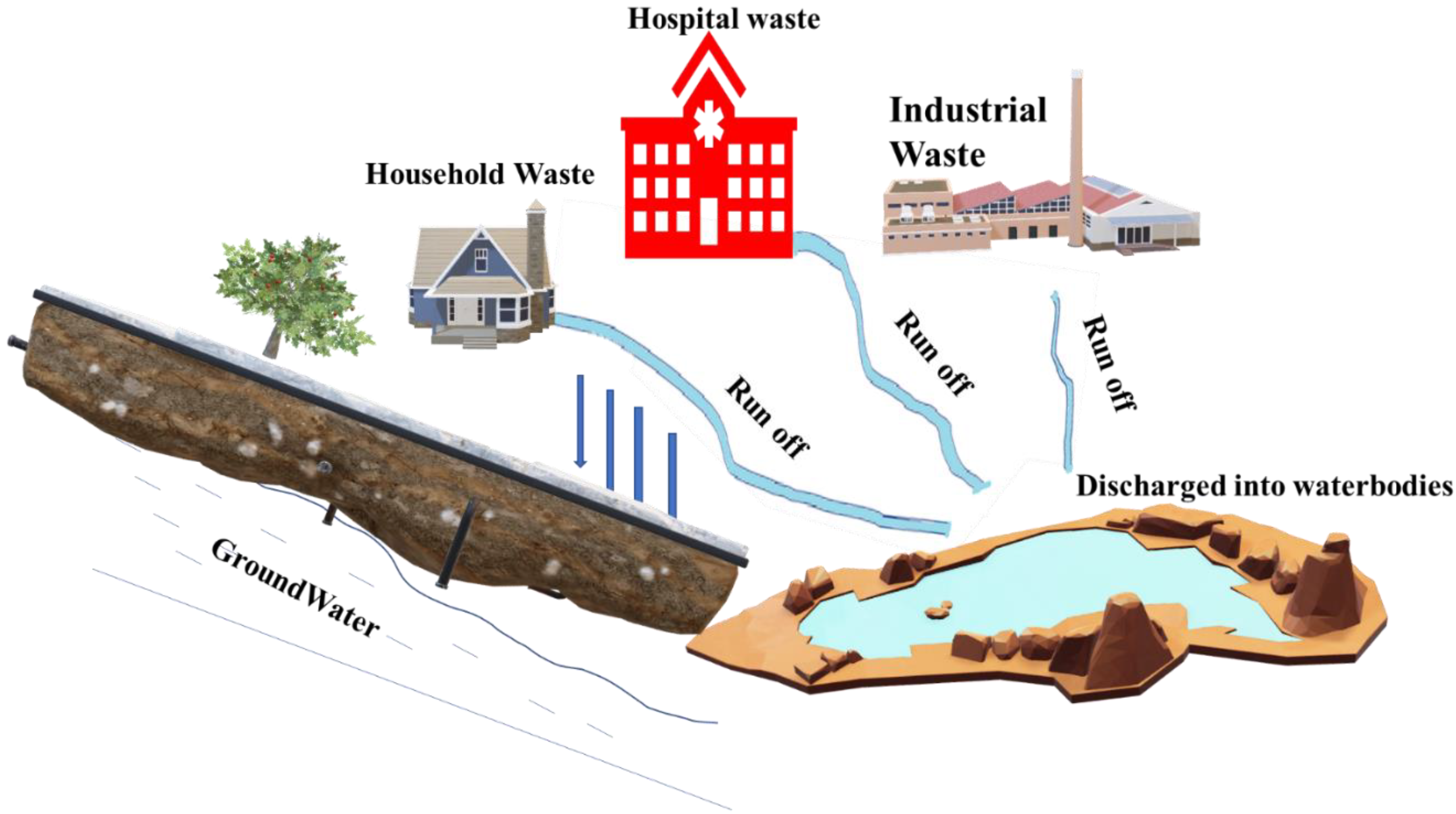
3. Runoffs
4. Impact of Antibiotics in Wastewater
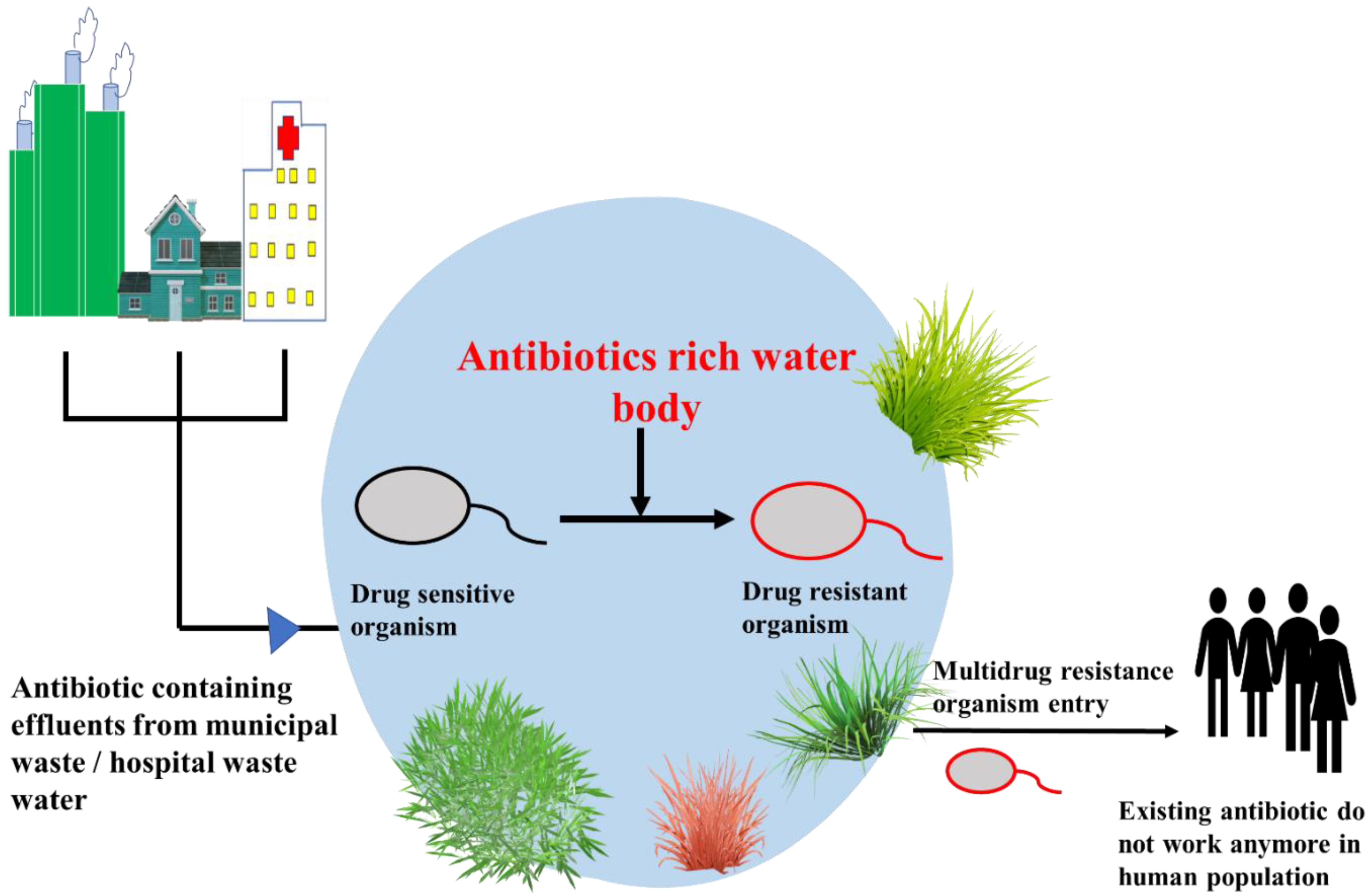
4.1. Emergence of Antibiotic Resistance
4.2. Emergence of New Disease
4.3. Immune Response
5. Strategies of Antibiotics Removal
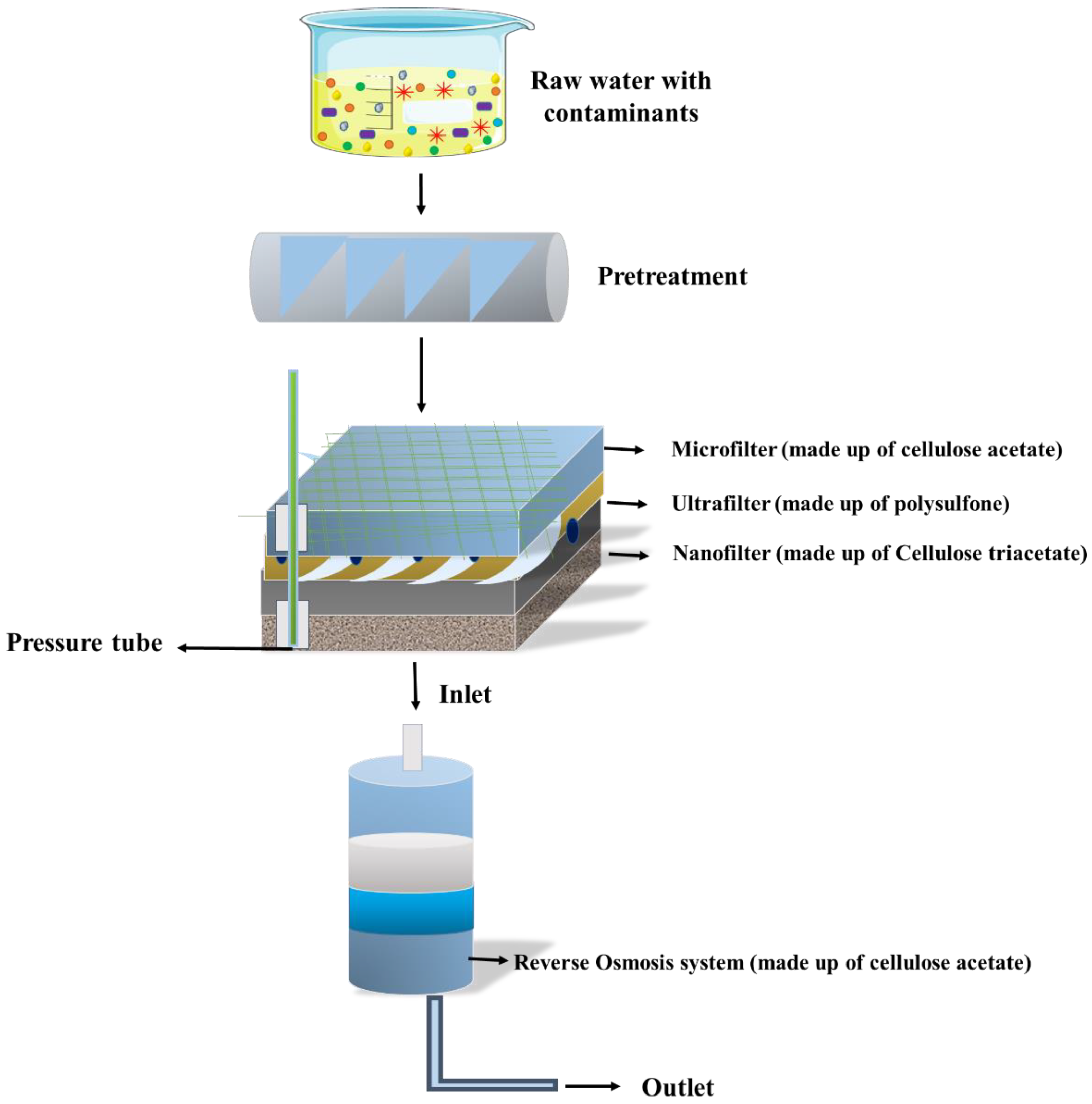
5.1. Membrane Technology
5.1.1. Biological Aerated Filter (BAF) Process
5.1.2. Ultrafiltration
5.1.3. Microfiltration
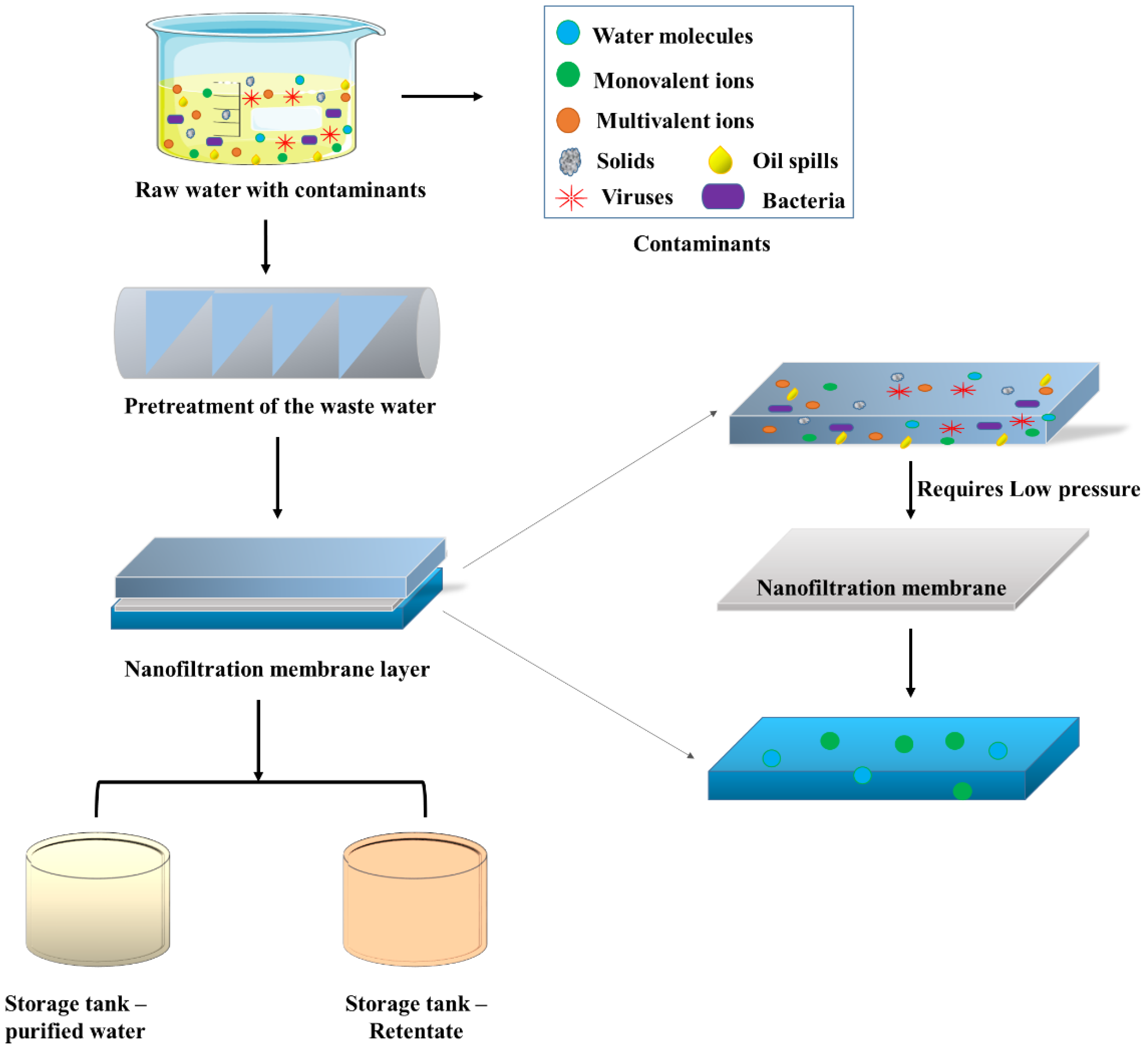
5.1.4. Nanofiltration
5.1.5. Reverse Osmosis
5.1.6. Membranous Biological Reactor
5.1.7. Advance Oxidation Process
5.2. Adsorption
Nanoparticle Interaction
6. Recycling Strategies
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Correa, E.M.C.; Maria Franco, A.; Gonzalez, C.F. Advanced Oxidation Processes for the Removal of Antibiotics from Water. An Overview. Water 2019, 12, 102. [Google Scholar] [CrossRef]
- Bateganya, N.L.; Nakalanzi, D.; Babu, M.; Hein, T. Buffering municipal wastewater pollution using urban wetlands in sub-Saharan Africa: A case of Masaka municipality, Uganda. Environ. Technol. 2015, 36, 2149–2160. [Google Scholar] [CrossRef] [PubMed]
- von Sperling, M.; de Lemos Chernicharo, C.A. Urban wastewater treatment technologies and the implementation of discharge standards in developing countries. Urban Water 2002, 4, 105–114. [Google Scholar] [CrossRef]
- Polprasert, C.; Ujang, Z.; Henze, M. Municipal Wastewater Management in Developing Countries-Principles and Engineering, 5th ed.; IWA Publishing: London, UK, 2006. [Google Scholar]
- Lien, L.T.Q.; Hoa, N.Q.; Chuc, N.T.K.; Thoa, N.T.M.; Phuc, H.D.; Diwan, V.; Dat, N.T.; Tamhankar, A.J.; Lundborg, C.S. Antibiotics in Wastewater of a Rural and an Urban Hospital before and after Wastewater Treatment, and the Relationship with Antibiotic Use—A One Year Study from Vietnam. Int. J. Environ. Res. Public Health 2016, 13, 588. [Google Scholar] [CrossRef]
- Burch, K.D.; Han, B.; Pichtel, J.; Zubkov, T. Removal efficiency of commonly prescribed antibiotics via tertiary wastewater treatment. Environ. Sci. Pollut. Res. 2019, 26, 6301–6310. [Google Scholar] [CrossRef]
- Aydin, S.; Aydin, M.E.; Ulvi, A.; Kilic, H. Antibiotics in hospital effluents: Occurrence, contribution to urban wastewater, removal in a wastewater treatment plant, and environmental risk assessment. Environ. Sci. Pollut. Res. 2018, 26, 544–558. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, J.; Zhuo, L.; Yan, X.; Cai, F.; Luo, W.; Ren, M.; Liu, Q.; Yu, Y. Antibiotics in wastewater from multiple sources and surface water of the Yangtze River in Chongqing in China. Environ. Monit. Assess. 2020, 192, 159. [Google Scholar] [CrossRef]
- Baquero, F.; Martinez, J.L.; Cantón, R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef]
- Calero-Cáceres, W.; Muniesa, M. Persistence of naturally occurring antibiotic resistance genes in the bacteria and bacteriophage fractions of wastewater. Water Res. 2016, 95, 11–18. [Google Scholar] [CrossRef]
- Chandler, C.I. Current accounts of antimicrobial resistance: Stabilisation, individualisation and antibiotics as infrastructure. Palgrave Commun. 2019, 5, 53. [Google Scholar] [CrossRef]
- Huang, A.; Yan, M.; Lin, J.; Xu, L.; Gong, H.; Gong, H. A Review of Processes for Removing Antibiotics from Breeding Wastewater. Int. J. Environ. Res. Public Health 2021, 18, 4909. [Google Scholar] [CrossRef] [PubMed]
- Borbély, G.; Nagy, E. Removal of zinc and nickel ions by complexation–membrane filtration process from industrial wastewater. Desalination 2009, 240, 218–226. [Google Scholar] [CrossRef]
- Tsakona, M.; Anagnostopoulou, E.; Gidarakos, E. Hospital waste management and toxicity evaluation: A case study. Waste Manag. 2007, 27, 912–920. [Google Scholar] [CrossRef]
- Azanu, D.; Styrishave, B.; Darko, G.; Weisser, J.J.; Abaidoo, R.C. Occurrence and risk assessment of antibiotics in water and lettuce in Ghana. Sci. Total. Environ. 2018, 622–623, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, K.; Meyer, M.T. Occurrence of antibiotics in wastewater treatment facilities in Wisconsin, USA. Sci. Total. Environ. 2006, 361, 196–207. [Google Scholar] [CrossRef]
- Carmona, E.; Andreu, V.; Picó, Y. Occurrence of acidic pharmaceuticals and personal care products in Turia River Basin: From waste to drinking water. Sci. Total. Environ. 2014, 484, 53–63. [Google Scholar] [CrossRef]
- Aghalari, Z.; Dahms, H.U.; Sillanpää, M.; Sosa-Hernandez, J.E.; Parra-Saldívar, R. Effectiveness of wastewater treatment systems in removing microbial agents: A systematic review. Glob. Health 2020, 16, 13. [Google Scholar] [CrossRef]
- Zhang, T.; Li, B. Occurrence, Transformation, and Fate of Antibiotics in Municipal Wastewater Treatment Plants. Crit. Rev. Environ. Sci. Technol. 2011, 41, 951–998. [Google Scholar] [CrossRef]
- Kim, S.; Carlson, K. Quantification of human and veterinary antibiotics in water and sediment using SPE/LC/MS/MS. Anal. Bioanal. Chem. 2007, 387, 1301–1315. [Google Scholar] [CrossRef]
- Al-Riyami, I.M.; Ahmed, M.; Al-Busaidi, A.; Choudri, B.S. Antibiotics in wastewaters: A review with focus on Oman. Appl. Water Sci. 2018, 8, 199. [Google Scholar] [CrossRef]
- Li, W.; Shi, Y.; Gao, L.; Liu, J.; Cai, Y. Occurrence of antibiotics in water, sediments, aquatic plants, and animals from Baiyangdian Lake in North China. Chemosphere 2012, 89, 1307–1315. [Google Scholar] [CrossRef] [PubMed]
- Rosal, R.; Rodríguez, A.; Perdigón-Melón, J.A.; Petre, A.; García-Calvo, E.; Gómez, M.J. Occurrence of emerging pol-lutants in urban wastewater and their removal through biological treatment followed by ozonation. Water Res. 2010, 44, 578–588. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhou, J. Occurrence and behavior of antibiotics in water and sediments from the Huangpu River, Shanghai, China. Chemosphere 2014, 95, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Carlson, K. Routine monitoring of antibiotics in water and wastewater with a radioimmunoassay technique. Water Res. 2004, 38, 3155–3166. [Google Scholar] [CrossRef] [PubMed]
- Hubeny, J.; Harnisz, M.; Korzeniewska, E.; Buta, M.; Zieliński, W.; Rolbiecki, D.; Giebułtowicz, J.; Nałęcz-Jawecki, G.; Płaza, G. Industrialization as a source of heavy metals and antibiotics which can enhance the antibiotic resistance in wastewater, sewage sludge and river water. PLoS ONE 2021, 16, e0252691. [Google Scholar] [CrossRef]
- Ebrahimi, S.M.; Reyhani, R.D.; Asghari-JafarAbadi, M.; Fathifar, Z. Diversity of antibiotics in hospital and municipal wastewaters and receiving water bodies and removal efficiency by treatment processes: A systematic review protocol. Environ. Evid. 2020, 9, 19. [Google Scholar] [CrossRef]
- Kumar, S.B.; Arnipalli, S.R.; Ziouzenkova, O. Antibiotics in Food Chain: The Consequences for Antibiotic Resistance. Antibiotics 2020, 9, 688. [Google Scholar] [CrossRef]
- Sun, S.; Yao, H.; Fu, W.; Xue, S.; Zhang, W. Enhanced degradation of antibiotics by photo-fenton reactive membrane filtration. J. Hazard. Mater. 2019, 386, 121955. [Google Scholar] [CrossRef]
- Gao, P.; Munir, M.; Xagoraraki, I. Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci. Total Environ. 2012, 421–422, 173–183. [Google Scholar] [CrossRef]
- Kumar, A.; Pal, D. Antibiotic resistance and wastewater: Correlation, impact and critical human health challenges. J. Environ. Chem. Eng. 2018, 6, 52–58. [Google Scholar] [CrossRef]
- Teixeira, P.; Costa, S.; Brown, B.; Silva, S.; Rodrigues, R.; Valério, E. Quantitative PCR Detection of Enteric Viruses in Wastewater and Environmental Water Sources by the Lisbon Municipality: A Case Study. Water 2020, 12, 544. [Google Scholar] [CrossRef]
- Magureanu, M.; Piroi, D.; Mandache, N.B.; David, V.; Medvedovici, A.; Bradu, C.; Parvulescu, V.I. Degradation of antibiotics in water by non-thermal plasma treatment. Water Res. 2011, 45, 3407–3416. [Google Scholar] [CrossRef] [PubMed]
- Magureanu, M.; Piroi, D.; Mandache, N.B.; David, V.; Medvedovici, A.; Parvulescu, V.I. Degradation of pharmaceutical compound pentoxifylline in water by non-thermal plasma treatment. Water Res. 2010, 44, 3445–3453. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Mozaz, S.; Vaz-Moreira, I.; Della Giustina, S.V.; Llorca, M.; Barceló, D.; Schubert, S.; Berendonk, T.U.; Michael-Kordatou, I.; Fatta-Kassinos, D.; Martinez, J.L.; et al. Antibiotic residues in final effluents of European wastewater treatment plants and their impact on the aquatic environment. Environ. Int. 2020, 140, 105733. [Google Scholar] [CrossRef] [PubMed]
- Leung, H.; Minh, T.; Murphy, M.; Lam, J.C.; So, M.; Martin, M.; Lam, P.K.; Richardson, B. Distribution, fate and risk assessment of antibiotics in sewage treatment plants in Hong Kong, South China. Environ. Int. 2012, 42, 1–9. [Google Scholar] [CrossRef]
- Mahmood, A.R.; Al-Haideri, H.H.; Hassan, F.M. Detection of Antibiotics in Drinking Water Treatment Plants in Baghdad City, Iraq. Adv. Public Health 2019, 2019, 7851354. [Google Scholar] [CrossRef]
- Mutiyar, P.K.; Mittal, A.K. Occurrences and fate of selected human antibiotics in influents and effluents of sewage treatment plant and effluent-receiving river Yamuna in Delhi (India). Environ. Monit. Assess. 2013, 186, 541–557. [Google Scholar] [CrossRef]
- Diwan, V.; Tamhankar, A.J.; Khandal, R.K.; Sen, S.; Aggarwal, M.; Marothi, Y.; Iyer, R.V.; Sundblad-Tonderski, K.; Lundborg, C.S. Antibiotics and antibiotic-resistant bacteria in waters associated with a hospital in Ujjain, India. BMC Public Health 2010, 10, 414. [Google Scholar] [CrossRef]
- Watkinson, A.; Murby, E.; Kolpin, D.; Costanzo, S. The occurrence of antibiotics in an urban watershed: From wastewater to drinking water. Sci. Total. Environ. 2009, 407, 2711–2723. [Google Scholar] [CrossRef]
- Faleye, A.C.; Adegoke, A.A.; Ramluckan, K.; Fick, J.; Bux, F.; Stenström, T.A. Concentration and reduction of antibiotic residues in selected wastewater treatment plants and receiving waterbodies in Durban, South Africa. Sci. Total Environ. 2019, 678, 10–20. [Google Scholar] [CrossRef]
- Kortesmäki, E.; Östman, J.R.; Meierjohann, A.; Brozinski, J.; Eklund, P.; Kronberg, L. Occurrence of Antibiotics in Influent and Effluent from 3 Major Wastewater-Treatment Plants in Finland. Environ. Toxicol. Chem. 2020, 39, 1774–1789. [Google Scholar] [CrossRef] [PubMed]
- Göbel, A.; Thomsen, A.; McArdell, C.S.; Joss, A.; Giger, W. Occurrence and Sorption Behavior of Sulfonamides, Macrolides, and Trimethoprim in Activated Sludge Treatment. Environ. Sci. Technol. 2005, 39, 3981–3989. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, R.H.; Wennberg, P.; Johansson, M.I.; Tysklind, M.; Andersson, B.A.V. Screening of Human Antibiotic Sub-stances and Determination of Weekly Mass Flows in Five Sewage Treatment Plants in Sweden. Environ. Sci. Technol. 2005, 39, 3421–3429. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cheng, W.; Xu, L.; Strong, P.J.; Chen, H. Antibiotic-resistant genes and antibiotic-resistant bacteria in the effluent of urban residential areas, hospitals, and a municipal wastewater treatment plant system. Environ. Sci. Pollut. Res. 2014, 22, 4587–4596. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cheng, W.; Xu, L.; Jiao, Y.; Baig, S.A.; Chen, H. Occurrence and removal of antibiotics and the corresponding resistance genes in wastewater treatment plants: Effluent’s influence to downstream water environment. Environ. Sci. Pollut. Res. 2015, 23, 6826–6835. [Google Scholar] [CrossRef] [PubMed]
- Dinh, Q.; Moreau-Guigon, E.; Labadie, P.; Alliot, F.; Teil, M.J.; Blanchard, M. Fate of antibiotics from hospital and domestic sources in a sewage network. Sci. Total Environ. 2017, 575, 758–766. [Google Scholar] [CrossRef]
- Choi, K.; Kim, Y.; Jung, J.; Kim, M.-H.; Kim, C.-S.; Kim, N.-H.; Park, J. Occurrences and ecological risks of roxithromycin, trimethoprim, and chloramphenicol in the Han River, Korea. Environ. Toxicol. Chem. 2008, 27, 711–719. [Google Scholar] [CrossRef]
- Ekwanzala, M.D.; Lehutso, R.F.; Kasonga, T.K.; Dewar, J.B.; Momba, M.N.B. Environmental Dissemination of Selected Antibiotics from Hospital Wastewater to the Aquatic Environment. Antibiotics 2020, 9, 431. [Google Scholar] [CrossRef]
- Rossmann, J.; Schubert, S.; Gurke, R.; Oertel, R.; Kirch, W. Simultaneous determination of most prescribed antibiotics in multiple urban wastewater by SPE-LC–MS/MS. J. Chromatogr. B 2014, 969, 162–170. [Google Scholar] [CrossRef]
- Bisognin, R.P.; Wolff, D.B.; Carissimi, E.; Prestes, O.D.; Zanella, R. Occurrence and fate of pharmaceuticals in effluent and sludge from a wastewater treatment plant in Brazil. Environ. Technol. 2019, 42, 2292–2303. [Google Scholar] [CrossRef]
- Verlicchi, P.; Al Aukidy, M.; Jelic, A.; Petrović, M.; Barceló, D. Comparison of measured and predicted concentrations of selected pharmaceuticals in wastewater and surface water: A case study of a catchment area in the Po Valley (Italy). Sci. Total Environ. 2014, 470–471, 844–854. [Google Scholar] [CrossRef] [PubMed]
- Verlicchi, P.; Galletti, A.; Petrovic, M.; Barceló, D. Hospital effluents as a source of emerging pollutants: An overview of micropollutants and sustainable treatment options. J. Hydrol. 2010, 389, 416–428. [Google Scholar] [CrossRef]
- Wegrzyn, A.; Machura, M.; Zabczynski, S. Possibilities for Removal of Contrast Agents from Wastewater. Ochro-Nasrodowiska 2015, 37, 55–63. [Google Scholar]
- Capoor, M.R.; Bhowmik, K.T. Cytotoxic Drug Dispersal, Cytotoxic Safety, and Cytotoxic Waste Management: Practices and Proposed India-specific Guidelines. Indian J. Med. Paediatr. Oncol. 2017, 38, 190–197. [Google Scholar]
- Wang, L.; Liu, Y.; Ma, J.; Zhao, F. Rapid degradation of sulphamethoxazole and the further transformation of 3-amino-5-methylisoxazole in a microbial fuel cell. Water Res. 2016, 88, 322–328. [Google Scholar] [CrossRef]
- Arslan, A.; Veli, S.; Bingöl, D. Use of response surface methodology for pretreatment of hospital wastewater by O3/UV and O3/UV/H2O2 processes. Sep. Purif. Technol. 2014, 132, 561–567. [Google Scholar] [CrossRef]
- Escher, B.I.; Baumgartner, R.; Koller, M.; Treyer, K.; Lienert, J.; McArdell, C.S. Environmental toxicology and risk assessment of pharmaceuticals from hospital wastewater. Water Res. 2011, 45, 75–92. [Google Scholar] [CrossRef]
- Paulus, G.K.; Hornstra, L.M.; Alygizakis, N.; Slobodnik, J.; Thomaidis, N.; Medema, G. The impact of on-site hospital wastewater treatment on the downstream communal wastewater system in terms of antibiotics and antibiotic resistance genes. Int. J. Hyg. Environ. Health 2019, 222, 635–644. [Google Scholar] [CrossRef]
- Orias, F.; Perrodin, Y. Characterisation of the ecotoxicity of hospital effluents: A review. Sci. Total. Environ. 2013, 454–455, 250–276. [Google Scholar] [CrossRef]
- Khan, N.A.; Ahmed, S.; Farooqi, I.H.; Ali, I.; Vambol, V.; Changani, F.; Yousefi, M.; Vambol, S.; Khan, S.U.; Khan, A.H. Occurrence, sources and conventional treatment techniques for various antibiotics present in hospital wastewaters: A critical review. TrAC Trends Anal. Chem. 2020, 129, 115921. [Google Scholar] [CrossRef]
- Carraro, E.; Bonetta, S.; Bertino, C.; Lorenzi, E.; Bonetta, S.; Gilli, G. Hospital effluents management: Chemical, physical, microbiological risks and legislation in different countries. J. Environ. Manag. 2016, 168, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Savin, M.; Bierbaum, G.; Hammerl, J.A.; Heinemann, C.; Parcina, M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. Antibiotic-resistant bacteria and antimicrobial residues in wastewater and process water from German pig slaughterhouses and their receiving municipal wastewater treatment plants. Sci. Total Environ. 2020, 727, 138788. [Google Scholar] [CrossRef] [PubMed]
- Savin, M.; Bierbaum, G.; Hammerl, J.A.; Heinemann, C.; Parcina, M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. ESKAPE Bacteria and Extended-Spectrum-β-Lactamase-Producing Escherichia coli Isolated from Wastewater and Process Water from German Poultry Slaughterhouses. Appl. Environ. Microbiol. 2020, 86, e02748-19. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Huang, H.; Xue, W.; Chang, Y.; Li, Y.; Guo, X.; Zhong, C. Rigidifying induced fluorescence enhancement in 2D porous covalent triazine framework nanosheets for the simultaneously luminous detection and adsorption removal of antibiotics. Chem. Eng. J. 2020, 384, 123382. [Google Scholar] [CrossRef]
- Homeier-Bachmann, T.; Heiden, S.; Lübcke, P.; Bachmann, L.; Bohnert, J.; Zimmermann, D.; Schaufler, K. Antibiotic-Resistant Enterobacteriaceae in Wastewater of Abattoirs. Antibiotics 2021, 10, 568. [Google Scholar] [CrossRef]
- Bustillo-Lecompte, C.; Mehrvar, M. Slaughterhouse wastewater: Treatment, management and resource recovery. In Physico-Chemical Wastewater Treatment and Resource Recovery; IntechOpen: London, UK, 2017; pp. 153–174. [Google Scholar] [CrossRef]
- Masse, M.T.; Aloys, R.J.S.; Betbui, B.T. Profile of Antibiotic Resistant Bacteria Isolated from Slaughterhouse Effluents of Etoudi-Yaounde and Its Receiving Waterbody. Int. J. Health Sci. Res. 2021, 11, 40–47. [Google Scholar] [CrossRef]
- Lucas, D.; Badia-Fabregat, M.; Vicent, T.; Caminal, G.; Rodríguez-Mozaz, S.; Balcázar, J.; Barceló, D. Fungal treatment for the removal of antibiotics and antibiotic resistance genes in veterinary hospital wastewater. Chemosphere 2016, 152, 301–308. [Google Scholar] [CrossRef]
- Beattie, R.E.; Walsh, M.; Cruz, M.C.; McAliley, L.R.; Dodgen, L.; Zheng, W.; Hristova, K.R. Agricultural contamination impacts antibiotic resistance gene abundances in riverbed sediment temporally. FEMS Microbiol. Ecol. 2018, 94, fiy131. [Google Scholar] [CrossRef]
- Davis, J.G.; Truman, C.C.; Kim, S.C.; Ascough, J.C.; Carlson, K. Antibiotic Transport via Runoff and Soil Loss. J. Environ. Qual. 2006, 35, 2250–2260. [Google Scholar] [CrossRef]
- Christy, M.L.; Sampson, M.; Edson, M.; Anthony, O. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar]
- Tasho, R.P.; Cho, J.Y. Veterinary antibiotics in animal waste, its distribution in soil and uptake by plants: A review. Sci. Total Environ. 2016, 563, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Hocquet, D.; Muller, A.; Bertrand, X. What happens in hospitals does not stay in hospitals: Antibiotic-resistant bacteria in hospital wastewater systems. J. Hosp. Infect. 2016, 93, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Anthony, A.A.; Adekunle, C.F.; Thor, A.S. Residual antibiotics, antibiotic resistant superbugs and antibiotic resistance genes in surface water catchments: Public health impact. Phys. Chem. Earth 2018, 105, 177–183. [Google Scholar] [CrossRef]
- Tarrass, F.; Benjelloun, M.; Benjelloun, O. Recycling Wastewater After Hemodialysis: An Environmental Analysis for Alternative Water Sources in Arid Regions. Am. J. Kidney Dis. 2008, 52, 154–158. [Google Scholar] [CrossRef] [PubMed]
- Meena, V.D.; Dotaniya, M.L.; Saha, J.K.; Patra, A. Antibiotics and antibiotic resistant bacteria in wastewater: Impact on environment, soil microbial activity and human health. Afr. J. Microbiol. Res. 2015, 9, 965–978. [Google Scholar]
- Niemi, L.; Taggart, M.; Boyd, K.; Zhang, Z.; Gaffney, P.P.; Pfleger, S.; Gibb, S. Assessing hospital impact on pharmaceutical levels in a rural ‘source-to-sink’water system. Sci. Total Environ. 2020, 737, 139618. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.M.P.T.; Silva, L.J.G.; Laranjeiro, C.S.M.; Meisel, L.M.; Lino, C.M.; Pena, A. Human pharmaceuticals in Portuguese rivers: The impact of water scarcity in the environmental risk. Sci. Total Environ. 2017, 609, 1182–1191. [Google Scholar] [CrossRef]
- Hernández, F.; Montory, M.; Gómez-Fuentes, C. Occurrence of antibiotics and bacterial resistance in wastewater and sea water from the Antarctic. In Proceedings of the 15th International Conference on Environmental Science and Technology, Rhodes, Greece, 31 August–2 September 2017. [Google Scholar]
- Cheng, D.; Ngo, H.; Guo, W.; Liu, Y.; Zhou, J.; Chang, S.; Nguyen, D.; Bui, X.; Zhang, X. Bioprocessing for elimination antibiotics and hormones from swine wastewater. Sci. Total. Environ. 2018, 621, 1664–1682. [Google Scholar] [CrossRef]
- Li, X.; Guo, P.; Shan, Y.; Ke, Y.; Li, H.; Fu, Q.; Wang, Y.; Liu, T.; Xia, X. Determination of 82 veterinary drugs in swine waste lagoon sludge by ultra-high performance liquid chromatography–tandem mass spectrometry. J. Chromatogr. A 2017, 1499, 57–64. [Google Scholar] [CrossRef]
- Sosa-Hernández, J.E.; Rodas-Zuluaga, L.I.; López-Pacheco, I.Y.; Melchor-Martínez, E.M.; Aghalari, Z.; Limón, D.S. Sources of antibiotics pollutants in the aquatic environment under SARS-CoV-2 pandemic situation. Case Stud. Chem. Environ. Eng. 2021, 4, 100127. [Google Scholar] [CrossRef]
- Vaz-Moreira, I.; Nunes, O.; Manaia, C.M. Bacterial diversity and antibiotic resistance in water habitats: Searching the links with the human microbiome. FEMS Microbiol. Rev. 2014, 38, 761–778. [Google Scholar] [CrossRef] [PubMed]
- Hultman, J.; Tamminen, M.; Pärnänen, K.; Cairns, J.; Karkman, A.; Virta, M. Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent. FEMS Microbiol. Ecol. 2018, 94, fiy038. [Google Scholar] [CrossRef]
- Cacace, D.; Fatta-Kassinos, D.; Manaia, C.M.; Cytryn, E.; Kreuzinger, N.; Rizzo, L.; Karaolia, P.; Schwartz, T.; Alexander, J.; Merlin, C.; et al. Antibiotic resistance genes in treated wastewater and in the receiving water bodies: A pan-European survey of urban settings. Water Res. 2019, 162, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Sharmaa, N.; Sharmab, S.K. Wastewater Treatment Plants as emerging source of antibiotic resistance. In Green Chemistry and Water Remediation: Research and Applications; Elsevier: Amsterdam, The Netherlands, 2020; p. 239. [Google Scholar] [CrossRef]
- Gwenzi, W.; Kanda, A.; Danha, C.; Muisa-Zikali, N.; Chaukura, N. Occurrence, human health risks, and removal of pharmaceuticals in aqueous systems: Current knowledge and future perspectives. In Applied Water Science Volume 1: Fundamentals and Applications; Scrivener Publishing LLC: Beverly, MA, USA, 2021; pp. 63–101. [Google Scholar] [CrossRef]
- Singh, R.; Singh, A.P.; Kumar, S.; Giri, B.S.; Kim, K.-H. Antibiotic resistance in major rivers in the world: A systematic review on occurrence, emergence, and management strategies. J. Clean. Prod. 2019, 234, 1484–1505. [Google Scholar] [CrossRef]
- Zhang, Y.; Marrs, C.F.; Simon, C.; Xi, C. Wastewater treatment contributes to selective increase of antibiotic resistance among Acinetobacter spp. Sci. Total. Environ. 2009, 407, 3702–3706. [Google Scholar] [CrossRef]
- Kraemer, S.A.; Ramachandran, A.; Perron, G.G. Antibiotic Pollution in the Environment: From Microbial Ecology to Public Policy. Microorganisms 2019, 7, 180. [Google Scholar] [CrossRef]
- Lupo, A.; Coyne, S.; Berendonk, T.U. Origin and Evolution of Antibiotic Resistance: The Common Mechanisms of Emergence and Spread in Water Bodies. Front. Microbiol. 2012, 3, 18. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Aga, D.S. Potential Ecological and Human Health Impacts of Antibiotics and Antibiotic-Resistant Bacteria from Wastewater Treatment Plants. J. Toxicol. Environ. Health Part B 2007, 10, 559–573. [Google Scholar] [CrossRef]
- Madigan, M.T.; Martinko, J.M.; Parker, J. Brock Biology of Microorganisms, 8th ed.; Prentice Hall International, Inc.: New York, NY, USA, 2021. [Google Scholar]
- Mwangi, M.M.; Wu, S.W.; Zhou, Y.; Sieradzki, K.; de Lencastre, H.; Richardson, P.; Bruce, D.; Rubin, E.; Myers, E.; Siggia, E.D.; et al. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 2007, 104, 9451–9456. [Google Scholar] [CrossRef]
- Khameneh, B.; Diab, R.; Ghazvini, K.; Bazzaz, B.S.F. Breakthroughs in bacterial resistance mechanisms and the potential ways to combat them. Microb. Pathog. 2016, 95, 32–42. [Google Scholar] [CrossRef]
- Golovko, O.; Kumar, V.; Fedorova, G.; Randak, T.; Grabic, R. Seasonal changes in antibiotics, antidepressants/psychiatric drugs, antihistamines and lipid regulators in a wastewater treatment plant. Chemosphere 2014, 111, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Sims, N.; Kasprzyk-Hordern, B. Future perspectives of wastewater-based epidemiology: Monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020, 139, 105689. [Google Scholar] [CrossRef] [PubMed]
- Morse, S. Factors and determinants of disease emergence. Rev. Sci. Tech.-Off. Int. Des Epizoot. 2004, 23, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Crittenden, J.C.; Trussell, R.R.; Hand, D.W.; Howe, K.J.; Tchobanoglous, G. MWH’s Water Treatment: Principles and Design; John Wiley & Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- Rafraf, I.D.; Lekunberri, I.; Sànchez-Melsió, A.; Aouni, M.; Borrego, C.M.; Balcázar, J.L. Abundance of antibiotic resistance genes in five municipal wastewater treatment plants in the Monastir Governorate, Tunisia. Environ. Pollut. 2016, 219, 353–358. [Google Scholar] [CrossRef]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; Salamat, M.K.F.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef]
- Rolain, J.; Canton, R.; Cornaglia, G. Emergence of antibiotic resistance: Need for a new paradigm. Clin. Microbiol. Infect. 2012, 18, 615–616. [Google Scholar] [CrossRef]
- Gouliouris, T.; Raven, K.E.; Moradigaravand, D.; Ludden, C.; Coll, F.; Blane, B. Detection of vancomycin-resistant Enterococcus faecium hospital-adapted lineages in municipal wastewater treatment plants indicates widespread distribution and release into the environment. Genome Res. 2019, 29, 626–634. [Google Scholar] [CrossRef]
- Cahill, N.; O’Connor, L.; Mahon, B.; Varley, Á.; McGrath, E.; Ryan, P. Hospital effluent: A reservoir for carbapenemase-producing Enterobacterales? Sci. Total Environ. 2019, 672, 618–624. [Google Scholar] [CrossRef]
- Burnham, J.P. Climate change and antibiotic resistance: A deadly combination. Ther. Adv. Infect. Dis. 2021, 8, 2049936121991374. [Google Scholar] [CrossRef]
- Chahal, C.; Akker, B.V.D.; Young, F.; Franco, C.; Blackbeard, J.; Monis, P. Pathogen and Particle Associations in Wastewater. Adv. Appl. Microbiol. 2016, 97, 63–119. [Google Scholar] [CrossRef]
- Farooq, R.; Ahmad, Z. Biological Wastewater Treatment and Resource Recovery Spreading of Antibiotic Resistance with Wastewater. In Biological Wastewater Treatment and Resource Recovery; IntechOpen: London, UK, 2017; pp. 73–98. [Google Scholar]
- Pandey, P.K.; Kass, P.H.; Soupir, M.L.; Biswas, S.; Singh, V.P. Contamination of water resources by pathogenic bacteria. AMB Express 2014, 4, 51. [Google Scholar] [CrossRef] [PubMed]
- Handel, A.; Margolis, E.; Levin, B.R. Exploring the role of the immune response in preventing antibiotic resistance. J. Theor. Biol. 2009, 256, 655–662. [Google Scholar] [CrossRef]
- Marshall, J.S.; Warrington, R.; Watson, W.; Kim, H.L. An introduction to immunology and immunopathology. Allergy Asthma Clin. Immunol. 2018, 14, 49. [Google Scholar] [CrossRef] [PubMed]
- Peterson, C.T.; Sharma, V.; Elmén, L.; Peterson, S.N. Immune homeostasis, dysbiosis and therapeutic modulation of the gut microbiota. Clin. Exp. Immunol. 2015, 179, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Miarov, O.; Tal, A.; Avisar, D. A critical evaluation of comparative regulatory strategies for monitoring pharmaceuticals in recycled wastewater. J. Environ. Manag. 2019, 254, 109794. [Google Scholar] [CrossRef]
- Bos, J.; Zhang, Q.; Vyawahare, S.; Rogers, E.; Rosenberg, S.M.; Austin, R.H. Emergence of antibiotic resistance from multinucleated bacterial filaments. Proc. Natl. Acad. Sci. USA 2014, 112, 178–183. [Google Scholar] [CrossRef]
- Gillissen, G. Side Effects of Antibiotics on Immune Response Parameters and their Possible Implications in Antimicrobial Chemotherapy. Zent. Bakteriol. Mikrobiol. Hyg. 1988, 270, 171–199. [Google Scholar] [CrossRef]
- Cotten, C.M.; Taylor, S.; Stoll, B.; Goldberg, R.N.; Hansen, N.I.; Sánchez, P.J.; Ambalavanan, N.; Benjamin, D.K., Jr.; NICHD Neonatal Research Network. Prolonged duration of initial empirical antibiotic treatment is associated with increased rates of necrotizing enterocolitis and death for extremely low birth weight infants. Pediatrics 2009, 123, 58–66. [Google Scholar] [CrossRef]
- Miller, J.E.; Wu, C.S.; Pedersen, L.H.; de Klerk, N.; Olsen, J.; Burgner, D.P. Maternal antibiotic exposure during pregnancy and hospitalization with infection in offspring: A population-based cohort study. Int. J. Epidemiol. 2018, 47, 561–571. [Google Scholar] [CrossRef]
- Gonzalez-Perez, G.; Hicks, A.L.; Tekieli, T.M.; Radens, C.M.; Williams, B.L.; Lamouse-Smith, E.S.N. Maternal antibiotic treatment impacts development of the neonatal intestinal microbiome and antiviral immunity. J. Immunol. 2016, 196, 3768–3779. [Google Scholar] [CrossRef]
- Derakhshan, Z.; Mokhtari, M.; Babaei, F.; Ahmadi, R.M.; Ehrampoush, M.H.; Faramarzian, M. Removal Methods of Antibiotic Compounds from Aqueous Environments—A Review. J. Environ Health Sustain Dev. 2016, 1, 43–62. [Google Scholar]
- Nasrollohai, N.; Vatanpour, V.; Khataee, A. Removal of antibiotics from wastewaters by membrane technology: Limitations, successes, and future improvements. Sci. Total Environ. 2022, 838, 156010. [Google Scholar] [CrossRef] [PubMed]
- Nqombolo, A.; Mpupa, A.; Moutloali, R.; Nomngongo, P. Wastewater Treatment Using Membrane Technology. In Wastewater and Water Quality; Yonar, T., Ed.; IntechOpen: London, UK, 2018; pp. 29–40. [Google Scholar]
- Schwermer, C.U.; Krzeminski, P.; Wennberg, A.C.; Vogelsang, C.; Uhl, W. Removal of antibiotic resistant E. coli in two Norwegian wastewater treatment plants and by nano- and ultra-filtration processes. Water Sci. Technol. 2017, 77, 1115–1126. [Google Scholar] [CrossRef]
- Liu, M.K.; Liu, Y.Y.; Bao, D.D.; Zhu, G.; Yang, G.H.; Geng, J.F.; Li, H.T. Effective Removal of Tetracycline Antibiotics from Water using Hybrid Carbon Membranes. Sci. Rep. 2017, 7, srep43717. [Google Scholar] [CrossRef]
- Sharma, V.; Kumar, R.V.; Pakshirajan, K.; Pugazhenthi, G. Integrated adsorption-membrane filtration process for antibiotic removal from aqueous solution. Powder Technol. 2017, 321, 259–269. [Google Scholar] [CrossRef]
- Ganiyu, S.O.; van Hullebusch, E.D.; Cretin, M.; Esposito, G.; Oturan, M.A. Coupling of membrane filtration and advanced oxidation processes for removal of pharmaceutical residues: A critical review. Sep. Purif. Technol. 2015, 156, 891–914. [Google Scholar] [CrossRef]
- Yin, F.; Lin, S.; Zhou, X.; Dong, H.; Zhan, Y. Fate of antibiotics during membrane separation followed by physical-chemical treatment processes. Sci. Total Environ. 2021, 759, 143520. [Google Scholar] [CrossRef]
- Arefi-Oskoui, S.; Khataee, A.; Jabbarvand Behrouz, S.; Vatanpour, V.; Haddadi Gharamaleki, S.; Orooji, Y.; Safarpour, M. Development of MoS2/O-MWCNTs/PES blended membrane for efficient removal of dyes, antibiotic, and protein. Sep. Purif. Technol. 2022, 280, 11982. [Google Scholar] [CrossRef]
- Javad, M.; Mohsen, S.; Vatanpour, V. Performance improvement of PES membrane decorated by Mil-125(Ti)/chitosan nanocomposite for removal of organic pollutants and heavy metal. Chemosphere 2022, 290, 133335. [Google Scholar]
- Phoon, B.L.; Ong, C.C.; Saheed, M.S.M.; Show, P.-L.; Chang, J.-S.; Ling, T.C.; Lam, S.S.; Juan, J.C. Conventional and emerging technologies for removal of antibiotics from wastewater. J. Hazard. Mater. 2020, 400, 122961. [Google Scholar] [CrossRef]
- Ezugbe, E.O.; Rathilal, S. Membrane Technologies in Wastewater Treatment: A Review. Membranes 2020, 10, 89. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Zhou, Y.; Sun, X.; Fu, L. The recent development of advanced wastewater treatment by ozone and biological aerated filter. Environ. Sci. Pollut. Res. 2018, 25, 8315–8329. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.-T.; Su, Z.-X.; Lai, W.-X.; Zhang, Y.-B.; Liu, Y.-W. Insights into the fate and removal of antibiotics and antibiotic resistance genes using biological wastewater treatment technology. Sci. Total Environ. 2021, 776, 145906. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Y.-S.; Zhang, J.-N.; Yang, Y.-Q.; Hu, L.-X.; Yang, Y.-Y.; Zhao, J.-L.; Chen, F.-R.; Ying, G.-G. Removal of antibiotics from piggery wastewater by biological aerated filter system: Treatment efficiency and biodegradation kinetics. Bioresour. Technol. 2017, 238, 70–77. [Google Scholar] [CrossRef]
- Wang, J.; Yang, Z.; Wang, H.; Wu, S.; Lu, H.; Wang, X. Decomposition process of cefotaxime sodium from antibiotic wastewater by Up-flow Blanket Filter (UBF) reactor: Reactor performance, sludge characteristics and microbial community structure analysis. Sci. Total Environ. 2021, 758, 143670. [Google Scholar] [CrossRef]
- Azimi, S.M.; Malakootian, M.; Yaghmaeian, K.; Ahmadian, M.; Rahimi, S. Studying the efficiency of a biological aerated filter (BAF) with oyster Media on improving the quality of effluent produced by treatment plants. Fresen. Environ. Bull. 2014, 23, 2401–2406. [Google Scholar]
- Li, S.-Z.; Li, X.-Y.; Wang, D.-Z. Membrane (RO-UF) filtration for antibiotic wastewater treatment and recovery of antibiotics. Sep. Purif. Technol. 2004, 34, 109–114. [Google Scholar] [CrossRef]
- Liang, C.; Wei, D.; Zhang, S.; Ren, Q.; Shi, J.; Liu, L. Removal of antibiotic resistance genes from swine wastewater by membrane filtration treatment. Ecotoxicol. Environ. Saf. 2021, 210, 111885. [Google Scholar] [CrossRef]
- Li, X.; Jiang, L.; Li, H. Application of ultrafiltration technology in water treatment. In IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2018; Volume 186. [Google Scholar] [CrossRef]
- Bourgeous, K.N.; Darby, J.L.; Tchobanoglous, G. Ultrafiltration of wastewater: Effects of particles, mode of operation, and backwash effectiveness. Water Res. 2000, 35, 77–90. [Google Scholar] [CrossRef]
- Bera, S.P.; Godhaniya, M.; Kothari, C. Emerging and advanced membrane technology for wastewater treatment: A review. J. Basic Microbiol. 2021, 62, 245–259. [Google Scholar] [CrossRef]
- Qiu, G.; Chen, H.; Srinivasa Raghavan, D.S.; Ting, Y.P. Removal behaviors of antibiotics in a hybrid microfiltration-forward osmotic membrane bioreactor for real municipal wastewater treatment. Chem. Eng. J. 2021, 417, 129146. [Google Scholar] [CrossRef]
- Samrot, A.V.; Sahithya, C.S.; Selvarani, A.J.; Pachiyappan, S.; Kumar, S. Surface-engineered Super-Paramagnetic Iron Oxide Nanoparticles for Chromium Removal. Int. J. Nanomed. 2019, 14, 8105–8119. [Google Scholar] [CrossRef] [PubMed]
- Samrot, A.V.; Sahithya, C.S.; Purayil, S.K.; Ponnaiah, P. A review on synthesis, characterization and potential biological applications of superparamagnetic iron oxide nanoparticles. Curr. Res. Green Sustain. Chem. 2020, 4, 100042. [Google Scholar] [CrossRef]
- Barancheshme, F.; Munir, M. Strategies to Combat Antibiotic Resistance in the Wastewater Treatment Plants. Front. Microbiol. 2018, 8, 2603. [Google Scholar] [CrossRef]
- Moarefian, A.; Golestani, H.A.; Bahmanpour, H. Removal of amoxicillin from wastewater by self-made Polyethersulfone membrane using nanofiltration. J. Environ. Health Sci. Eng. 2014, 12, 127. [Google Scholar] [CrossRef]
- Abdel-Fatah, M.A. Nanofiltration systems and applications in wastewater treatment: Review article. Ain Shams Eng. J. 2018, 9, 3077–3092. [Google Scholar] [CrossRef]
- Homem, V.; Santos, L. Degradation and removal methods of antibiotics from aqueous matrices—A review. J. Environ. Manag. 2011, 92, 2304–2347. [Google Scholar] [CrossRef] [PubMed]
- Gholami, M.; Mirzaei, R.; Kalantary, R.R.; Sabzali, A.; Gatei, F. Performance evaluation of reverse osmosis technology for selected antibiotics removal from synthetic pharmaceutical wastewater. Iran. J. Environ. Health Sci. Eng. 2012, 9, 19. [Google Scholar] [CrossRef]
- Dolar, D.; Vuković, A.; Ašperger, D.; Košutić, K. Efficiency of RO/NF membranes at the removal of veterinary antibiotics. Water Sci. Technol. 2012, 65, 317–323. [Google Scholar] [CrossRef]
- Sahar, E.; David, I.; Gelman, Y.; Chikurel, H.; Aharoni, A.; Messalem, R.; Brenner, A. The use of RO to remove emerging micropollutants following CAS/UF or MBR treatment of municipal wastewater. Desalination 2011, 273, 142–147. [Google Scholar] [CrossRef]
- Govardhan, B.; Fatima, S.; Madhumala, M.; Sridhar, S. Modification of used commercial reverse osmosis membranes to nan-ofiltration modules for the production of mineral-rich packaged drinking water. Appl. Water Sci. 2020, 10, 230. [Google Scholar] [CrossRef]
- Cheng, H.; Hong, P.Y. Removal of Antibiotic-Resistant Bacteria and Antibiotic Resistance Genes Affected by Varying Degrees of Fouling on Anaerobic Microfiltration Membranes. Environ. Sci. Technol. 2017, 51, 12200–12209. [Google Scholar] [CrossRef]
- Dave, S.; Das, J. Technological model on advanced stages of oxidation of wastewater effluent from food industry. In Advanced Oxidation Processes for Effluent Treatment Plants; Elsevier: Amsterdam, The Netherlands, 2021; pp. 33–49. [Google Scholar] [CrossRef]
- Mahdi, M.H.; Mohammed, T.J.; Al-Najar, J. Advanced Oxidation Processes (AOPs) for treatment of antibiotics in wastewater: A review. IOP Conf. Series Earth Environ. Sci. 2021, 779, 012109. [Google Scholar] [CrossRef]
- Akbari, M.Z.; Xu, Y.; Lu, Z.; Peng, L. Review of antibiotics treatment by advance oxidation processes. Environ. Adv. 2021, 5, 100111. [Google Scholar] [CrossRef]
- Ahmed, M.B.; Zhou, J.L.; Ngo, H.H.; Guo, W. Adsorptive removal of antibiotics from water and wastewater: Progress and challenges. Sci. Total. Environ. 2015, 532, 112–126. [Google Scholar] [CrossRef]
- Lakherwal, D. Adsorption of Heavy Metals: A Review. Int. J. Environ. Res. Devel-Opment 2014, 4, 41–48. [Google Scholar]
- Farhana, A.; Selvarani, A.J.; Samrot, A.V.; Alsrhani, A.; Raji, P.; Sahithya, C.S.; Cypriyana, P.J.J.; Senthilkumar, P.; Ling, M.P.; Yishak, S. Utilization of Superparamagnetic Iron Oxide Nanoparticles (SPIONs) Impregnated Activated Carbon for Removal of Hexavalent Chromium. J. Nanomater. 2022, 2022, 4326939. [Google Scholar] [CrossRef]
- Samrot, A.V.; Shobana, N. Citrus sinensis cellulose fibres incorporated with SPIONs for effective removal of crystal violet dye. Biocatal. Agric. Biotechnol. 2021, 39, 102211. [Google Scholar] [CrossRef]
- Wang, X.; Yin, R.; Zeng, L.; Zhu, M. A review of graphene-based nanomaterials for removal of antibiotics from aqueous environments. Environ. Pollut. 2019, 253, 100–110. [Google Scholar] [CrossRef]
- Eniola, J.O.; Kumar, R.; Barakat, M.A. Adsorptive removal of antibiotics from water over natural and modified adsorbents. Environ. Sci. Pollut. Res. 2019, 26, 34775–34788. [Google Scholar] [CrossRef]
- Yang, S.-F.; Lin, C.-F.; Wu, C.-J.; Ng, K.-K.; Lin, A.Y.-C.; Hong, P.-K.A. Fate of sulfonamide antibiotics in contact with activated sludge—Sorption and biodegradation. Water Res. 2012, 46, 1301–1308. [Google Scholar] [CrossRef]
- Samrot, A.V.; Angalene, J.L.A.; Roshini, S.M.; Raji, P.; Stefi, S.M.; Preethi, R.; Selvarani, A.J.; Madankumar, A. Bioactivity and Heavy Metal Removal Using Plant Gum Mediated Green Synthesized Silver Nanoparticles. J. Clust. Sci. 2019, 30, 1599–1610. [Google Scholar] [CrossRef]
- Samrot, A.V.; Senthilkumar, P.; Rashmitha, S.; Veera, P.; Sahithya, C.S. Azadirachta indica influenced biosynthesis of super-paramagnetic iron-oxide nanoparticles and their applications in tannery water treatment and X-ray imaging. J. Nanostructure Chem. 2018, 8, 343–351. [Google Scholar] [CrossRef]
- Samrot, A.V.; Shobana, N.; Sruthi, P.D.; Sahithya, C.S. Utilization of chitosan-coated superparamagnetic iron oxide nanoparticles for chromium removal. Appl. Water Sci. 2018, 8, 192. [Google Scholar] [CrossRef]
- Stafiej, A.; Pyrzynska, K. Adsorption of heavy metal ions with carbon nanotubes. Sep. Purif. Technol. 2005, 58, 49–52. [Google Scholar] [CrossRef]
- Cong, Q.; Yuan, X.; Qu, J. A review on the removal of antibiotics by carbon nanotubes. Water Sci. Technol. 2013, 68, 1679–1687. [Google Scholar] [CrossRef]
- Yu, F.; Ma, J.; Bi, D. Enhanced adsorptive removal of selected pharmaceutical antibiotics from aqueous solution by activated graphene. Environ. Sci. Pollut. Res. 2015, 22, 4715–4724. [Google Scholar] [CrossRef]
- Asano, T.; Levine, A.D. Wastewater reclamation, recycling and reuse: Past, present, and future. Water Sci. Technol. 1996, 33, 1–14. [Google Scholar] [CrossRef]
- Ajibode, O.M. Effect of Residence Time on Microbial and Chemical Quality of Reclaimed Water in Urban Infrastructures. Ph.D. Thesis, The University of Arizona, Tucson, AZ, USA, 2012. [Google Scholar]
- Kihila, J.; Mtei, K.M.; Njau, K.N. Wastewater treatment for reuse in urban agriculture; the case of Moshi Municipality, Tanzania. Phys. Chem. Earth Parts A/B/C 2014, 72–75, 104–110. [Google Scholar] [CrossRef]
- Angelakis, A.; Durham, B. Water recycling and reuse in EUREAU countries: Trends and challenges. Desalination 2008, 218, 3–12. [Google Scholar] [CrossRef]
- Yang, H.; Abbaspour, K.C. Analysis of wastewater reuse potential in Beijing. Desalination 2007, 212, 238–250. [Google Scholar] [CrossRef]
- Manouchehri, M.; Kargari, A. Water recovery from laundry wastewater by the cross flow microfiltration process: A strategy for water recycling in residential buildings. J. Clean. Prod. 2017, 168, 227–238. [Google Scholar] [CrossRef]
- Al-Hashimia, M.A.I.; Abbas, T.R.; Jasema, Y.I. Performance of sequencing anoxic/anaerobic membrane bioreactor (SAM) system in hospital wastewater treatment and reuse. Eur. Sci. J. 2013, 9, 169–180. [Google Scholar]
- Tiwari, B.; Sellamuthu, B.; Ouarda, Y.; Drogui, P.; Tyagi, R.D.; Buelna, G. Review on fate and mechanism of removal of pharmaceutical pollutants from wastewater using biological approach. Bioresour. Technol. 2017, 224, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Mozaz, S.; Lucas, D.; Barceló, D. Full-Scale Plants for Dedicated Treatment of Hospital Effluents. Hosp. Wastewaters Charact. Manag. Treat. Environ. Risks 2017, 60, 189–208. [Google Scholar] [CrossRef]
- Boehler, M.; Zwickenpflug, B.; Hollender, J.; Ternes, T.; Joss, A.; Siegrist, H. Removal of micropollutants in municipal wastewater treatment plants by powder-activated carbon. Water Sci. Technol. 2012, 66, 2115–2121. [Google Scholar] [CrossRef]
- Golovko, O.; de Brito Anton, L.; Cascone, C.; Ahrens, L.; Lavonen, E.; Köhler, S.J. Sorption Characteristics and Removal Efficiency of Organic Micropollutants in Drinking Water Using Granular Activated Carbon (GAC) in Pilot-Scale and Full-Scale Tests. Water 2020, 12, 2053. [Google Scholar] [CrossRef]
- Polianciuc, S.I.; Gurzău, A.E.; Kiss, B.; Ştefan, M.G.; Loghin, F. Antibiotics in the environment: Causes and consequences. Med. Pharm. Rep. 2020, 93, 231–240. [Google Scholar] [CrossRef]
- Akhil, D.; Lakshmi, D.; Kumar, P.S.; Vo, D.-V.N.; Kartik, A. Occurrence and removal of antibiotics from industrial wastewater. Environ. Chem. Lett. 2021, 19, 1477–1507. [Google Scholar] [CrossRef]
- Wu, H.; Niu, X.; Yang, J.; Wang, C.; Lu, M. Retentions of bisphenol A and norfloxacin by three different ultrafiltration membranes in regard to drinking water treatment. Chem. Eng. J. 2016, 294, 410–416. [Google Scholar] [CrossRef]
- Baran, W.; Adamek, E.; Jajko, M.; Sobczak, A. Removal of veterinary antibiotics from wastewater by electrocoagulation. Chemosphere 2018, 194, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Anjali, R.; Shanthakumar, S. Insights on the current status of occurrence and removal of antibiotics in wastewater by advanced oxidation processes. J. Environ. Manag. 2019, 246, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Carbajo, J.B.; Petre, A.L.; Rosal, R.; Herrera, S.; Letón, P.; García-Calvo, E.; Fernández-Alba, A.R.; Perdigón-Melón, J.A. Continuous ozonation treatment of ofloxacin: Transformation products, water matrix effect and aquatic toxicity. J. Hazard. Mater. 2015, 292, 34–43. [Google Scholar] [CrossRef] [PubMed]

| S. No. | Antibiotics | Concentration (ng/L) | Region | References |
|---|---|---|---|---|
| 1. | Cefalexin | 38.4 | WWTP of Portugal | Mozaz et al. [35] |
| Trimethoprim | 69.1 | |||
| Ciprofloxacin | 231.4–584.9 | |||
| Ofloxacin | 89.7–184.9 | |||
| Clindamycin | 8.5–86.6 | |||
| Azithromycin | 178.9–597.5 | |||
| Clarithromycin | 74.2–313.2 | |||
| Pipemidic Acid | 10.8–20.1 | |||
| Sulfamethoxazole | 7.1–30.2 | |||
| Sulfapyridine | 4.7–84.5 | |||
| Tetracycline | 147.5–231.2 | |||
| 2. | Tetracycline | 70–370 | Wastewater treatment facilities Wisconsin, USA | Riyami et al. [21] |
| Trimethoprim | 120–550 | |||
| Sulfamethoxazole | 50–370 | |||
| Macrolides | 300 | |||
| Fluoroquinolones, in the form of ciprofloxacin | 40–140 | |||
| 3 | Ofloxacin, | 96–7870 | Sewage treatment plants (STPs), Hong Kong, South China | Leung et al. [36]; Riyami et al. [21] |
| Norfloxacin | 35–4000 | |||
| Cefalexin | 180–4000 | |||
| Erythromycin | 250–4000 | |||
| Sulfamethoxazole | 5–300 | |||
| Trimethoprim | 60–450 | |||
| 4. | Ciprofloxacin | 1270 | Al-Wihda (W1) and Al-Rasheed water treatment plants (R1) Baghdad City, Iraq | Mahmood et al. [37] |
| Levofloxacin | 177 | |||
| Amoxicillin | 1500 | |||
| 5. | Metronidazole | 2600 | Rural hospital in Vietnam | Lien et al. [5] |
| Sulfamethoxazole | 9800 | |||
| Trimethoprim | 7700 | |||
| Ceftazidime | 1200 | |||
| Ciprofloxacin | 42,800 | |||
| Ofloxacin | 4600 | |||
| Spiramycin | 1700 | |||
| 6. | Ampicillin | 12,680 | Yamuna, Delhi, India | Mutiyar and Mittal. [38] |
| Ciprofloxacin | 8000 | |||
| Gatifloxacin | 1220 | |||
| Sparfloxacin | 140 | |||
| Cefuroxime | 220 | |||
| 7. | Ciprofloxacin | 2200–236,600 | Hospital in Ujjain, India | Diwan et al. [39] |
| Norfloxacin | 6400–29,600 | |||
| Levofloxacin | 5000–8800 | |||
| Ofloxacin | 4500–7500 | |||
| 8. | Amoxycillin | 90 | Watersheds of Southeast Queensland, Australia | Watkinson et al. [40] |
| Cephalexin | 4100–10,000 | |||
| Nalidixic acid | 20–40 | |||
| Enrofloxacin | 60–100 | |||
| Clindamycin | 4–90 | |||
| Lincomycin | 6–1700 | |||
| Roxithromycin | 50–400 | |||
| Doxycycline | 130–200 | |||
| Sulfamethoxazole | 100–300 | |||
| Trimethoprim | 300 | |||
| 9. | Metronidazole | 0.17 | WWTPs in Durban, South Africa | Faleye et al. [41] |
| Erythromycin | 0.14 | |||
| Ofloxacin | 0.22 | |||
| Trimethoprim | 0.11 | |||
| Azithromycin | 0.11 | |||
| 10. | Tetracycline | 37 | WWTPs located in Turku, Tampere and Helsinki, Finland | Kortesmäki et al. [42] |
| Carbamazepine | 47–417 | |||
| Sulfadiazine | 326–1069 | |||
| Trimethoprim | 170–490 | |||
| Clarithromycin | 50–327 | |||
| Roxithromycin | 60–145 | |||
| 11. | Sulfapyridine | 90–150 | Wastewater treatment plant Kloten-Opfikon Zurich, Switzerland | Göbel et al. [43] |
| Azithromycin | 170–380 | |||
| Clarithromycin | 380–600 | |||
| Sulfamethoxazole | 430–570 | |||
| 12. | Norfloxacin | 72–155 | Sewage treatment plants, Sweden | Lindberg et al. [44] |
| Ofloxacin | 287 | |||
| Ciprofloxacin | 90–205 | |||
| Sulfamethoxazole | 144–674 | |||
| Doxycycline | 2480 | |||
| 13. | Tetracycline | 286.7–582.5 | Linan City, Eastern China | Li et al. [45] |
| Sulfonamides | 909.9–2741.3 | |||
| Sulfamethazine | 1159 | |||
| 14. | Sulfamethoxazole | 136.7–426.0 | WWTPs and the downstream water adjacent to WWTP, Eastern China | Li et al. [46] |
| 15. | Amoxicillin | 0.11 | Essonne district, France | Dinh et al. [47] |
| Sulfamethoxazole | 2.1 | |||
| Ofloxacin | 17.9 | |||
| Vancomycin | 3.6 | |||
| Norfloxacin | 12.1 | |||
| Ciprofloxacin | 5.8 | |||
| 16. | Roxithromycin | 44.7–130.2 | Han River, Republic of Korea | Choi et al. [48] |
| Trimethoprim | 27–89 | |||
| Chloramphenicol | 54 | |||
| 17. | Sulfamethoxazole | 65–200 | Girona Catalonia, Spain | Ekwanzala et al. [49] |
| Clindamycin | 184–1465 | |||
| Azithromycin | 85–113 | |||
| Ciprofloxacin | 5329–7494 | |||
| 18. | Clindamycin | 41 | Sewage Treatment Plant in Dresden Kaditz, Germany | Ekwanzala et al. [49]; Rossmann et al. [50] |
| Doxycycline | 249 | |||
| Azithromycin | 285 | |||
| Ciprofloxacin | 422 | |||
| 19. | Oxytetracycline | 0.641 | WWTPs in metropolitan regions of Porto Alegre, Capital of Rio Grande do Sul State, Brazil | Bisognin et al. [51] |
| Metronidazole | 0.023 | |||
| Sulfamethoxazole | 0.980 | |||
| Paracetamol | 13.64 | |||
| Tetracycline | 0.042 | |||
| Ciprofloxacin | 0.385 | |||
| 20. | Carbamazepine | 379 | WWTPs in the Po Valley, in Northern Italy | Verlicchi et al. [52] |
| Metronidazole | 42 | |||
| Ofloxacin | 400 | |||
| Norfloxacin | 210 |
| S. No | Strategy Used | Antibiotics Removed | Efficiency | References |
|---|---|---|---|---|
| 1. | Vertical flow constructed wetlands | Sulfamethazine | 68–73% | Huang et al. [134] |
| Ciprofloxacin | 91–95% | |||
| Oxytetracycline | 82–85% | |||
| 2. | Horizontal subsurface flow constructed wetlands | Sulfamethoxazole | 4–59% | Huang et al. [134]; Liu et al. [123] |
| 3. | Photocatalytic degradation | Tetracycline | 94.96% | Akhil et al. [181] |
| Cephalexin | 96% | |||
| Metronidazole | 94.5% | |||
| Sulfamethazine | 78% | |||
| 4. | Sonocatalytic irradiation | Norfloxacin | 69.07% | Akhil et al. [181] |
| Sulfanilamide | 95.64% | |||
| Rifampicin | 95.3% | |||
| Azithromycin | 98.4% | |||
| 5. | Ultrafiltration using PVC membrane | Norfloxacin | 80% | Bao et al. [129]; Wu et al. [182] |
| 6. | Adsorption by activated carbon | Cephalexin | 74–88% | Ahmed et al. [156] |
| Ciprofloxacin | 100% | |||
| Amoxicillin | 95% | |||
| Tetracycline | 74–88% | |||
| Ornidazole | 90% | |||
| 7. | Adsorption by carbon nanotubes | Sulfamethoxazole | 80% | Ahmed et al. [156] |
| Sulfamethoxazole | 96% | |||
| Lincomycin | >90% | |||
| Amoxicillin | 86.5% | |||
| 8. | Electro-coagulation | Ampicillin | 3.6% | Baran et al. [183] |
| Doxycycline | 96.4% | |||
| Sulfathiazole | 3.3% | |||
| Tylosin | 3.1% | |||
| 9. | Advanced oxidation processes in combination of UV/hydrogen peroxide | Metronidazole | 92% | Anjali and Shanthakumar, [184] |
| Ciprofloxacin | 93% | |||
| 10. | Advanced oxidation processes using Fenton process | Ofloxacin | 100% | Carbajo et al. [185] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samrot, A.V.; Wilson, S.; Sanjay Preeth, R.S.; Prakash, P.; Sathiyasree, M.; Saigeetha, S.; Shobana, N.; Pachiyappan, S.; Rajesh, V.V. Sources of Antibiotic Contamination in Wastewater and Approaches to Their Removal—An Overview. Sustainability 2023, 15, 12639. https://doi.org/10.3390/su151612639
Samrot AV, Wilson S, Sanjay Preeth RS, Prakash P, Sathiyasree M, Saigeetha S, Shobana N, Pachiyappan S, Rajesh VV. Sources of Antibiotic Contamination in Wastewater and Approaches to Their Removal—An Overview. Sustainability. 2023; 15(16):12639. https://doi.org/10.3390/su151612639
Chicago/Turabian StyleSamrot, Antony V., Samraj Wilson, Ram Singh Sanjay Preeth, Pandurangan Prakash, Mahendran Sathiyasree, Subramanian Saigeetha, Nagarajan Shobana, Senthilkumar Pachiyappan, and Vinod Vincent Rajesh. 2023. "Sources of Antibiotic Contamination in Wastewater and Approaches to Their Removal—An Overview" Sustainability 15, no. 16: 12639. https://doi.org/10.3390/su151612639
APA StyleSamrot, A. V., Wilson, S., Sanjay Preeth, R. S., Prakash, P., Sathiyasree, M., Saigeetha, S., Shobana, N., Pachiyappan, S., & Rajesh, V. V. (2023). Sources of Antibiotic Contamination in Wastewater and Approaches to Their Removal—An Overview. Sustainability, 15(16), 12639. https://doi.org/10.3390/su151612639






