Effect of Nitrification Inhibitors on Photosynthesis and Nitrogen Metabolism in ‘Sweet Sapphire’ (V. vinifera L.) Grape Seedlings
Abstract
:1. Introduction
2. Materials and Methods
2.1. Overview of the Experimental Site
2.2. Experimental Design
2.3. Assay Items and Methods
2.3.1. Seedlings Biomass Analysis
2.3.2. Roots Phenotype Characteristics Analysis
2.3.3. Photosynthetic Pigment Content Analysis
2.3.4. Photosynthetic Parameters Measurement
2.3.5. Chlorophyll Fluorescence Parameters Measurement
2.3.6. RNA Extraction and Transcriptome Sequencing
2.4. Statistical Analysis
3. Results
3.1. Effect of DMPP on Grape Seedlings Biomass
3.2. Effect of DMPP on Grape Roots Characteristics
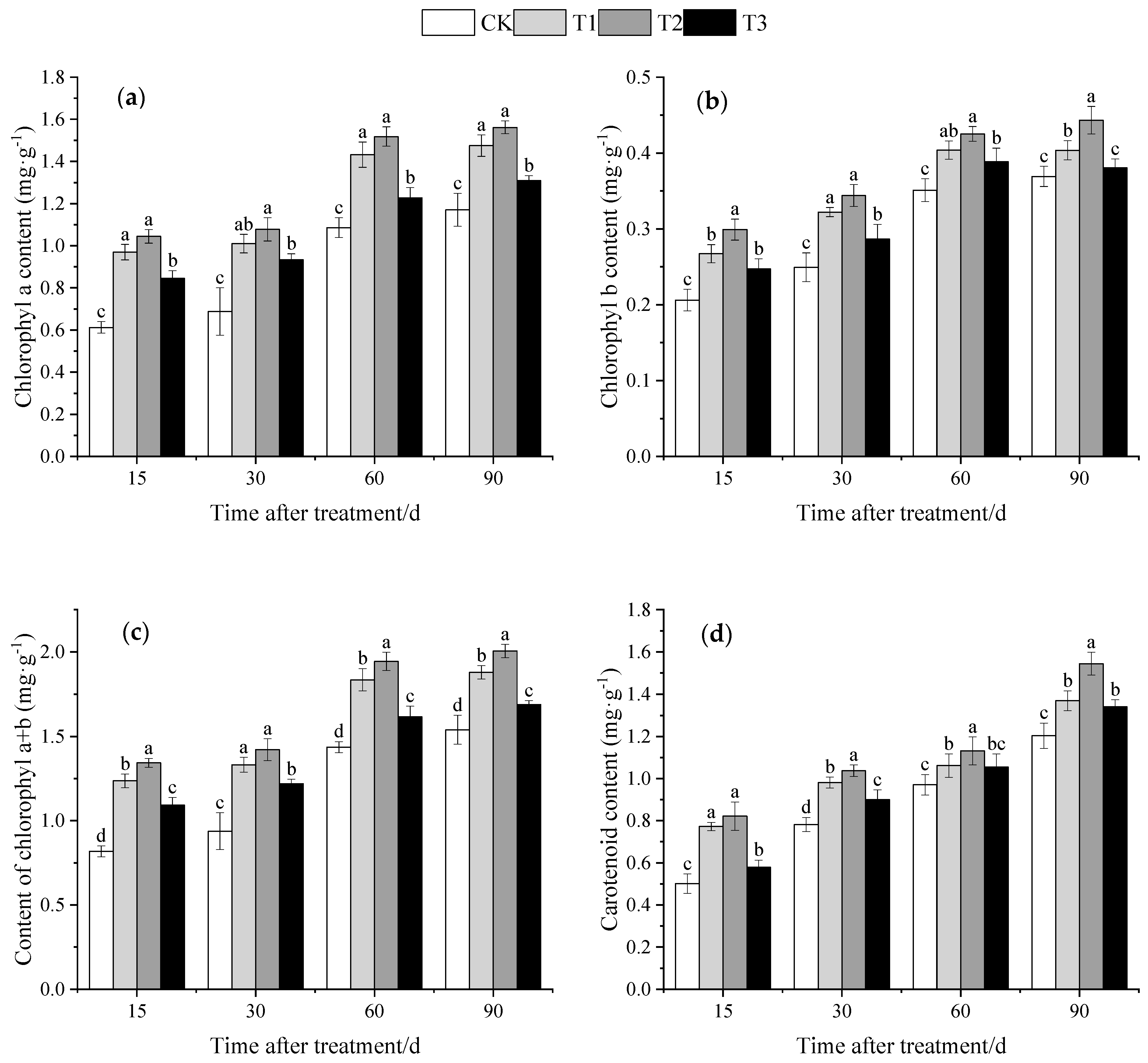
3.3. Effect of DMPP on Photosynthetic Pigment Contents of Grape Leaves
3.4. Effect of DMPP on Photosynthetic Parameters of Grape Leaves
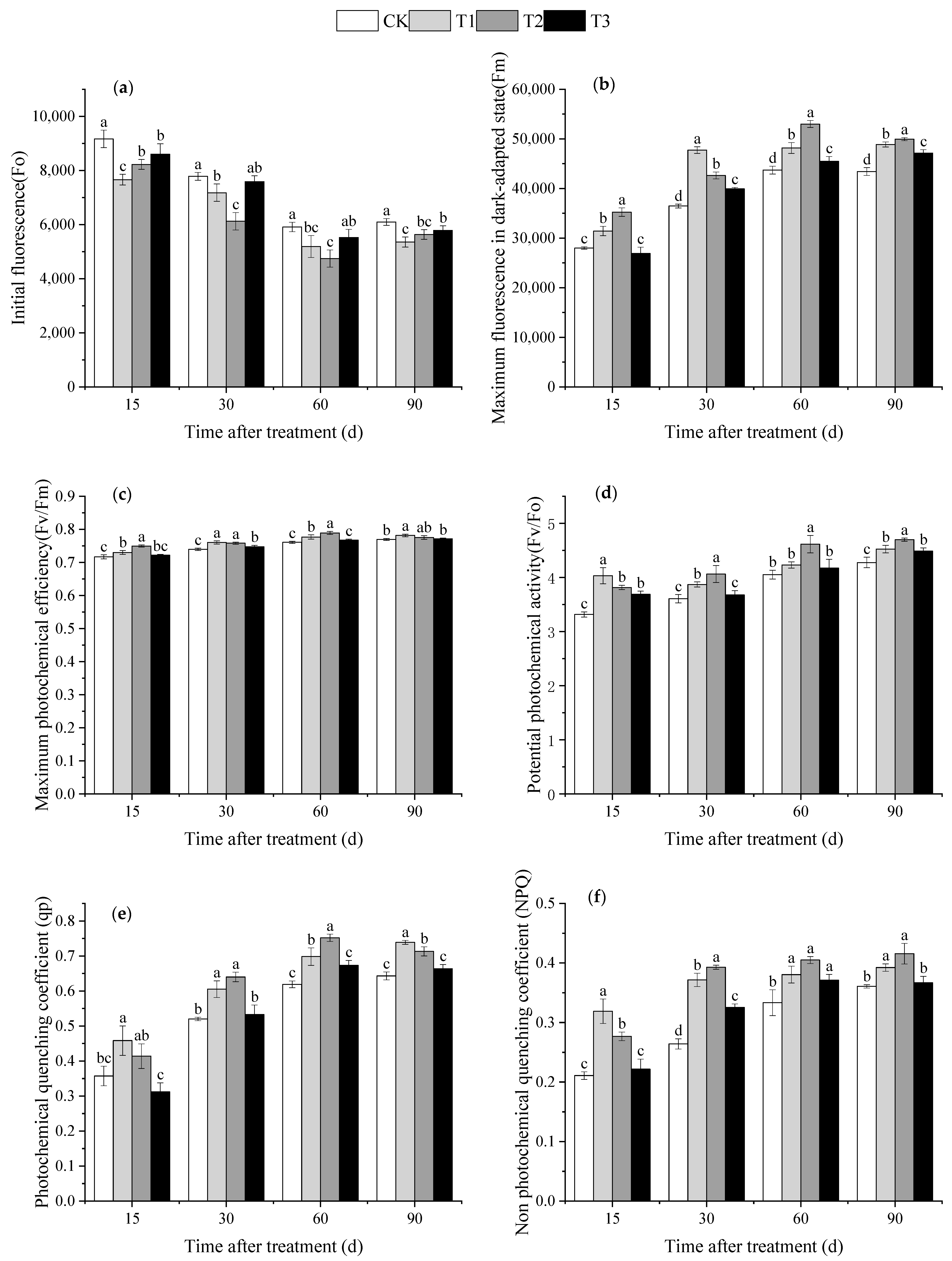
3.5. Effect of DMPP on Chlorophyll Fluorescence Parameters in Grape Leaves
3.6. Effect of DMPP on the Expression of Photosynthesis and Nitrogen Metabolism-Related Genes in Grape Seedlings
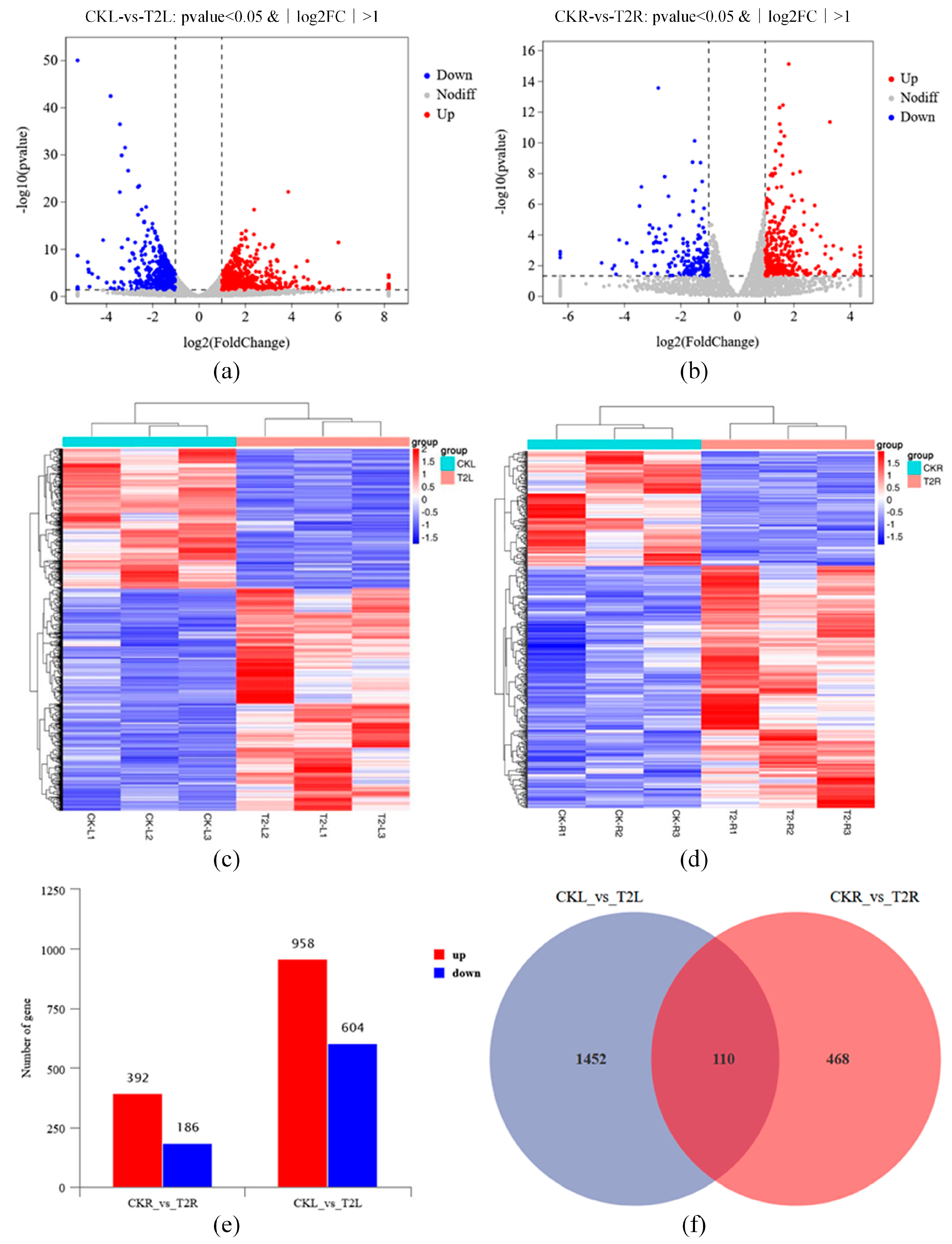
3.6.1. Global Analysis of Grapevine RNA-Seq Data with Application of Urea Nitrogen Content 1% of DMPP
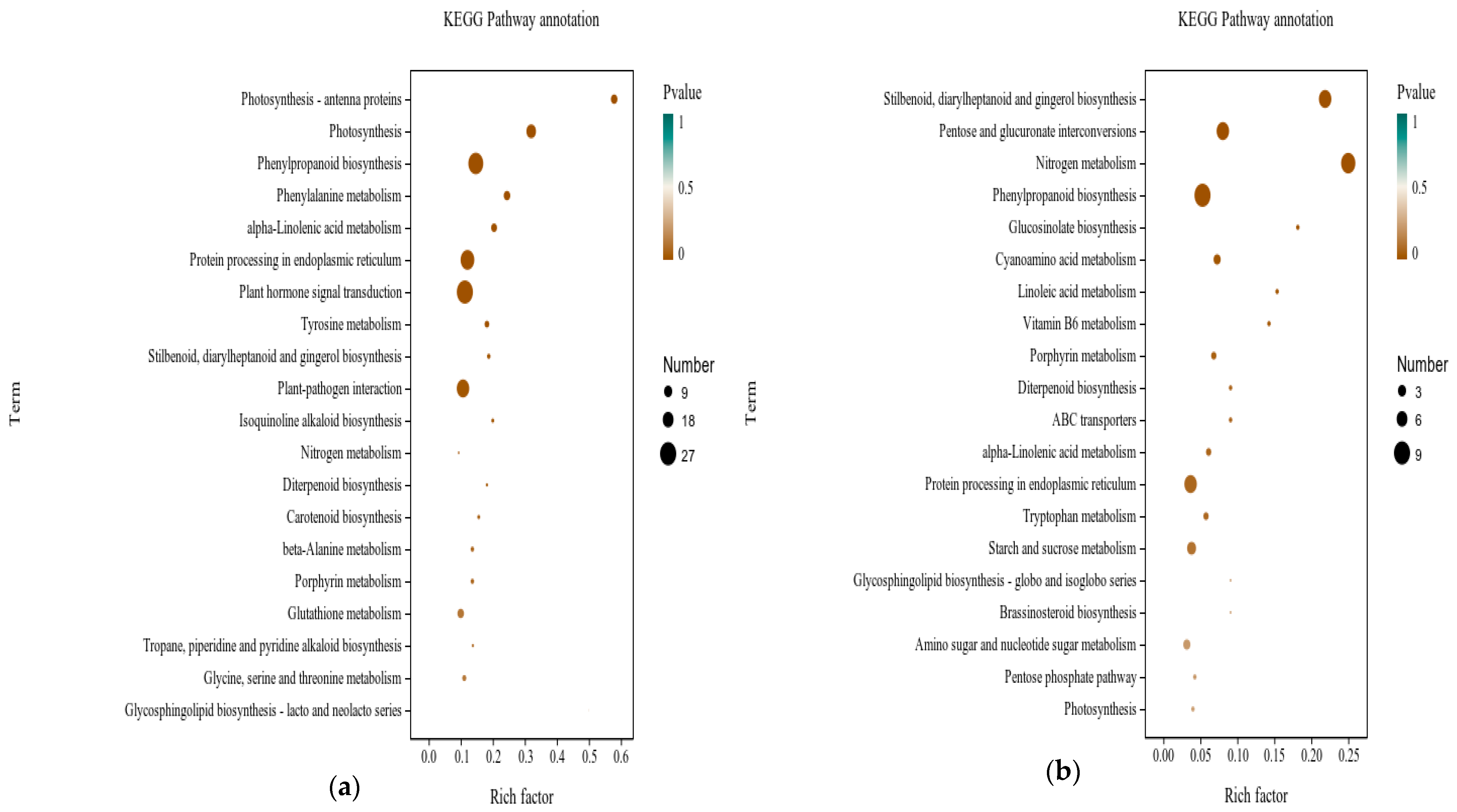
3.6.2. Functional Analysis of DEGs in Grape with Application of Urea Nitrogen Content 1% of DMPP
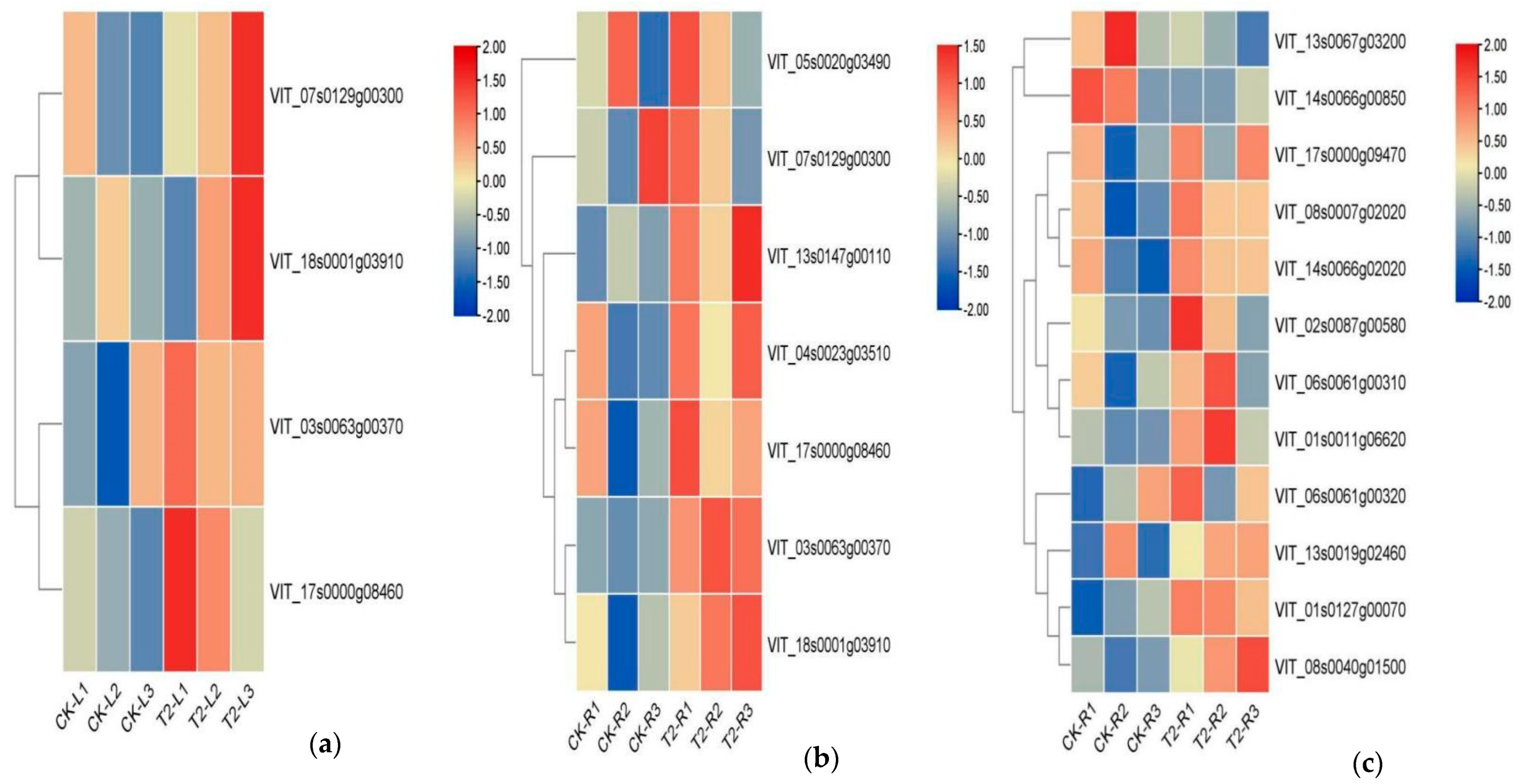
3.6.3. Analysis of Gene Families Associated with Metabolic Process
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- de Freitas Laiber Pascoal, G.; de Almeida Sousa Cruz, M.A.; Pimentel de Abreu, J.; Santos, M.C.B.; Bernardes Fanaro, G.; Junior, M.R.M.; Freitas Silva, O.; Moreira, R.F.A.; Cameron, L.C.; Simoes Larraz Ferreira, M.; et al. Evaluation of the antioxidant capacity, volatile composition and phenolic content of hybrid Vitis vinifera L. varieties sweet sapphire and sweet surprise. Food Chem. 2022, 366, 130644. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Pareek, N.K.; Kumar, U.; Javed, T.; Al-Huqail, A.A.; Rathore, V.S.; Nangia, V.; Choudhary, A.; Nanda, G.; Ali, H.M.; et al. Coupling Effects of Nitrogen and Irrigation Levels on Growth Attributes, Nitrogen Use Efficiency, and Economics of Cotton. Front. Plant Sci. 2022, 13, 890181. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Zhang, G.; Zhang, Y.; Xu, W.; Xie, R.; Ming, B.; Hou, P.; Wang, K.; Xue, J.; Li, S. Effect of the Rate of Nitrogen Application on Dry Matter Accumulation and Yield Formation of Densely Planted Maize. Sustainability 2022, 14, 14940. [Google Scholar] [CrossRef]
- Fu, H.; Cui, D.; Shen, H. Effects of Nitrogen Forms and Application Rates on Nitrogen Uptake, Photosynthetic Characteristics and Yield of Double-Cropping Rice in South China. Agronomy 2021, 11, 158. [Google Scholar] [CrossRef]
- Ren, B.; Ma, Z.; Zhao, B.; Liu, P.; Zhang, J. Influences of split application and nitrification inhibitor on nitrogen losses, grain yield, and net income for summer maize production. Front. Plant Sci. 2022, 13, 982373. [Google Scholar] [CrossRef]
- Bakass, M.; Mokhlisse, A.; Lallemant, M. Absorption and desorption of liquid water by a superabsorbent polymer: Effect of polymer in the drying of the soil and the quality of certain plants. J. Appl. Polym. Sci. 2002, 83, 234–243. [Google Scholar] [CrossRef]
- Gonzalez, M.E.; Cea, M.; Medina, J.; Gonzalez, A.; Diez, M.C.; Cartes, P.; Monreal, C.; Navia, R. Evaluation of biodegradable polymers as encapsulating agents for the development of a urea controlled-release fertilizer using biochar as support material. Sci. Total Environ. 2015, 505, 446–453. [Google Scholar] [CrossRef]
- Schoninger, E.L.; González-Villalba, H.A.; Bendassolli, J.A.; Ocheuze Trivelin, P.C. Fertilizer Nitrogen and Corn Plants: Not all Volatilized Ammonia is Lost. Agron. J. 2018, 110, 1111–1118. [Google Scholar] [CrossRef]
- Mahmud, K.; Panday, D.; Mergoum, A.; Missaoui, A. Nitrogen Losses and Potential Mitigation Strategies for a Sustainable Agroecosystem. Sustainability 2021, 13, 2400. [Google Scholar] [CrossRef]
- Corrochano-Monsalve, M.; Gonzalez-Murua, C.; Bozal-Leorri, A.; Lezama, L.; Artetxe, B. Mechanism of action of nitrification inhibitors based on dimethylpyrazole: A matter of chelation. Sci. Total Environ. 2021, 752, 141885. [Google Scholar] [CrossRef]
- Li, H.; Liang, X.; Chen, Y.; Lian, Y.; Tian, G.; Ni, W. Effect of nitrification inhibitor DMPP on nitrogen leaching, nitrifying organisms, and enzyme activities in a rice-oilseed rape cropping system. J. Environ. Sci. 2008, 20, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Huérfano, X.; Fuertes-Mendizábal, T.; Fernández-Diez, K.; Estavillo, J.M.; González-Murua, C.; Menéndez, S. The new nitrification inhibitor 3,4-dimethylpyrazole succinic (DMPSA) as an alternative to DMPP for reducing N2O emissions from wheat crops under humid Mediterranean conditions. Eur. J. Agron. 2016, 80, 78–87. [Google Scholar] [CrossRef]
- Shi, X.; Hu, H.W.; Zhu-Barker, X.; Hayden, H.; Wang, J.; Suter, H.; Chen, D.; He, J.Z. Nitrifier-induced denitrification is an important source of soil nitrous oxide and can be inhibited by a nitrification inhibitor 3,4-dimethylpyrazole phosphate. Environ. Microbiol 2017, 19, 4851–4865. [Google Scholar] [CrossRef] [PubMed]
- Vilarrasa-Nogue, M.; Teira-Esmatges, M.R.; Pascual, M.; Villar, J.M.; Rufat, J. Effect of N dose, fertilisation duration and application of a nitrification inhibitor on GHG emissions from a peach orchard. Sci. Total Environ. 2020, 699, 134042. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-f.; Wu, L.-h.; Shi, Q.-w.; Wang, Z.-q.; Chen, X.-y.; Li, Y.-s. Effects of a new nitrification inhibitor 3,4-dimethylpyrazole phosphate (DMPP) on nitrate and potassium leaching in two soils. J. Environ. Sci. 2007, 19, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Qiao, C.; Liu, L.; Hu, S.; Compton, J.E.; Greaver, T.L.; Li, Q. How inhibiting nitrification affects nitrogen cycle and reduces environmental impacts of anthropogenic nitrogen input. Global. Chang. Biol. 2015, 21, 1249–1257. [Google Scholar] [CrossRef] [PubMed]
- Kirschke, T.; Spott, O.; Vetterlein, D. Impact of urease and nitrification inhibitor on NH4+ and NO3− dynamic in soil after urea spring application under field conditions evaluated by soil extraction and soil solution sampling. J. Plant Nutr. Soil Sci. 2019, 182, 441–450. [Google Scholar] [CrossRef]
- Thapa, R.; Chatterjee, A. Wheat Production, Nitrogen Transformation, and Nitrogen Losses as Affected by Nitrification and Double Inhibitors. Agron. J. 2017, 109, 1825–1835. [Google Scholar] [CrossRef]
- Sha, Z.; Ma, X.; Wang, J.; Lv, T.; Li, Q.; Misselbrook, T.; Liu, X. Effect of N stabilizers on fertilizer-N fate in the soil-crop system: A meta-analysis. Agric. Ecosyst. Environ. 2020, 290. [Google Scholar] [CrossRef]
- Dawar, K.; Khan, A.; Sardar, K.; Fahad, S.; Saud, S.; Datta, R.; Danish, S. Effects of the nitrification inhibitor nitrapyrin and mulch on N2O emission and fertilizer use efficiency using 15N tracing techniques. Sci. Total Environ. 2021, 757, 143739. [Google Scholar] [CrossRef]
- Li, W.; Wang, Y.; Xu, Q.; Cao, G.; Guo, X.; Zhou, H.; Du, Y. Global analysis of nitrification inhibitors on grasslands nitrous oxide emission rates. Biochem. Syst. Ecol. 2021, 97, 104289. [Google Scholar] [CrossRef]
- Alhaj Hamoud, Y.; Shaghaleh, H.; Sheteiwy, M.; Guo, X.; Elshaikh, N.A.; Ullah Khan, N.; Oumarou, A.; Rahim, S.F. Impact of alternative wetting and soil drying and soil clay content on the morphological and physiological traits of rice roots and their relationships to yield and nutrient use-efficiency. Agric. Water Manag. 2019, 223. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Plant cell wall extensibility: Connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J. Exp. Bot. 2016, 67, 463–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Li, W.; Xu, Z.; Zhang, H.; Sun, G.; Zhang, H.; Yang, C.; Liu, G. Effects of different nitrogen forms and concentrations on seedling growth traits and physiological characteristics of Populus simonii × P. nigra. J. For. Res. 2022, 33, 1593–1606. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefifield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Hu, H.-W.; Chen, Q.-L.; Chen, D.; He, J.-Z. Growth of comammox Nitrospira is inhibited by nitrification inhibitors in agricultural soils. J. Soils Sediments 2020, 20, 621–628. [Google Scholar] [CrossRef]
- Keerio, H.A.; Bae, W.; Panhwar, S. Nitrite Accumulation at Low Ammonia Concentrations in Wastewater Treatment Plants. Sustainability 2022, 14, 16449. [Google Scholar] [CrossRef]
- Rufty, T.W.; Volk, R.J. Alteration in enrichment of NO3−and reduced-N in xylem exudate during and after extended plant exposure to 15NO3−. Plant Soil 1986, 91, 329–332. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, W.; Beebout, S.S.; Zhang, H.; Liu, L.; Yang, J.; Zhang, J. Grain yield, water and nitrogen use efficiencies of rice as influenced by irrigation regimes and their interaction with nitrogen rates. Field Crop. Res. 2016, 193, 54–69. [Google Scholar] [CrossRef]
- Xu, G.-w.; Lu, D.-K.; Wang, H.-Z.; Li, Y. Morphological and physiological traits of rice roots and their relationships to yield and nitrogen utilization as influenced by irrigation regime and nitrogen rate. Agric. Water Manag. 2018, 203, 385–394. [Google Scholar] [CrossRef]
- Andrews, M.; Raven, J.A.; Lea, P.J. Do plants need nitrate? The mechanisms by which nitrogen form affects plants. Ann. Appl. Biol. 2013, 163, 174–199. [Google Scholar] [CrossRef]
- Gao, Y.; Li, Y.; Yang, X.; Li, H.; Shen, Q.; Guo, S. Ammonium nutrition increases water absorption in rice seedlings (Oryza sativa L.) under water stress. Plant Soil 2009, 331, 193–201. [Google Scholar] [CrossRef]
- Wang, P.; Wang, Z.-k.; Sun, X.-c.; Mu, X.-h.; Chen, H.; Chen, F.-j.; Lixing, Y.; Mi, G.-h. Interaction effect of nitrogen form and planting density on plant growth and nutrient uptake in maize seedlings. J. Integr. Agric. 2019, 18, 1120–1129. [Google Scholar] [CrossRef]
- Simkin, A.J.; Kapoor, L.; Doss, C.G.P.; Hofmann, T.A.; Lawson, T.; Ramamoorthy, S. The role of photosynthesis related pigments in light harvesting, photoprotection and enhancement of photosynthetic yield in planta. Photosynth. Res. 2022, 152, 23–42. [Google Scholar] [CrossRef]
- Lin, Z.H.; Chen, L.S.; Chen, R.B.; Zhang, F.Z.; Jiang, H.X.; Tang, N. CO2 assimilation, ribulose-1,5-bisphosphate carboxylase/oxygenase, carbohydrates and photosynthetic electron transport probed by the JIP-test, of tea leaves in response to phosphorus supply. BMC Plant Biol. 2009, 9, 43. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Chen, X.; Ge, Q.; Li, B.; Tong, Y.; Zhang, A.; Li, Z.; Kuang, T.; Lu, C. Tolerance of photosynthesis to photoinhibition, high temperature and drought stress in flag leaves of wheat: A comparison between a hybridization line and its parents grown under field conditions. Plant Sci. 2006, 171, 389–397. [Google Scholar] [CrossRef]
- Liu, G.; Du, Q.; Li, J. Interactive effects of nitrate-ammonium ratios and temperatures on growth, photosynthesis, and nitrogen metabolism of tomato seedlings. Sci. Hortic. 2017, 214, 41–50. [Google Scholar] [CrossRef] [Green Version]
- Collado-Gonzalez, J.; Pinero, M.C.; Otalora, G.; Lopez-Marin, J.; Del Amor, F.M. Effects of Different Nitrogen Forms and Exogenous Application of Putrescine on Heat Stress of Cauliflower: Photosynthetic Gas Exchange, Mineral Concentration and Lipid Peroxidation. Plants 2021, 10, 152. [Google Scholar] [CrossRef]
- Ogawa, T.; Sonoike, K. Screening of mutants using chlorophyll fluorescence. J. Plant Res. 2021, 134, 653–664. [Google Scholar] [CrossRef]
- Ahammed, G.J.; Xu, W.; Liu, A.; Chen, S. COMT1 Silencing Aggravates Heat Stress-Induced Reduction in Photosynthesis by Decreasing Chlorophyll Content, Photosystem II Activity, and Electron Transport Efficiency in Tomato. Front. Plant Sci. 2018, 9, 998. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, H.; He, T.; Gao, R.; Guo, G.; Lu, R.; Chen, Z.; Liu, C. Comparative Analysis of Morphology, Photosynthetic Physiology, and Transcriptome Between Diploid and Tetraploid Barley Derived From Microspore Culture. Front. Plant Sci. 2021, 12, 626916. [Google Scholar] [CrossRef] [PubMed]
- Tietz, S.; Hall, C.C.; Cruz, J.A.; Kramer, D.M. NPQ(T): A chlorophyll fluorescence parameter for rapid estimation and imaging of non-photochemical quenching of excitons in photosystem-II-associated antenna complexes. Plant Cell Environ. 2017, 40, 1243–1255. [Google Scholar] [CrossRef] [PubMed]
- GUO, H.X.; LIU, W.Q.; SHI, Y.C. Effects of different nitrogen forms on photosynthetic rate and the chlorophyll fluorescence induction kinetics of flue-cured tobacco. Photosynthetica 2006, 44, 140–142. [Google Scholar] [CrossRef]
- Croucher, N.J.; Fookes, M.C.; Perkins, T.T.; Turner, D.J.; Marguerat, S.B.; Keane, T.; Quail, M.A.; He, M.; Assefa, S.; Bahler, J.; et al. A simple method for directional transcriptome sequencing using Illumina technology. Nucleic Acids Res. 2009, 37, e148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senevirathna, J.D.M.; Yonezawa, R.; Saka, T.; Igarashi, Y.; Funasaka, N.; Yoshitake, K.; Kinoshita, S.; Asakawa, S. Transcriptomic Insight into the Melon Morphology of Toothed Whales for Aquatic Molecular Developments. Sustainability 2021, 13, 13997. [Google Scholar] [CrossRef]
- Wen, B.; Gong, X.; Chen, X.; Tan, Q.; Li, L.; Wu, H. Transcriptome analysis reveals candidate genes involved in nitrogen deficiency stress in apples. J. Plant Physiol. 2022, 279, 153822. [Google Scholar] [CrossRef]
- Morosinotto, T.; Breton, J.; Bassi, R.; Croce, R. The nature of a chlorophyll ligand in Lhca proteins determines the far red fluorescence emission typical of photosystem I. J. Biol. Chem. 2003, 278, 49223–49229. [Google Scholar] [CrossRef] [Green Version]
- Wientjes, E.; van Stokkum, I.H.; van Amerongen, H.; Croce, R. The role of the individual Lhcas in photosystem I excitation energy trapping. Biophys. J. 2011, 101, 745–754. [Google Scholar] [CrossRef] [Green Version]
- Wientjes, E.; Oostergetel, G.T.; Jansson, S.; Boekema, E.J.; Croce, R. The role of Lhca complexes in the supramolecular organization of higher plant photosystem I. J. Biol. Chem. 2009, 284, 7803–7810. [Google Scholar] [CrossRef] [Green Version]
- Amunts, A.; Toporik, H.; Borovikova, A.; Nelson, N. Structure determination and improved model of plant photosystem I. J. Biol. Chem. 2010, 285, 3478–3486. [Google Scholar] [CrossRef] [Green Version]
- Fristedt, R.; Trotta, A.; Suorsa, M.; Nilsson, A.K.; Croce, R.; Aro, E.M.; Lundin, B. PSB33 sustains photosystem II D1 protein under fluctuating light conditions. J. Exp. Bot. 2017, 68, 4281–4293. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Yokono, M.; Akimoto, S.; Takabayashi, A.; Tanaka, A.; Tanaka, R. Deficiency of the Stroma-Lamellar Protein LIL8/PSB33 Affects Energy Transfer Around PSI in Arabidopsis. Plant Cell Physiol. 2017, 58, 2026–2039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vargas-Suarez, M.; Castro-Sanchez, A.; Toledo-Ortiz, G.; Gonzalez de la Vara, L.E.; Garcia, E.; Loza-Tavera, H. Protein phosphorylation regulates in vitro spinach chloroplast petD mRNA 3′-untranslated region stability, processing, and degradation. Biochimie 2013, 95, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Huarancca Reyes, T.; Scartazza, A.; Pompeiano, A.; Ciurli, A.; Lu, Y.; Guglielminetti, L.; Yamaguchi, J. Nitrate Reductase Modulation in Response to Changes in C/N Balance and Nitrogen Source in Arabidopsis. Plant Cell Physiol. 2018, 59, 1248–1254. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Zhang, H.; Zhang, H.; Deng, X.W.; Wei, N. HY5 regulates nitrite reductase 1 (NIR1) and ammonium transporter1;2 (AMT1;2) in Arabidopsis seedlings. Plant Sci. 2015, 238, 330–339. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Zhang, W.; Yang, Y.; Li, Z.; Li, N.; Qi, S.; Crawford, N.M.; Wang, Y. The Arabidopsis NLP7 gene regulates nitrate signaling via NRT1.1-dependent pathway in the presence of ammonium. Sci. Rep. 2018, 8, 1487. [Google Scholar] [CrossRef] [Green Version]
- Teng, Y.; Liang, Y.; Wang, M.; Mai, H.; Ke, L. Nitrate Transporter 1.1 is involved in regulating flowering time via transcriptional regulation of FLOWERING LOCUS C in Arabidopsis thaliana. Plant Sci. 2019, 284, 30–36. [Google Scholar] [CrossRef]
- Zhang, X.; Cui, Y.; Yu, M.; Su, B.; Gong, W.; Baluska, F.; Komis, G.; Samaj, J.; Shan, X.; Lin, J. Phosphorylation-Mediated Dynamics of Nitrate Transceptor NRT1.1 Regulate Auxin Flux and Nitrate Signaling in Lateral Root Growth. Plant Physiol. 2019, 181, 480–498. [Google Scholar] [CrossRef] [Green Version]
- Medici, A.; Szponarski, W.; Dangeville, P.; Safi, A.; Dissanayake, I.M.; Saenchai, C.; Emanuel, A.; Rubio, V.; Lacombe, B.; Ruffel, S.; et al. Identification of Molecular Integrators Shows that Nitrogen Actively Controls the Phosphate Starvation Response in Plants. Plant Cell 2019, 31, 1171–1184. [Google Scholar] [CrossRef]



| Treatments | Total Biomass (g) | Above-Ground Biomass (g) | Roots Biomass (g) |
|---|---|---|---|
| CK | 130.77 ± 1.57 d | 96.62 ± 1.02 d | 34.15 ± 0.63 d |
| T1 | 154.86 ± 1.62 b | 114.31 ± 1.06 b | 40.55 ± 0.80 b |
| T2 | 199.07 ± 2.07 a | 138.38 ± 1.49 a | 60.69 ± 1.25 a |
| T3 | 140.95 ± 1.23 c | 103.55 ± 0.97 c | 37.40 ± 1.47 c |
| Treatment | Total Root Length (cm) | Total Surface Area (cm2) | Total Root Volume (cm3) | Root Vigour (mg/g.h) |
|---|---|---|---|---|
| CK | 4942.67 ± 109.64 d | 2413.73 ± 121.62 b | 69.33 ± 0.63 c | 4869.60 ± 70.41 b |
| T1 | 6051.69 ± 93.30 b | 2755.79 ± 167.24 ab | 74.09 ± 1.65 ab | 5471.67 ± 391.29 ab |
| T2 | 6648.38 ± 79.38 a | 2880.42 ± 104.43 a | 75.32 ± 0.74 a | 5785.72 ± 230.64 a |
| T3 | 5581.79 ± 211.85 c | 2573.53 ± 161.97 ab | 71.36 ± 1.54 bc | 4825.60 ± 347.30 b |
| GO Category | Term | DEGs-Up | DEGs-Down | |
|---|---|---|---|---|
| Leaf | Molecular function | Molecular function | 696 | 433 |
| Biological process | Biological process | 648 | 430 | |
| Cellular processes | Cellular component | 578 | 372 | |
| Cellular processes | Cellular anatomical entity | 561 | 366 | |
| Root | Molecular function | Molecular function | 295 | 108 |
| Biological process | Biological process | 285 | 100 | |
| Cellular processes | Cellular component | 245 | 95 | |
| Cellular processes | Cellular anatomical entity | 243 | 95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, J.; Dou, F.; Phillip, F.O.; Liu, G.; Liu, H. Effect of Nitrification Inhibitors on Photosynthesis and Nitrogen Metabolism in ‘Sweet Sapphire’ (V. vinifera L.) Grape Seedlings. Sustainability 2023, 15, 4130. https://doi.org/10.3390/su15054130
Zhu J, Dou F, Phillip FO, Liu G, Liu H. Effect of Nitrification Inhibitors on Photosynthesis and Nitrogen Metabolism in ‘Sweet Sapphire’ (V. vinifera L.) Grape Seedlings. Sustainability. 2023; 15(5):4130. https://doi.org/10.3390/su15054130
Chicago/Turabian StyleZhu, Jingjing, Feifei Dou, Fesobi Olumide Phillip, Gang Liu, and Huaifeng Liu. 2023. "Effect of Nitrification Inhibitors on Photosynthesis and Nitrogen Metabolism in ‘Sweet Sapphire’ (V. vinifera L.) Grape Seedlings" Sustainability 15, no. 5: 4130. https://doi.org/10.3390/su15054130
APA StyleZhu, J., Dou, F., Phillip, F. O., Liu, G., & Liu, H. (2023). Effect of Nitrification Inhibitors on Photosynthesis and Nitrogen Metabolism in ‘Sweet Sapphire’ (V. vinifera L.) Grape Seedlings. Sustainability, 15(5), 4130. https://doi.org/10.3390/su15054130





