1. Introduction
Light Detection and Ranging (LiDAR) remote sensing has demonstrated its advantages over traditional remote sensing (e.g., multispectral and radar) for forest inventory and vegetation structure characterization [
1,
2,
3]. With technological advances, commercial airborne LiDAR data such as small footprint full waveform (FW) LiDAR data have become available to wider remote sensing and ecological communities. Compared to conventional LiDAR systems, FW LiDAR can record the whole echo scattered from intercepted objects to inform their spatial arrangements [
4,
5,
6,
7]. Theoretically, such an advantage can enable FW LiDAR systems to better characterize vegetation structure with fine details. Past experience of FW LiDAR applications has also demonstrated their superior capacities for measuring vegetation structure [
8,
9,
10,
11]. Yet, FW LiDAR remote sensing of vegetation structure has not reached as well-established a stage as the discrete-return (DR) LiDAR remote sensing. Developing alternative ways to analyze FW LiDAR data in relevant applications are critical to facilitating the widespread use of FW LiDAR data, as well as improving the accuracy of characterizing the three-dimensional structure of vegetation.
Increased availability of FW LiDAR data over the past decades has spurred various applications targeted at estimating vegetation structural parameters and biomass through decoding the structural information inherent in waveforms [
11,
12,
13,
14]. Early applications of FW LiDAR data mainly focus on estimating elevation, canopy height, crown depth, canopy cover and biomass at the footprint level over large scale using large-footprint profilers such as the Laser Vegetation Imaging Sensor (LVIS) and the Geoscience Laser Altimeter System (GLAS) [
6,
15,
16]. More recent applications of FW LiDAR centered on individual tree analyses with small-footprint FW LiDAR data such as understory detection [
17], stem volume estimation [
18], individual tree detection and tree species classification with advanced statistical methods [
8]. There are also some studies that employed FW LiDAR to extract canopy fuel parameters [
19], classify land cover [
20] and predict biomass [
12,
21,
22] with the synthetic use of other remote sensing data [
23]. These studies have accentuated the great potential of FW LiDAR for characterizing vegetation structure and demonstrate their advantages compared to DR LiDAR data. Despite the envisioned advantages, fewer practical FW LiDAR applications are available as compared to DR LiDAR applications. A notable challenge of FW LiDAR data application is related to technical barriers such as complicated processing steps or approaches and the lack of available software tools for handling large data volumes [
4,
17]. Moreover, currently available tools or software for LiDAR data analysis are primarily oriented to processing point cloud data, while the commonly available FW LiDAR data are stored in the wave format. This conflict essentially makes FW LiDAR data more difficult to adopt than DR LiDAR data, but it also leaves us considerable room to further investigate possible approaches for interpreting information from FW LiDAR data. In addition, directly visualizing complete FW LiDAR data is still hardly available to the remote sensing and ecological communities.
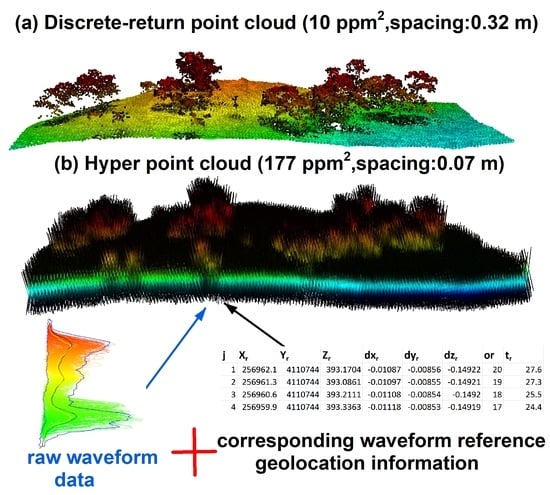
Current endeavors of FW LiDAR data processing can mainly be categorized into two types. First, part of the waveform signals that represent objects are converted into points through the decomposition or deconvolution methods using dedicated algorithms and functions [
24,
25,
26]. Simultaneously, these methods could render additional information such as echo widths, the number of peaks and amplitudes for corresponding peaks, which provide valuable insights into vegetation structure characterization and dynamics [
9,
17,
27]. Essentially, this approach treats FW LiDAR data as DR LiDAR data to extract useful metrics for the subsequent analysis. Despite more information with higher density point cloud having been obtained with this approach as compared to DR LiDAR data, the intensity information embedded in waveforms, which is the most conspicuous advantage of FW LiDAR data, is still insufficiently studied. Through this intensity information, we can gain unique insights into the energy or intensity distribution of vegetation and individual trees, but this is rarely being investigated. To make the most use of information contained in raw waveforms and enable practitioners to easily adopt FW LiDAR data, we deliver a new concept or product named the Hyper Point Cloud (HPC), with a density approximately 20 times that of DR LiDAR data, through directly transforming all waveform intensities into points with the aid of georeferenced data. This process will be detailed later in
Section 2.3.1. Second, the other type of FW LiDAR data processing directly extracts vegetation’s vertical information from waveforms as waveform signatures or features for possible application. Their effectiveness has been demonstrated in previous studies [
6,
19,
28]. This second concept was first used in large-footprint waveforms and it was introduced into small-footprint waveforms recently with the aid of the tree crown boundary. The main reason for the necessity of the tree crown boundary is that an individual small-footprint waveform only captures the small portion of tree crowns that are intercepted by the laser beam [
28]. More useful and representative vegetation information can be extracted by employing all waveforms within the tree crown boundary, rather than only using an individual waveform as the large-footprint waveform. Actually, the demand for additional information such as the tree crown boundary gives rise to another concern of directly using small-footprint FW LiDAR data. Waveform decomposition needs to be carried out to obtain the Canopy Height Model (CHM) as the input to tree crown segmentation, which possibly requires users to have a deep understanding of complicated waveform processing methods and precludes practitioners’ willingness to explore FW LiDAR data’s potential. Therefore, we introduce the CHM-like products derived from the HPC without the aid of dedicated waveform processing algorithms, namely the HPC-based intensity and percentile height (PH) surfaces, for tree segmentation, and further explore their potential applications.
This study mainly aims to propose a new and convenient way to visualize and process FW LiDAR data and explore their potential for characterizing vegetation structure. More specifically, we attempt to: (1) introduce a new concept or product, named the HPC, to relax subsequent FW LiDAR data analysis through converting all waveforms into points that are easy to reconcile with existing LiDAR processing tools well known by practitioners; (2) exemplify potential applications of the HPC such as the HPC-based PH CHMs and the HPC-based intensity surfaces for tree crown segmentation by exploring height and intensity information inherent in waveforms; and (3) evaluate the effectiveness of the HPC-based surfaces for estimating fundamental tree dimensional variables such as tree location, crown width and tree height. The logic of these three objectives is that we start from proposing a new product, the HPC, and then explore its applications, such as the HPC-based height and intensity surfaces, in a practical way, to ultimately demonstrate their usefulness for extracting tree attributes from the height and intensity information. Of particular note, the concepts or products of the HPC and the HPC-based surfaces are new to the remote sensing and ecological communities. It is anticipated that the novelty of these concepts or products could alleviate technical impediments for the extensive use of FW LiDAR data and could inspire more potential and innovative applications of FW LiDAR data.
4. Discussion
The emphasis of this study was on expanding the potential uses of FW LiDAR data from a new perspective and enhancing their value for vegetation characterization. We introduced the HPC and HPC-based products or concepts by directly using FW LiDAR data without complicated processing algorithms, and exemplified their uses for characterizing vegetation structure.
4.1. Hyper Point Cloud
A plethora of information inherent in FW LiDAR data is stored in the wave format, which cannot be easily processed by most practitioners to extract useful vegetation biophysical parameters. We proposed an approach to directly convert waveform data into a point cloud with a structure that is well known to remote sensing and ecological scientists. To some extent, this will alleviate the difficulty we experienced in understanding and employing sophisticated algorithms for extracting information from FW LiDAR data [
24,
25]. Moreover, our approach also provides a competitive alternative way to the conventional method for processing FW LiDAR data. Previous studies have explored potential uses of FW LiDAR for vegetation characterization such as tree species identification and global canopy height mapping using features extracted from waveforms [
15,
28]. However, some amount of raw information will automatically be lost during these existing procedures. With the advent of the HPC, all original information contained in FW LiDAR data is preserved by converting every signal of the waveforms into a point. However, the shape of the waveforms cannot be directly obtained, as with the Gaussian decomposition method, which can interpret the shape of waveforms through the estimated echo widths and amplitudes.
Additionally, how to directly visualize complete FW LiDAR data over a region like DR LiDAR data is still an open question that challenges the remote sensing community. The HPC provides a solution to this question and enables users to straightforwardly visualize FW LiDAR data using available handy tools designed for DR LiDAR data, as shown over a subset region (
Figure 3) and individual trees (
Figure 8). This point cloud is analogous in format to terrestrial LiDAR data, to some extent, with detailed information in the mid-story of the canopy. As anticipated, the HPC is approximately 18 times or more, on average (varies on the overlap of flight lines), denser than corresponding DR LiDAR data by keeping all signal intensities along the pulse path. It is worth noting that the vegetation or canopy parts of the HPC are composed of a substantial number of points instead of sparsely sampling points from the top of the canopy for the DR LiDAR point cloud (
Figure 8). This could overcome the disadvantage of airborne DR LiDAR data that cannot provide detailed information in the mid-story of vegetation. Meanwhile, it also leaves us considerable room and flexibility to exploit their potential applications such as semantic segmentation and vegetation structure reconstruction.
Overall, the HPC, with its point cloud format and the rich information it contains, gives interpreters or users more accessibility and flexibility to investigate the potentials of FW LiDAR data for multiple purposes. Certainly, the HPC requires more storage space and calls for more efforts to exploit and interpret the ample information inherent in waveforms. However, there is little doubt that the HPC can render new perspectives on investigating vegetation structure and facilitate extensive applications of FW LiDAR data with the continued technological advances.
4.2. HPC-Based Intensity and Height Surfaces
To enhance the practical value and use efficiency of the HPC, we exemplify a grid-based statistical method to generate the thinned version of point cloud for characterizing vegetation structure. The HPC-based intensity (MAXI and MI) DSMs exhibit promising prospects for distinguishing vegetation and ground as shown in
Figure 5. Interestingly, higher MAXI and MI are more likely to present at the ground sections rather than vegetation sections. The vegetation has a complex porous structure, while the ground is an impenetrable surface. Consequently, more energy loss occurs during the process of the pulse passing through the gaps of vegetation layers compared to directly interacting with ground given the same amount of pulse energy. This is also substantiated by the fact that the first peak intensity of the waveform (amplitude) generally has larger values than the subsequent amplitudes [
24]. However, the vegetation could give rise to more signal returns that result in higher TI and NI for a given region. Surprisingly, the spatial patterns of the HPC-based TI and NI DSMs are not as evident as the HPC-based MI and MAXI DSMs (
Figure 5). A closer examination of these HPC-based intensity DSMs reveals that a different number of overlapping flight lines mainly contributed to this discrepancy. At this point, the vegetation and ground can be well captured by the HPC-based intensity products.
LiDAR-derived CHM is deemed as the most fundamental and valuable model to characterize vegetation parameters such as tree height, crown width and biomass for complementing forest inventory [
14,
30,
32,
33]. The motivation for rescaling the original intensity information is to generate a CHM-like product and further assess the feasibility of the HPC-based intensity surface for augmenting or substituting CHM in the study of forest structure and improve the accuracy of estimating individual tree attributes. To generalize the rescaling steps and preserve original intensity information inherent in the HPC, all steps are done with simple linear conversion. Among four representative HPC-based intensity surfaces, the HPC-based MAXI surface is more likely to give us a better separation of vegetation and ground. The possible reason for this is that the MAXIs (amplitude) of waveforms generally correspond to the objects such as trees and ground that the pulses interact with, and different objects give different amplitude values. For instance, the waveforms coming from ground sections yield similar maximum intensities, while they differ significantly from the maximum intensities from vegetation components, which results in the discernible boundary between ground and vegetation. Of particular note, multiple possible variables or features such as intensity percentiles can also be extracted from the HPC. We encourage researchers to further exploit other variables or features from the HPC to further test their utilities and validity over various types of forest.
As expected, the HPC-based PH CHMs demonstrate the promising prospects of the PHs as surrogates for canopy structure variables from the HPC. These CHMs show consistent or identical spatial distribution with the DR-based and waveform-based CHMs. Overall, the proposed approaches for obtaining the HPC-based intensity and height surface not only expand existing approaches for processing FW LiDAR data, but also provide a new insight into fully using FW LiDAR data for characterizing canopy structure.
4.3. Individual Tree Intensity Distribution
Along with the HPC-based intensity and height surface results, the detailed energy distribution at the individual tree level is also examined (
Figure 8). The waveform intensity contours of three representative trees provide a theoretical justification for potential applications of the HPC-based surfaces. For instance, the most inner contour of an individual tree is more prone to be the tree top locations and the outer contour is similar to the shape of a tree crown boundary.
The intensity contours such as the MAXI, MI, TI and NI at the individual tree level provide insight into the changing pattern of energy over the vegetation at a high level of detail. Both the MAXI and MI contours reveal that the intensities are increasing from the center of tree crown to the boundary of the tree. This can be explained by the fact that more energy loss occurs during the penetration process over the center of tree crown with taller vegetation, where more dense vegetation is present, compared to the boundary of the tree in general [
34]. As a consequence, smaller amplitudes or maximum intensities were returned from the region with dense vegetation. Analogous to the TI and NI WISs, the TI and NI contours at the individual tree level exhibit decreasing pattern from the center of tree crown to the boundary of the tree. More signal returns are generated from the center of tree crowns or dense vegetation areas can attribute to this pattern. Additionally, there is a pronounced difference of contour center and changing patterns between the MAXI and MI, and the TI and NI contours. These inconsistencies are possibly due to the TI and NI contours being obtained from more “noise” in the point clouds resulted from multiple flight lines compared to the MAXI and MI contours. In addition, the lack of calibration or proper normalization steps for the TI and NI products also potentially counteract their usefulness for subsequent tree analysis. Of course, the calibration of the MAXI and MI products is also needed to enhance their capability for vegetation characterization or extend new potential applications. However, it is beyond the scope of this study to investigate proper intensity calibration procedures. Our further research will aim to explore the proper calibration or normalization steps for efficiently using the HPC or the HPC-based surfaces.
Although we only examine these four variables for the HPC, there is little doubt that other variables or features extracted from the HPC will be useful as competitive alternatives for solving potential research problems, such as using contour boundaries or intensity percentiles to estimate fuel loads of forests, crown base height, or to better characterize the horizontal and vertical vegetation structure. Overall, the contours of intensity distribution at the individual tree level provide a theoretical justification to extract attributes from the HPC-based surfaces for characterizing tree structure and ultimately promote the extensive use of FW LiDAR data.
4.4. Potential Applications
The HPC-based intensity and height surfaces provide a new perspective for employing intensity information to characterize vegetation structure and exhibits the HPC’s potential usefulness, which is expected to spur more efforts to explore these products for practical and scientific applications. We exemplify the utility of these surfaces for delineating tree crowns, identifying tree top locations, estimating crown width and tree height in this study. These applications offer an alternative way to extract information from FW LiDAR data and boost the use of information inherent in waveforms.
Tree crown segmentation is the fundamental procedure for forest inventory and monitoring, which is commonly conducted on the LiDAR-derived CHM [
13,
28,
31]. Our tree detection comparisons suggest that the fine resolution HPC-based MAXI surfaces can generate comparable results to the DR and waveform-based CHMs with the tree detection rates being around 90%. The accurate tree detection and crown segmentation results render promising prospects for subsequent tree attribute estimation. Our results also suggest that the HPC-based intensity surfaces with various resolution are prone to detect additional vegetation areas compared to the CHMs. As noted from the blue circles in
Figure 7, the WISs appear to have some “noise” compared to CHMs, but a closer examination reveals that these regions are covered with low vegetation (<3 m) such as California sagebrush. This finding may shed light on detecting understory layers or studying savanna ecosystems using intensity information of FW LiDAR data [
24]. Overall, the performance of the WIS’s tree crown segmentation is comparable to the CHM and demonstrate the potential of the HPC-based MAXI surface as an alternative product to the CHM for vegetation characterization.
Results of individual tree attributes estimated from the HPC-based intensity and height surfaces further confirmed the promising prospect of the HPC. Compared to the tree location identification and crown width estimation, the tree height estimation from the HPC-based MAXI surface is less direct and more challenging to derive than the CHMs. We developed two methods to explore the potential of tree height estimation directly from the HPC-based MAXI surface. The motivation of the first method was to examine whether a simple relationship between the height and normalized intensity exists. Despite the fact that a parsimonious linear relationship does not exist in this study (low R-square), a general trend between the height and normalized intensity is observed without the field-based intensity calibration step. Nevertheless, this method holds promise for enhancing the tree height predictive capability after conducting proper calibrations. Compared to the results of the first method, the second method can achieve better results through additional processing steps. We integrated the HPC-based MAXI surface with the HPC to search possible points containing ground and tree top through the criteria such as the radius around the estimated tree locations and intensity ranges of these points. Moreover, our results may be improved with other possible tree height estimation methods such as combing the tree locations identified by the HPC-based MAXI surface with the height from the HPC-based PH CHM. Ultimately, these potential applications of the WIS exhibit promising prospects of HPC and provide significant implications for measuring fine-scale vegetation attributes with intensity information. As anticipated, the HPC-based PH CHMs outperform the HPC-based MAXI surface for estimating tree height and generate similar tree height estimation to the DR and waveform-based CHMs in terms of the MD and RMSE.
To the best of our knowledge, the concepts or products of the HPC, the HPC-based intensity and height surfaces are new to remote sensing and ecological communities. It is anticipated that these new concepts and products can pave a new way to make the most use of waveform intensity information, facilitate extensive use of FW LiDAR data and expand their promising applications for characterizing vegetation structure. A wealth of original information is contained in the HPC, but practical applications such as biomass estimation or fuel load assessment require us to identify representative and critical variables or features. We hope our potential application examples could encourage more efforts to recognize and draw out significant information from the HPC for characterizing vegetation structure. Moreover, there are also opportunities to advance the understanding the energy or radiative transfer modeling of vegetation through integrating optical earth observations with these products. Certainly, careful calibration of the intensity is needed to ensure the consistent performance or accuracy of models. Overall, the HPC and the HPC-based intensity and height surfaces presented in this study represent a new means for studying vegetation structure using FW LiDAR data and offer promising prospects for serving different user groups with a deep understanding of waveform intensity and height information.
5. Conclusions
Conventional ways of extracting FW LiDAR information, such as using the decomposition or waveform signatures, require sophisticated algorithms and deep understanding of waveform data, which curbs the widespread use of FW LiDAR data and further obstruct their potential and extensive applications for vegetation characterization. The HPC proposed in this study not only presents a direct way to visualize FW LiDAR data with available software, but also mitigates the technical barriers for most potential practitioners and further enables FW LiDAR data processing to become more accessible to remote sensing and ecological communities. Simultaneously, this product preserves as much information of raw FW LiDAR data as possible, offering users more flexibility to interpret and decode the information inherent in waveforms. The HPC-based intensity and height surfaces exemplify the potential applications of the HPC. These applications not only expand existing ways or products for vegetation characterization but may also provide an exciting new direction for vegetation structure characterization using intensity information of FW LiDAR data. The intensity distribution of vegetation at the tree level with contours provide the theoretical justification for subsequent individual tree analysis using the HPC-based MAXI surfaces. Individual tree attribute analysis, such as tree location identification, crown width and tree height estimation, provide initial proofs for the usefulness of the HPC and the HPC-based intensity and height surfaces from the practical perspective rather than just a conceptual approach. Despite that the HPC-based MAXI surface can achieve comparable or better results compared to the CHMs in some aspects, we made no attempt to substitute the CHM with the HPC-based MAXI surface but instead to complement each other for accurately characterizing vegetation structure using both height and intensity information. Given the high density and mid-story detection capacity of the HPC, it is feasible to analyze the detailed internal structure of individual trees or vegetation over a large region. Additionally, the contours of individual trees’ intensity distributions also provide insights into other potential applications, such as estimating the crown base heights and forest surface fuel load or even identifying tree species. Actually, the HPC-based intensity and height surfaces provides promising prospects in the 2D perspective, further research is also needed to develop approaches to obtain vegetation structure or biophysical attributes with detailed 3D reconstruction models using FW LiDAR data.