Methane Anaerobic Oxidation Potential and Microbial Community Response to Sulfate Input in Coastal Wetlands of the Yellow River Delta
Abstract
:1. Introduction
2. Materials and Methods
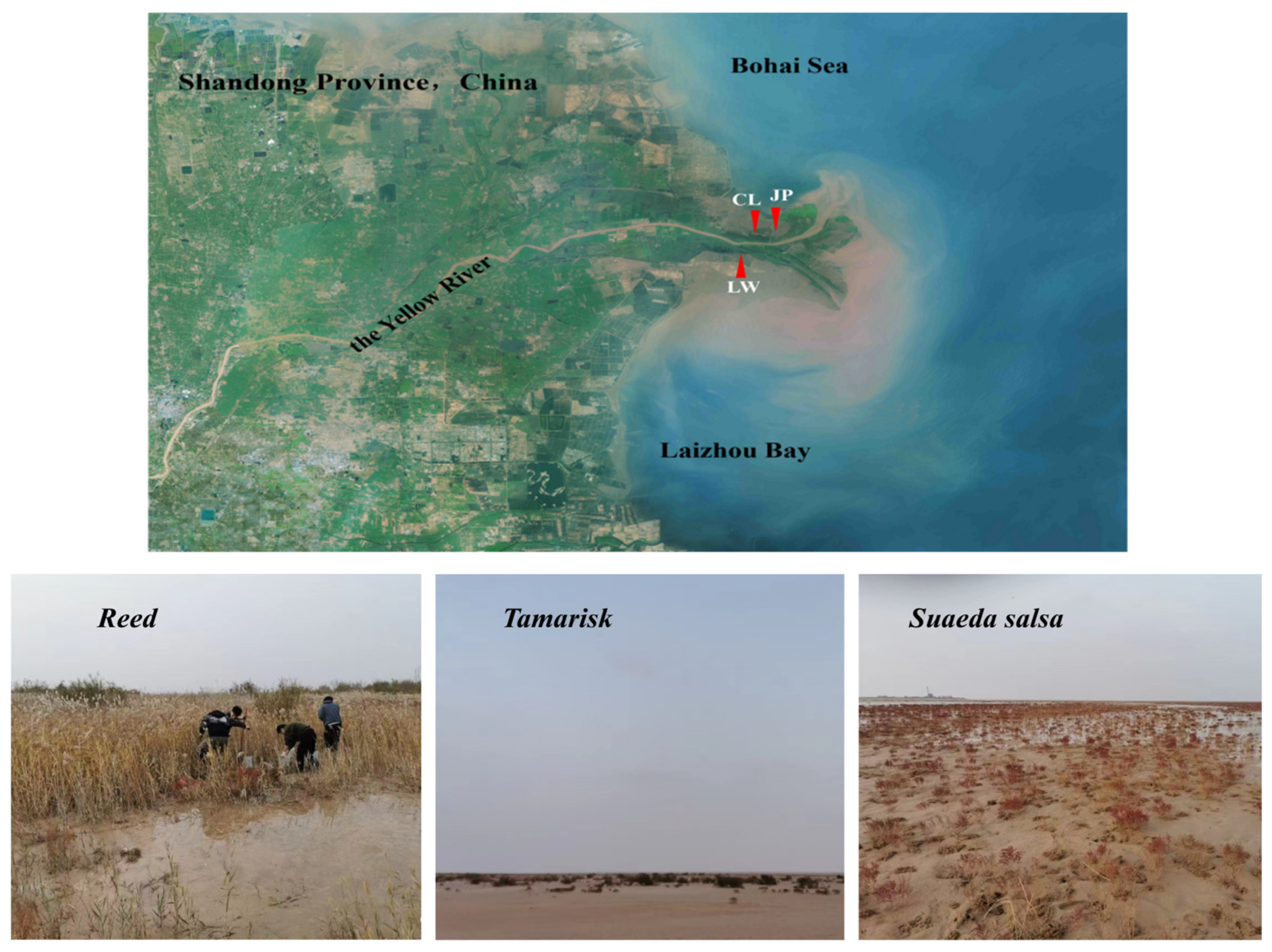
2.1. Study Area Overview
2.2. Sample Collection and Culture Methods
2.2.1. Sample Collection
2.2.2. Determination of Physical and Chemical Factors
2.2.3. Indoor Culture Experiments
2.2.4. Methane Concentration Analysis Method
2.3. DNA Extraction and PCR Amplification
2.4. PCR Products Quantification and Qualification
2.5. Library Construction and Illumina Miseq Sequencing
2.6. Data Analysis
3. Results
3.1. Physicochemical Properties Determination Results
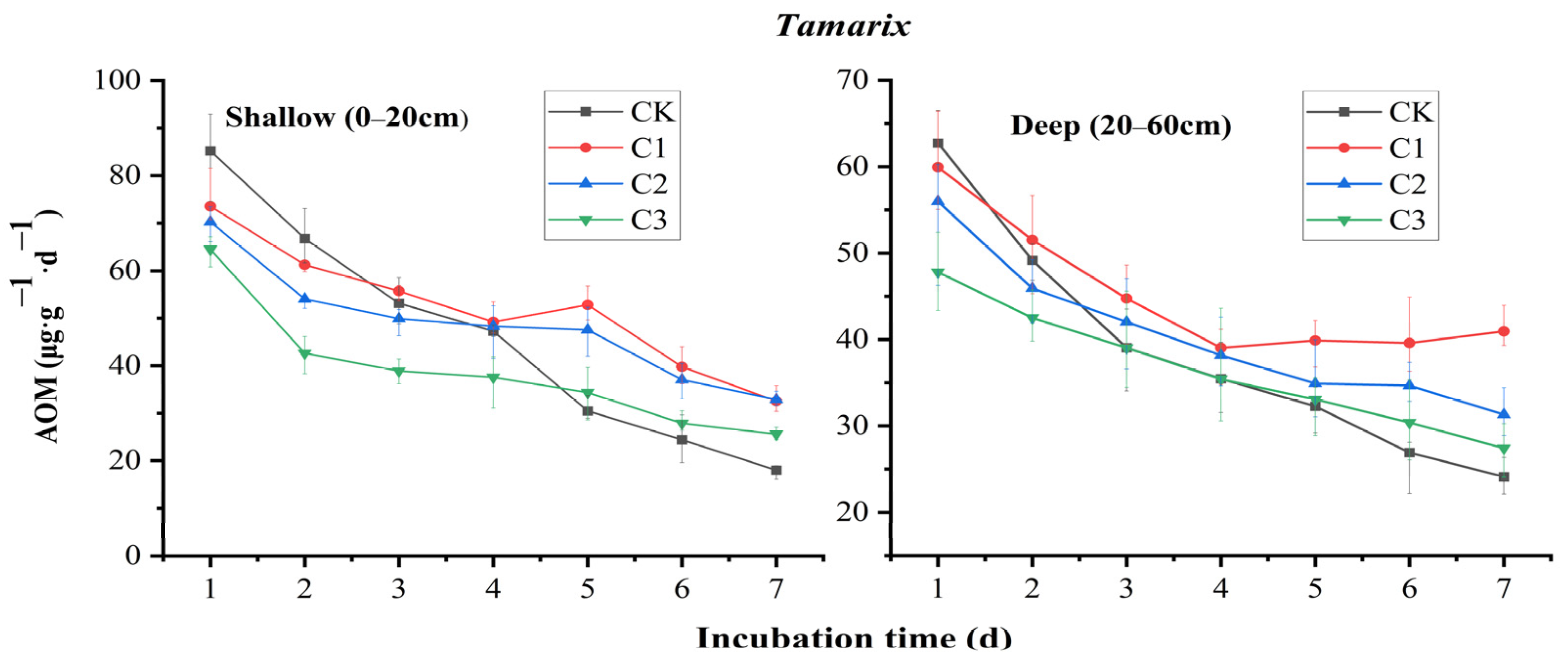
3.2. Effect of SO42− Input on AOM Potential
3.3. Effect of SO42− Input on Microorganisms
3.3.1. Changes in Microbial Diversity
3.3.2. Changes in the Structural Composition of Microbial Communities
4. Discussion
4.1. Response of Soil AOM Potential to SO42− Input
4.2. Microbial Response to SO42− Input
5. Conclusions
- (1)
- The response of AOM potential to SO42− input was somewhat different in different vegetation types and soils at different depths. Only significant SAMO processes were found in Tamarix soil, indicating that not all types of soil can undergo significant SAMO reactions after SO42− input. This may be due to the significant differences in soil microorganisms caused by different soil vegetation types; this can also better explain why the AOM potential changes vary after inputting SO42− in different wetland types;
- (2)
- SO42− input caused changes in the abundance and homogeneity of soil microorganisms, and the relationship between the changes and their input concentrations was correlated. Moreover, a peak in the increase of AOM potential was found in tamarisk soils, indicating that there is a certain threshold of SO42− addition in tamarisk soils to make the SAMO effect strongest;
- (3)
- The relative abundance of Methylophaga, Methylomonas, Methylotenera, Methylobcater, Desulfobacterota, and Crenarchaeota showed significant changes after the input of SO42−, indicating that these bacteria may be involved in the process of methane consumption and sulfur cycling in the soil.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Shukla, P.R.; Skea, J.; Slade, R. IPCC, 2022: Climate Change 2022: Mitigation of Climate Change. Contribution of Working Group III to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; IPCC: Cambridge, UK; New York, NY, USA, 2022. [Google Scholar]
- Islam, M.M.; Chowdhury, M.A.M.; Begum, R.A.; Amir, A.A. A bibliometric analysis on the research trends of climate change effects on economic vulnerability. Environ. Sci. Pollut. Res. Int. 2022, 29, 59300–59315. [Google Scholar] [CrossRef]
- Raihan, A.; Tuspekova, A. The nexus between economic growth, renewable energy use, agricultural land expansion, and carbon emissions: New insights from Peru. Energy Nexus 2022, 6, 100067. [Google Scholar] [CrossRef]
- Whiticar, M.J. The Biogeochemical Methane Cycle. In Hydrocarbons, Oils and Lipids Diversity, Origin, Chemistry and Fate; Springer: Cham, Switzerland, 2020; pp. 669–746. [Google Scholar]
- IPCC. Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovern-Mental Panel on Climate Change; Cambridge University Press: Cambridge, UK, 2013. [Google Scholar]
- Zeng, L.-L.; Tian, J.-Q.; Chen, H.; Wu, N.; Yan, Z.-Y.; Du, L.-F.; Shen, Y.; Wang, X. Changes in methane oxidation ability and methanotrophic community composition across different climatic zones. J. Soils Sediments 2018, 19, 533–543. [Google Scholar] [CrossRef]
- Li, T.; Lu, Y.; Yu, L.; Sun, W.; Zhang, R. Evaluation of CH4MODwetland and Terrestrial Ecosystem Model (TEM) used to estimate global CH4 emissions from natural wetlands. Geosci. Model Dev. 2020, 13, 3769–3788. [Google Scholar] [CrossRef]
- Liu, J.-X.; Chen, H.; Xue, D.; Gao, Y.-H.; Liu, J.-L.; Yang, J. Advances in microbial mediated anaerobic oxidation of methane and its influencing factors. Chin. J. Ecol. 2020, 39, 1033–1044. [Google Scholar] [CrossRef]
- Segarra, K.-E.-A.; Schubotz, F.; Samarkin, V.; Yoshinaga, M.-Y.; Hinrichs, K.-U.; Joye, S.-B. High rates of anaerobic methane oxidation in freshwater wetlands reduce potential atmospheric methane emissions. Nat. Commun. 2015, 6, 7477. [Google Scholar] [CrossRef]
- Conrad, R. The global methane cycle: Recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 2009, 1, 285–292. [Google Scholar] [CrossRef]
- Hu, S.; Zeng, R.-J.; Keller, J.; Lant, P.-A.; Yuan, Z. Effect of nitrate and nitrite on the selection of microorganisms in the denitrifying anaerobic methane oxidation process. Environ. Microbiol. Rep. 2011, 3, 315–319. [Google Scholar] [CrossRef]
- Sheng, L.-D.; Jin, J.-H.; Jin, X. Research progreess on anaerobic methanotrophs in inland wetlands and freshwater aquatic systems. Acta Ecol. Sin. 2022, 53, 3842–3855. [Google Scholar] [CrossRef]
- Buckley, D.H.; Baumgartner, L.K.; Visscher, P.T. Vertical distribution of methane metabolism in microbial mats of the Great Sippewissett Salt Marsh. Environ. Microbiol. 2008, 10, 967–977. [Google Scholar] [CrossRef]
- Huang, S.R. Study on the Characteristics of Rhizospheric Microbial Communities in the Heterogeneous Habitats of Coastal Wetlands in the Yellow River Delta. Master’s Thesis, Shandong University, Jinan, China, 2020. [Google Scholar]
- Wang, Z.Y.; Xin, Y.Z.; Li, F.M.; Gao, D.M. Microbial Community Characteristics in a Degraded Wetland of the Yellow River Delta. Period. Ocean. Univ. China 2009, 39, 1005–1012. [Google Scholar] [CrossRef]
- Douterelo, I.; Goulder, R.; Lillie, M. Soil microbial community response to land-management and depth, related to the degradation of organic matter in English wetlands: Implications for the in situ preservation of archaeological remains. Appl. Soil Ecol. 2010, 44, 219–227. [Google Scholar] [CrossRef]
- Liu, F.; Ye, S.Y.; Tang, Y.Q. Analysis of Microbial Community Structure in Coastal Wetland Soil of the Yellow River Delta. Chin. J. Appl. Environ. Biol. 2007, 13, 691–696. [Google Scholar]
- Shi, Y.; Wang, Z.-Q.; He, C.-G.; Zhang, X.-Y.; Sheng, L.-X.; Ren, X.-D. Using (13)C isotopes to explore denitrification-dependent anaerobic methane oxidation in a paddy-peatland. Sci. Rep. 2017, 7, 40848. [Google Scholar] [CrossRef]
- Zhu, B.; Van, D.-G.; Fritz, C.; Smolders, A.-J.; Pol, A.; Jetten, M.-S.; Ettwig, K.-F. Anaerobic oxidization of methane in a minerotrophic peatland: Enrichment of nitrite-dependent methane-oxidizing bacteria. Appl. Environ. Microbiol. 2012, 78, 8657–8665. [Google Scholar] [CrossRef]
- McCullough, L.L. Examining Anaerobic Oxidation of Methane in a Northern Peat Bog. Ph.D. Thesis, University of Oregon, Eugene, OR, USA, 2019. [Google Scholar]
- Zhang, M.; Dai, Y.; Dai, P.; Sun, Z.; Lin, X.; He, Z.; Tian, Y. Finding the pieces for the anaerobic methane oxidation jigsaw puzzle in mangrove wetlands. Int. Biodeterior. Biodegrad. 2022, 168, 105375. [Google Scholar] [CrossRef]
- Fan, L.-C.; Dippold, M.-A.; Ge, T.; Wu, J.; Thiel, V.; Kuzyakov, Y.; Dorodnikov, M. Anaerobic oxidation of methane in paddy soil: Role of electron acceptors and fertilization in mitigating CH4 fluxes. Soil Biol. Biochem. 2020, 141, 107685. [Google Scholar] [CrossRef]
- Luo, D.; Meng, X.; Zheng, N.; Li, Y.; Yao, H.; Chapman, S.-J. The anaerobic oxidation of methane in paddy soil by ferric iron and nitrate, and the microbial communities involved. Sci. Total Environ. 2021, 788, 147773. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, Y.; Long, X.-E.; Guo, J.; Zhu, G. High abundance and diversity of nitrite-dependent anaerobic methane-oxidizing bacteria in a paddy field profile. FEMS Microbiol. Lett. 2014, 360, 33–41. [Google Scholar] [CrossRef]
- Li, X.-F.; Lin, X.-B.; Cheng, L.; Hou, L.-J.; Liu, J.; Hu, X.-T. Extracellular Enzyme Activity and Greenhouse Gases Emisssion Potential in Sediments of Chongming Dongtan Wetland and Their Influencing Factors. J. Soil Water Conserv. 2017, 29, 202–207. [Google Scholar] [CrossRef]
- Zhou, J.-Y. Effect of different plant communities on anaerobic methantrophic oxidation and the functonal microbes in Chongxi wetland. Master’s Thesis, Shanghai University, Shanghai, China, 2014. [Google Scholar]
- Wang, W.-Q.; Zeng, C.-S.; Tong, C. Methane Production and Oxidation Capacities of Soil from the Reed Marsh of the Min jiang River Estuary. Wetl. Sci. 2008, 6, 9. [Google Scholar] [CrossRef]
- Gao, J.-Y.; Wang, W.-Q.; Yan, E.; Liao, J.; Yao, S. Influences of Tide on Physical and Chemical Properties of Soil in the Estuarine Wetland of Minjiang River. Wetl. Sci. Manag. 2012, 7, 48–52. [Google Scholar]
- Li, J.Y.; Chen, Q.F.; Li, Q.; Zhao, C.S.; Feng, Y.; Li, L. Analysis of microbial diversity and driving factors in coastal wetlands of the Yellow River Delta. Acta Ecol. Sin. 2021, 41, 12. [Google Scholar] [CrossRef]
- Moran, J.-J.; Beal, E.-J.; Vrentas, J.-M.; Orphan, V.-J.; Freeman, K.-H.; House, C.-H. Methyl sulfides as intermediates in the anaerobic oxidation of methane. Environ. Microbiol. 2008, 10, 162–173. [Google Scholar] [CrossRef]
- Tu, C.-X. Community Structure and Abundance of Methanogens, Sulfate-Reducing Bacteria in Tidal Maresh Soil in the Min River Estuary. Ph.D. Thesis, Fujian Normal University, Fujian, China, 2014. [Google Scholar]
- Lin, L.; Song, X.; Xi, J. Anaerobic oxidation of methane coupled to sulfate reduction: Consortium characteristics and application in co-removal of H2S and methane. J. Environ. Sci. 2019, 76, 238–248. [Google Scholar]
- Yavitt, J.B.; Burtis, J.C.; Smemo, K.A.; Welsch, M. Plot-scale spatial variability of methane, respiration, and net nitrogen mineralization in muck-soil wetlands across a land use gradient. Geoderma 2018, 315, 11–19. [Google Scholar] [CrossRef]
- Wang, Y.-N.; Wang, Y.; Zhang, Z.-M. Effcets of tidal creek morphology on succession of wetland plant communities in the Yellow River Delta. Ecol. Sci. 2019, 39, 33–41. [Google Scholar] [CrossRef]
- GB 15618-2018; Soil Environment Quality Risk Control Standard for Soilcontamination of Agriculture Land. Ministry of Ecology and Environment: Beijing, China, 2018.
- Wang, Z.-H.; Chen, Q.-F.; Li, J.-Y.; Si, G.-R.; Zhao, W.-Y.; Liu, T. Methane anaerobic oxidation potential and microbial community response to Fe and Mn input in a salt marsh wetland of the Yellow River Delta. Acta Sci. Circumstantiae 2022, 42, 452–461. [Google Scholar] [CrossRef]
- Hu, W.F.; Zeng, C.S.; Gao, J.Y.; Zhang, W.L.; Zhang, L.H.; Wang, W.Q.; Huang, J.F.; Yan, Y.Y. Response of methane production and oxidation potential to exogenous substances in the Phragmites australis marsh sediments of Shanyutan wetland in the Min River estuary. Acta Sci. Circumstantiae 2015, 35, 1116–1124. [Google Scholar] [CrossRef]
- Wang, J.-J. Study on Influence Factors on Methane Emission Flux and Methane Production Capacity in Poyang Lake Wetland. Master’s Thesis, Nanchang University, Nanchang, China, 2016. [Google Scholar]
- Liu, D.; Mo, Y.-L.; Peng, C.; Wang, B.-Z.; Wang, J.-J.; Tang, Y.; Lu, L. The impact of different electron acceptors on methane oxidation and the key microbial players in the soil of the Jialing River riparian wetland. Resour. Environ. Yangtze Basin 2022, 3, 2230–2245. [Google Scholar]
- Wang, D.; White, J.-R.; Delaune, R.-D.; Yu, Z.; Hu, Y. Peripheral freshwater deltaic wetlands are hotspots of methane flux in the coastal zone. Sci. Total Environ. 2021, 775, 145784. [Google Scholar] [CrossRef]
- Weber, H.S.; Thamdrup, B.; Habicht, K.S. High sulfur isotope fractionation associated with anaerobic oxidation of methane in a low-sulfate, iron-rich environment. Front. Earth Sci. 2016, 4, 61. [Google Scholar] [CrossRef]
- Wang, W.-Q. Anaerobic Oxidation of Methane and Its Mechanism in the Minjiang River Estuarine Wetland. Ph.D. Thesis, Fujian Normal University, Fuzhou, China, 2014. [Google Scholar]
- Zeng, C.-S.; Wang, W.-Q.; Tong, C. Effects of different exogenous electron acceptors and salt import on methane production potential of estuarine marsh soil. Geograp. Hical. Res. 2008, 27, 1321–1330. [Google Scholar]
- Gupta, V.; Smemo, K.-A.; Yavitt, J.-B.; Fowle, D.; Branfireun, B.; Basiliko, N. Stable isotopes reveal widespread anaerobic methane oxidation across latitude and peatland type. Environ. Sci. Technol. 2013, 47, 8273–8279. [Google Scholar] [CrossRef]
- Hu, B.L.; Shen, L.D.; Lian, X.; Zhu, Q.; Liu, S.; Huang, Q.; He, Z.F.; Geng, S.; Cheng, D.Q.; Lou, L.P.; et al. Evidence for nitrite-dependent anaerobic methane oxidation as a previously overlooked microbial methane sink in wetlands. Proc. Natl. Acad. Sci. USA 2014, 111, 4495–4500. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, B.; Zhang, W.; Hu, C.; An, S. Effects of plant and influent C:N:P ratio on microbial diversity in pilot-scale constructed wetlands. Ecol. Eng. 2010, 36, 441–449. [Google Scholar] [CrossRef]
- Chen, W.F.; Shi, Y.X. Distribution Characteristics of Microbes in New-born Wetlands of the Yellow River Delta. Acta Agrestia Sin. 2010, 18, 860–864. [Google Scholar]
- Zheng, M.; Tian, Y.; Liu, T.; Ma, T.; Li, L.; Li, C.; Ahmad, M.; Chen, Q.; Ni, J. Minimization of nitrous oxide emission in a pilot-scale oxidation ditch: Generation, spatial variation and microbial interpretation. Bioresour. Technol. 2015, 179, 510–517. [Google Scholar] [CrossRef]
- Vigneron, A.; Cruaud, P.; Alsop, E.; de Rezende, J.R.; Head, I.M.; Tsesmetzis, N. Beyond the tip of the iceberg; a new view of the diversity of sulfite-and sulfate-reducing microorganisms. ISME J. 2018, 12, 2096–2099. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Zhang, L.; Ji, Z.; Shao, Y.; Zhou, H.; Bao, Y.; Qu, Y.; Liu, L. Comparison of rhizosphere bacterial communities of reed and Suaeda in Shuangtaizi River Estuary, Northeast China. Mar. Pollut. Bull. 2019, 140, 171–178. [Google Scholar] [CrossRef]
- Zhu, P.; Wang, Y.; Shi, T.; Zhang, X.; Huang, G.; Gong, J. Intertidal zonation affects diversity and functional potentials of bacteria in surface sediments: A case study of the Golden Bay mangrove, China. Appl. Soil Ecol. 2018, 130, 159–168. [Google Scholar] [CrossRef]
- Zhang, B.; Wu, X.; Zhang, G.; Li, S.; Zhang, W.; Chen, X.; Sun, L.; Zhang, B.; Liu, G.; Chen, T. The diversity and biogeography of the communities of Actinobacteria in the forelands of glaciers at a continental scale. Environ. Res. Lett. 2016, 11, 054012. [Google Scholar] [CrossRef]
- Xian, W.D.; Zhang, X.T.; Li, W.J. Research status and prospect on bacterial phylum Chloroflexi. Acta Microbiol. Sin. 2020, 60, 1801–1820. [Google Scholar]
- Li, Y.; Lei, W.; Zhang, W.; Fu, X.; Le, Y. The variability of soil microbial community composition of different types of tidal wetland in Chongming Dongtan and its effect on soil microbial respiration. Ecol. Eng. 2011, 37, 1276–1282. [Google Scholar] [CrossRef]
- Arunrat, N.; Sansupa, C.; Kongsurakan, P.; Sereenonchai, S.; Hatano, R. Soil Microbial Diversity and Community Composition in Rice–Fish Co-Culture and Rice Monoculture Farming System. Biology 2022, 11, 1242. [Google Scholar] [CrossRef]
- Zhang, M.Z.; Li, L.; Liu, J.X. The parth way and methanotroph of anaerobic methane oxidation driven by nitrate or sulfate. Microbiol. China 2012, 39, 702–710. [Google Scholar] [CrossRef]





| Depth (cm) | pH | EC (mS·m−1) | NO3− (g·kg−1) | SO42− (g·kg−1) | TK (g·kg−1) | Na (g·kg−1) | TP (g·kg−1) | |
|---|---|---|---|---|---|---|---|---|
| Reed | 0~20 | 8.08 ± 0.01 | 49.63 ± 0.5 | 0.0018 ± 0.0005 | 0.025 ± 0.001 | 13.74 ± 2 | 17.98 ± 4 | 0.48 ± 0.02 |
| 20~60 | 8.05 ± 0.03 | 99.27 ± 0.3 | 0.0015 ± 0.00005 | 0.042 ± 0.002 | 14.74 ± 1 | 25.02 ± 3 | 0.40 ± 0.04 | |
| Suaeda salsa | 0~20 | 8.47 ± 0.02 | 143.50 ± 1 | 0.0016 ± 0.0003 | 0.071 ± 0.002 | 14.18 ± 2 | 21.51 ± 2 | 0.41 ± 0.05 |
| 20~60 | 8.45 ± 0.01 | 211.00 ± 5 | 0.0012 ± 0.0001 | 0.140 ± 0.003 | 12.60 ± 2 | 18.68 ± 4 | 0.47 ± 0.02 | |
| Tamarisk | 0~20 | 8.38 ± 0.03 | 120.73 ± 7 | 0.0014 ± 0.00003 | 0.079 ± 0.006 | 13.50 ± 1 | 22.17 ± 1 | 0.31 ± 0.04 |
| 20~60 | 8.43 ± 0.01 | 223.33 ± 5 | 0.0020 ± 0.0002 | 0.154 ± 0.001 | 15.15 ± 3 | 20.34 ± 0.5 | 0.47 ± 0.02 |
| Sample | Shannon | Simpson | Chao1 | ACE | OTU |
|---|---|---|---|---|---|
| RS-B | 10.147 | 0.998 | 4254.463 | 4328.002 | 3844 |
| RS-A1 | 9.995 | 0.996 | 4183.045 | 4259.318 | 3752 |
| RS-A2 | 9.872 | 0.995 | 3769.677 | 3915.421 | 3685 |
| RS-A3 | 9.822 | 0.995 | 3187.556 | 3162.027 | 2982 |
| RD-B | 9.716 | 0.995 | 4170.815 | 4240.972 | 3904 |
| RD-A1 | 9.276 | 0.993 | 3762.857 | 3984.751 | 3646 |
| RD-A2 | 9.413 | 0.980 | 3993.598 | 4034.976 | 3235 |
| RD-A3 | 9.222 | 0.995 | 3794.023 | 3813.640 | 3815 |
| SS-B | 9.046 | 0.988 | 4206.106 | 4369.964 | 3686 |
| SS-B1 | 6.669 | 0.933 | 2569.234 | 2749.548 | 2265 |
| SS-B2 | 6.534 | 0.932 | 3192.553 | 3209.625 | 3149 |
| SS-B3 | 6.442 | 0.925 | 3114.881 | 3238.852 | 2664 |
| SD-B | 7.731 | 0.964 | 3342.878 | 3602.431 | 2887 |
| SD-B1 | 7.635 | 0.958 | 3276.699 | 3308.590 | 2763 |
| SD-B2 | 7.414 | 0.972 | 2804.496 | 2855.120 | 2563 |
| SD-B3 | 6.651 | 0.937 | 2606.657 | 2654.131 | 2060 |
| TS-B | 8.172 | 0.982 | 3434.352 | 3602.298 | 2817 |
| TS-C1 | 8.665 | 0.986 | 3772.355 | 3941.136 | 3340 |
| TS-C2 | 8.352 | 0.982 | 3721.120 | 4007.182 | 3210 |
| TS-C3 | 8.264 | 0.983 | 3574.635 | 3727.044 | 3023 |
| TD-B | 6.741 | 0.962 | 3361.134 | 3387.750 | 2106 |
| TD-C1 | 8.589 | 0.985 | 3490.333 | 3570.662 | 3170 |
| TD-C2 | 7.474 | 0.965 | 3214.266 | 3286.921 | 2910 |
| TD-C3 | 7.414 | 0.972 | 2804.496 | 2855.120 | 2563 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Chen, Q.; Wang, X.; Tan, Y.; Li, L.; Zhang, B.; Guo, B.; Zhao, C. Methane Anaerobic Oxidation Potential and Microbial Community Response to Sulfate Input in Coastal Wetlands of the Yellow River Delta. Sustainability 2023, 15, 7053. https://doi.org/10.3390/su15097053
Li J, Chen Q, Wang X, Tan Y, Li L, Zhang B, Guo B, Zhao C. Methane Anaerobic Oxidation Potential and Microbial Community Response to Sulfate Input in Coastal Wetlands of the Yellow River Delta. Sustainability. 2023; 15(9):7053. https://doi.org/10.3390/su15097053
Chicago/Turabian StyleLi, Jun, Qingfeng Chen, Xinghua Wang, Yu Tan, Luzhen Li, Bowei Zhang, Beibei Guo, and Changsheng Zhao. 2023. "Methane Anaerobic Oxidation Potential and Microbial Community Response to Sulfate Input in Coastal Wetlands of the Yellow River Delta" Sustainability 15, no. 9: 7053. https://doi.org/10.3390/su15097053
APA StyleLi, J., Chen, Q., Wang, X., Tan, Y., Li, L., Zhang, B., Guo, B., & Zhao, C. (2023). Methane Anaerobic Oxidation Potential and Microbial Community Response to Sulfate Input in Coastal Wetlands of the Yellow River Delta. Sustainability, 15(9), 7053. https://doi.org/10.3390/su15097053





