The Development of Near Real-Time Biomass and Cover Estimates for Adaptive Rangeland Management Using Landsat 7 and Landsat 8 Surface Reflectance Products
Abstract
:1. Introduction
2. Materials and Methods
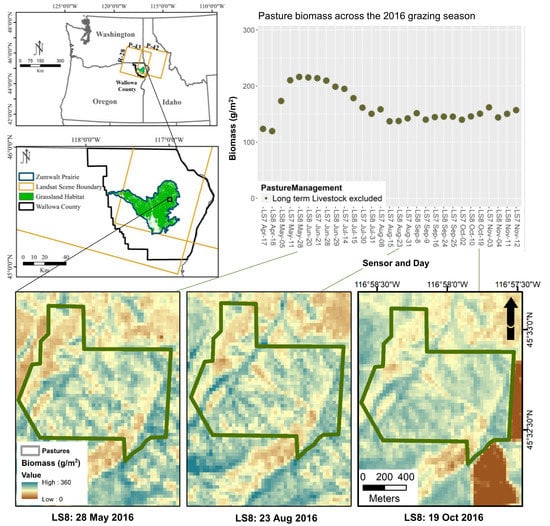
2.1. Study Area
2.2. Sampling Design
2.3. Data
2.3.1. Field Data
2.3.2. Remotely Sensed Data
2.4. Statistical Analysis
2.4.1. Variable Selection
2.4.2. Model Creation
2.4.3. Model Comparison across Landsat 7 and Landsat 8 Scenes for Summer and Fall
2.4.4. Exploring Pixel-Wise Phenology-Driven Model Application
2.4.5. Analysis of Model Residuals
3. Results
3.1. Biomass and Cover Field Data
3.2. Variable Selection
3.3. Candidate Model Comparisons and Model Selection
3.4. Relative Differences in Modeled Vegetation across Paired Landsat 7 and Landsat 8 Scenes
3.5. Assessing a Pixel-Wise Phenologically (NDVI) Driven Model Application across the Grazing Season
3.6. Correlation of NDVI Threshold Model Error with Sensor, Sampling, and Field Variables
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Asner, G.P.; Elmore, A.J.; Olander, L.P.; Martin, R.E.; Harris, T.A. Grazing Systems, Ecosystem Responses, and Global Change. Annu. Rev. Environ. Resour. 2004, 29, 261–299. [Google Scholar] [CrossRef]
- Fleishchner, T.L. Ecological Costs of Livestock Grazing in Western North America. Soc. Conserv. Biol. 1994, 8, 629–644. [Google Scholar] [CrossRef]
- Brunson, M.W.; Huntsinger, L. Ranching as a Conservation Strategy: Can Old Ranchers Save the New West? Rangel. Ecol. Manag. 2008, 61, 137–147. [Google Scholar] [CrossRef]
- Sullins, M.J.; Theobald, D.T.; Jones, J.R.; Burgess, L.M.; Knight, R.L.; Gilgert, W.C.; Marston, E. Lay of the Land: In Ranching West of the 100th Meridian: Culture, Ecology, and Economics; Island Press: Washington, DC, USA, 2002; ISBN 1-55963-826-5. [Google Scholar]
- Sayre, N.F.; McAllister, R.R.; Bestelmeyer, B.T.; Moritz, M.; Turner, M.D. Earth Stewardship of rangelands: Coping with ecological, economic, and political marginality. Front. Ecol. Environ. 2013, 11, 348–354. [Google Scholar] [CrossRef]
- Huntsinger, B.L.; Sayre, N.F. Introduction: The Working Landscapes Special Issue. Rangelands 2007, 29, 3–4. [Google Scholar] [CrossRef]
- Stafford Smith, D.M.; McKeon, G.M.; Watson, I.W.; Henry, B.K.; Stone, G.S.; Hall, W.B.; Howden, S.M. Learning from episodes of degradation and recovery in variable Australian rangelands. Proc. Natl. Acad. Sci. USA 2007, 104, 20690–20695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mckeon, G.; Day, K.; Howden, S.; Mott, J.; Orr, D.; Scattini, W.; Weston, E. Australian savannas: Management for pastoral production. J. Biogeogr. 1990, 17, 355–372. [Google Scholar] [CrossRef]
- Joyce, L.A.; Briske, D.D.; Brown, J.R.; Polley, W.H.; McCarl, B.A.; Bailey, D.W. Climate Change and North American Rangelands: Assessment of Mitigation and Adaptation Strategies. Rangel. Ecol. Manag. 2013, 66, 512–528. [Google Scholar] [CrossRef] [Green Version]
- Sayre, N.F.; deBuys, W.; Bestelmeyer, B.T.; Havstad, K.M. “The Range Problem” After a Century of Rangeland Science: New Research Themes for Altered Landscapes. Rangel. Ecol. Manag. 2012, 65, 545–552. [Google Scholar] [CrossRef]
- Bestelmeyer, B.T.; Briske, D.D. Grand Challenges for Resilience-Based Management of Rangelands. Rangel. Ecol. Manag. 2012, 65, 654–663. [Google Scholar] [CrossRef]
- Washington-Allen, R.A.; West, N.E.; Ramsey, R.D.; Efroymson, R.A. A Protocol for Retrospective Remote Sensing—Based Ecological Monitoring of. Rangel. Ecol. Manag. 2006, 59, 19–29. [Google Scholar] [CrossRef]
- Briske, D.D.; Fuhlendorf, S.D.; Smeins, F.E. State-and-Transition Models, Thresholds, and Rangeland Health: A Synthesis of Ecological Concepts and Perspectives. Rangel. Ecol. Manag. 2005, 58, 1–10. [Google Scholar] [CrossRef]
- Pyke, D.; Herrick, J.; Shaver, P.; Pellant, M. Rangeland health attributes and indicators for qualitative assessment. J. Range Manag. 2002, 55, 584–597. [Google Scholar] [CrossRef]
- Weltz, M.A.; Dunn, G.; Reeder, J.; Frasier, G. Ecological Sustainability of Rangelands. Arid L. Res. Manag. 2003, 369–388. [Google Scholar] [CrossRef]
- West, N.E. History of Rangeland Monitoring in the USA. Arid Land Res. Manag. 2003, 17, 495–545. [Google Scholar] [CrossRef]
- Briske, D.D.; Washington-Allen, R.A.; Johnson, C.R.; Lockwood, J.A. Catastrophic Thresholds: A Synthesis of Concepts, Perspectives, and Applications. Ecol. Soc. 2010, 15, 37. [Google Scholar] [CrossRef]
- Herrick, J.E.; Lessard, V.C.; Spaeth, K.E.; Shaver, P.L.; Dayton, R.S.; Pyke, D.A.; Jolley, L.; Goebel, J.J. National ecosystem assessments supported by scientific and local knowledge. Front. Ecol. Environ. 2010, 8, 403–408. [Google Scholar] [CrossRef]
- Hagen, S.C.; Heilman, P.; Marsett, R.; Torbick, N.; Salas, W.; van Ravensway, J.; Qi, J. Mapping Total Vegetation Cover across Western Rangelands with Moderate-Resolution Imaging Spectroradiometer Data. Rangel. Ecol. Manag. 2012, 65, 456–467. [Google Scholar] [CrossRef]
- Ikeda, H.; Okamoto, K.; Fukuhara, M. Estimation of aboveground grassland phytomass with a growth model using Landsat TM and climate data. Int. J. Remote Sens. 1999, 20, 2283–2294. [Google Scholar] [CrossRef]
- Jansen, V.S.; Kolden, C.A.; Taylor, R.V.; Newingham, B.A. Quantifying livestock effects on bunchgrass vegetation with Landsat ETM+ data across a single growing season. Int. J. Remote Sens. 2016, 37, 150–175. [Google Scholar] [CrossRef]
- Roy, D.P.; Kovalskyy, V.; Zhang, H.K.; Vermote, E.F.; Yan, L.; Kumar, S.S.; Egorov, A. Characterization of Landsat-7 to Landsat-8 reflective wavelength and normalized difference vegetation index continuity. Remote Sens. Environ. 2016, 185, 57–70. [Google Scholar] [CrossRef]
- Holden, C.E.; Woodcock, C.E. An analysis of Landsat 7 and Landsat 8 under flight data and the implications for time series investigations. Remote Sens. Environ. 2016. [Google Scholar] [CrossRef]
- Butterfield, H.S.; Malmström, C.M. The effects of phenology on indirect measures of aboveground biomass in annual grasses. Int. J. Remote Sens. 2009, 30, 3133–3146. [Google Scholar] [CrossRef]
- Xu, D.; Guo, X.; Li, Z.; Yang, X.; Yin, H. Measuring the dead component of mixed grassland with Landsat imagery. Remote Sens. Environ. 2014, 142, 33–43. [Google Scholar] [CrossRef]
- Van Leeuwen, W.J.D.; Huete, A.R. Effects of standing litter on the biophysical interpretation of plant canopies with spectral indices. Remote Sens. Environ. 1996, 55, 123–138. [Google Scholar] [CrossRef]
- Huete, A.R.; Jackson, R.D. Suitability of spectral indices for evaluating vegetation characteristics on arid rangelands. Remote Sens. Environ. 1987, 23, 213–218. [Google Scholar] [CrossRef]
- Todd, S.W.; Hoffer, R.M.; Milchunas, D.G. Biomass estimation on grazed and ungrazed rangelands using spectral indices. Int. J. Remote Sens. 1998, 19, 427–438. [Google Scholar] [CrossRef]
- Elvidge, C. Visible and near infrared reflectance characteristics of dry plant materials. Int. J. Remote Sens. 1990, 11, 1775–1795. [Google Scholar] [CrossRef]
- Jacques, D.C.; Kergoat, L.; Hiernaux, P.; Mougin, E.; Defourny, P. Monitoring dry vegetation masses in semi-arid areas with MODIS SWIR bands. Remote Sens. Environ. 2014, 153, 40–49. [Google Scholar] [CrossRef]
- Marsett, R.C.; Qi, J.; Heilman, P.; Biedenbender, S.H.; Watson, M.C.; Amer, S.; Weltz, M.; Goodrich, D.; Marsett, R. Remote Sensing for Grassland Management in the Arid Southwest. Rangel. Ecol. Manag. 2006, 59, 530–540. [Google Scholar] [CrossRef] [Green Version]
- Guerschman, J.P.; Scarth, P.F.; Mcvicar, T.R.; Renzullo, L.J.; Malthus, T.J.; Stewart, J.B.; Rickards, J.E.; Trevithick, R. Assessing the effects of site heterogeneity and soil properties when unmixing photosynthetic vegetation, non-photosynthetic vegetation and bare soil fractions from Landsat and MODIS data. Remote Sens. Environ. 2015, 161, 12–26. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Hansen, M.C.; Potapov, P.V.; Moore, R.; Hancher, M.; Turubanova, S.A.; Tyukavina, A.; Thau, D.; Stehman, S.V.V.; Goetz, S.J.; Loveland, T.R.; et al. High-Resolution Global Maps of 21st-Century Forest Cover Change. Science 2013, 342, 850–854. [Google Scholar] [CrossRef] [PubMed]
- Huntington, J.L.; Hegewisch, K.C.; Daudert, B.; Morton, C.G.; Abatzoglou, J.T.; McEvoy, D.J.; Erickson, T. Climate engine: Cloud computing and visualization of climate and remote sensing data for advanced natural resource monitoring and process understanding. Bull. Am. Meteorol. Soc. 2017, 98, 2397–2409. [Google Scholar] [CrossRef]
- Schmalz, H.J.; Taylor, R.V.; Johnson, T.N.; Kennedy, P.L.; DeBano, S.J.; Newingham, B.A.; McDaniel, P.A. Soil Morphologic Properties and Cattle Stocking Rate Affect Dynamic Soil Properties. Rangel. Ecol. Manag. 2013, 66, 445–453. [Google Scholar] [CrossRef]
- Kagan, J.; Ohmann, J.; Gregory, M.; Tobalske, C.; Hak, J.; Fried, J. Final Report on Land Cover Mapping Methods: Map Zones 8 and 9, PNW ReGAP; Institute for Natural Resources, Oregon State University: Corvallis, OR, USA, 2006. [Google Scholar]
- Herrick, J.E.; Van Zee, J.W.; Havstad, K.M.; Burkett, L.M.; Whitford, W.G.; Pyke, D.A.; Remmenga, M.D.; Shaver, P.L. Monitoring Manual for Grassland, Shrubland and Savanna Ecosystems; USDA-ARS Jornada Experimental Range: Las Cruces, NM, USA, 2005; p. 236. [Google Scholar]
- Friedel, M.H.; Bastin, G.N. Photographic standards for estimating compariative yield in arid rangelands. Aust. Rangel. J. 1988, 10, 34–38. [Google Scholar] [CrossRef]
- Parsons, C.T.; Momont, P.A.; Delcurto, T.; Mcinnis, M.; Porath, L.; Marni, L. Cattle distribution patterns and vegetation use in mountain riparian areas. J. Range Manag. 2003, 56, 334–341. [Google Scholar] [CrossRef]
- Masek, J.G.; Vermote, E.F.; Saleous, N.E.; Wolfe, R.; Hall, F.G.; Huemmrich, K.F.; Gao, F.; Kutler, J.; Lim, T. A Landsat Surface Reflectance Dataset. IEEE Geosci. Remote Sens. Lett. 2006, 3, 68–72. [Google Scholar] [CrossRef]
- Vermote, E.; Justice, C.; Claverie, M.; Franch, B. Preliminary analysis of the performance of the Landsat 8/OLI land surface reflectance product. Remote Sens. Environ. 2016. [Google Scholar] [CrossRef]
- Zhu, Z.; Woodcock, C.E. Object-based cloud and cloud shadow detection in Landsat imagery. Remote Sens. Environ. 2012, 118, 83–94. [Google Scholar] [CrossRef]
- Kauth, R.J.; Thomas, G.S. The Tasselled Cap—A Graphic Description of the Spectral-Temporal Development of Agricultural Crops as Seen by LANDSAT. In Proceedings of the Symposium on Machine Processing of Remotely Sensed Data, Purdue University, West LaFayette, IN, USA, 29 June–1 July 1976. [Google Scholar]
- Crist, E.P. A TM Tasseled Cap equivalent transformation for reflectance factor data. Remote Sens. Environ. 1985, 17, 301–306. [Google Scholar] [CrossRef]
- Hudak, A.T.; Crookston, N.L.; Evans, J.S.; Falkowski, M.J.; Smith, A.M.S.; Gessler, P.E.; Morgan, P. Regression modeling and mapping of coniferous forest basal area and tree density from discrete-return lidar and multispectral satellite data. Can. J. Remote Sens. 2006, 32, 126–138. [Google Scholar] [CrossRef]
- Crowley, P.H. Resampling methods for computation-intensive data analysis in ecology and evolution. Annu. Rev. Ecol. Syst. 1992, 23, 405–447. [Google Scholar] [CrossRef]
- Thomas, L. R package Version 3.0. Leaps: Regression Subset Selection. 2017. Available online: https://cran.r-project.org/package=leaps (accessed on 4 July 2018).
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Found. Stat. Computing: Vienna, Austria, 2016; ISBN 3-900051-07-0. Available online: http://www.R-project.org/ (accessed on 4 July 2018).
- Pineiro, G.; Perelman, S.; Guerschman, J.P.; Jose, P.M. How to evaluate models: Observed vs. predicted or predicted vs. observed? Ecol. Modell. 2008, 216, 316–322. [Google Scholar] [CrossRef]
- Lilliefors, H. On the Kolmogorov-Smirnov test for normality with mean and variance unknown. J. Am. Stat. Assoc. 1967, 62, 399–402. [Google Scholar] [CrossRef]
- Graham, M.H. Confronting multicollinearity in ecological Multiple Regression. Ecology 2003, 84, 2809–2815. [Google Scholar] [CrossRef]
- Vescovo, L.; Gianelle, D. Using the MIR bands in vegetation indices for the estimation of grassland biophysical parameters from satellite remote sensing in the Alps region of Trentino (Italy). Adv. Sp. Res. 2008, 41, 1764–1772. [Google Scholar] [CrossRef]
- Malmstrom, C.M.; Butterfield, H.S.; Barber, C.; Dieter, B.; Harrison, R.; Qi, J.; Riaño, D.; Schrotenboer, A.; Stone, S.; Stoner, C.J.; et al. Using Remote Sensing to Evaluate the Influence of Grassland Restoration Activities on Ecosystem Forage Provisioning Services. Restor. Ecol. 2009, 17, 526–538. [Google Scholar] [CrossRef]
- Svoray, T.; Perevolotsky, A.; Atkinson, P.M. Ecological sustainability in rangelands: The contribution of remote sensing. Int. J. Remote Sens. 2013, 34, 6216–6242. [Google Scholar] [CrossRef]
- Roberts, D.A.; Smith, M.O.; Adams, J.B. Green vegetation, nonphotosynthetic vegetation, and soils in AVIRIS data. Remote Sens. Environ. 1993, 44, 255–269. [Google Scholar] [CrossRef]
- Guerschman, J.P.; Hill, M.J.; Renzullo, L.J.; Barrett, D.J.; Marks, A.S.; Botha, E.J. Estimating fractional cover of photosynthetic vegetation, non-photosynthetic vegetation and bare soil in the Australian tropical savanna region upscaling the EO-1 Hyperion and MODIS sensors. Remote Sens. Environ. 2009, 113, 928–945. [Google Scholar] [CrossRef]
- Renier, C.; Waldner, F.; Jacques, D.C.; Babah Ebbe, M.A.; Cressman, K.; Defourny, P. A dynamic vegetation senescence indicator for near-real-time desert locust habitat monitoring with MODIS. Remote Sens. 2015, 7, 7545–7570. [Google Scholar] [CrossRef]
- Cramer, M.D.; Barger, N.N. Are mima-like mounds the consequence of long-term stability of vegetation spatial patterning? Palaeogeogr. Palaeoclimatol. Palaeoecol. 2014, 409, 72–83. [Google Scholar] [CrossRef]
- Rodríguez-Caballero, E.; Knerr, T.; Weber, B. Importance of biocrusts in dryland monitoring using spectral indices. Remote Sens. Environ. 2015, 170, 32–39. [Google Scholar] [CrossRef]
- Zheng, B.; Campbell, J.B.; Serbin, G.; Daughtry, C.S.T. Multitemporal remote sensing of crop residue cover and tillage practices: A validation of the minNDTI strategy in the United States. J. Soil Water Conserv. 2013, 68, 120–131. [Google Scholar] [CrossRef]
- McNairn, H.; Protz, R. Mapping Corn Residue Cover on Agricultural Fields in Oxford County, Ontario, Using Thematic Mapper. Can. J. Remote Sens. 1993, 19, 152–159. [Google Scholar] [CrossRef]
- Daughtry, C.S.T. Agroclimatology: Discriminating crop residues from soil by shortwave infrared reflectance. Agron. J. 2001, 93, 125–131. [Google Scholar] [CrossRef]
- Daughtry, C.S.T.; Hunt, E.R.; Doraiswamy, P.C.; McMurtrey, J.E. Remote Sensing the Spatial Distribution of Crop Residues. Agron. J. 2005, 97, 864. [Google Scholar] [CrossRef] [Green Version]
- Daughtry, C.S.T.; Hunt, E.R. Mitigating the effects of soil and residue water contents on remotely sensed estimates of crop residue cover. Remote Sens. Environ. 2008, 112, 1647–1657. [Google Scholar] [CrossRef]
- Knipling, E. Physical and physiological basis for the reflectance of visible and near-infrared radiation from vegetation. Remote Sens. Environ. 1970, 1, 155–159. [Google Scholar] [CrossRef]
- Tucker, C. Remote sensing of leaf water content in the near infrared. Remote Sens. Environ. 1980, 10, 23–32. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Feng, Q.; Liang, T.; Liu, B.; Zhang, W.; Xie, H. Modeling grassland above-ground biomass based on artificial neural network and remote sensing in the Three-River Headwaters Region. Remote Sens. Environ. 2018, 204, 448–455. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, X.; Zhu, X.; Dong, Z.; Guo, W. Estimation of biomass in wheat using random forest regression algorithm and remote sensing data. Crop J. 2016, 4, 212–219. [Google Scholar] [CrossRef]
- Li, J.; Roy, D.P. A Global Analysis of Sentinel-2A, Sentinel-2B and Landsat-8 Data Revisit Intervals and Implications for Terrestrial Monitoring. Remote Sens. 2017, 9, 902. [Google Scholar] [CrossRef]
- Gao, F.; Masek, J.; Schwaller, M.; Hall, F. On the blending of the Landsat and MODIS surface reflectance: Predicting daily Landsat surface reflectance. IEEE Trans. Geosci. Remote Sens. 2006, 44, 2207–2218. [Google Scholar] [CrossRef]
- Hilker, T.; Wulder, M.A.; Coops, N.C.; Linke, J.; McDermid, G.; Masek, J.G.; Gao, F.; White, J.C. A new data fusion model for high spatial- and temporal-resolution mapping of forest disturbance based on Landsat and MODIS. Remote Sens. Environ. 2009, 113, 1613–1627. [Google Scholar] [CrossRef]
- Knapp, C.N.; Fernandez-Gimenez, M.E. Knowledge in Practice: Documenting Rancher Local Knowledge in Northwest Colorado. Rangel. Ecol. Manag. 2009, 62, 500–509. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef] [Green Version]
- Roujean, J.; Breon, F. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Haboudane, D. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- hang, C.; Guo, X. Monitoring northern mixed prairie health using broadband satellite imagery. Int. J. Remote Sens. 2008, 29, 2257–2271. [Google Scholar] [CrossRef]
- Merzlyak, M.N.; Gitelson, A.A.; Chivkunova, O.B.; Rakitin, Y. Non-destructive optical detection of pigment changes during leaf senescence and fruit ripening. Physiol. Plant. 1999, 106, 135–141. [Google Scholar] [CrossRef]
- Hardisky, M.A.; Smart, R.M.; Klemas, V. Seasonal Spectral Characteristics and Aboveground Biomass of the Tidal Marsh Plant, Spartina alterniflora. Photogramm. Eng. Remote Sens. 1983, 49, 85–92. [Google Scholar]
- Key, C.H.; Benson, N.C. Landscape Assessment (LA) Sampling and Analysis Methods. In FIREMON: Fire Effects Monitoring and Inventory System; Lutes, D.C., Keane, R.E., Carati, J.F., Key, C.H., Benson, N.C., Gangi, L.J., Eds.; General Technical Report RMRS-GTR-164-CD; Rocky Mountains Research Station, USDA Forest Service: Fort Collins, CO, USA, 2006; p. 51. [Google Scholar]
- Gao, B. NDWI A Normalized Difference Water Index for Remote Sensing of Vegetation Liquid Water from Space. Remote Sens. Environ. 1996, 3, 257–266. [Google Scholar] [CrossRef]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Deventer, A.P.; Ward, A.D.; Gowda, P.H.; Lyon, J.G. Using thematic mapper data to identify contrasting soil plains and tillage practices. Photogramm. Eng. Remote Sens. 1997, 63, 87–93. [Google Scholar]
- Liu, H.Q.; Huete, A. Feedback based modification of the NDVI to minimize canopy background and atmospheric noise. IEEE Trans. Geosci. Remote Sens. 1995, 33, 457–465. [Google Scholar] [CrossRef]









| Biomass (g/m2) | Cover (%) | |||||
|---|---|---|---|---|---|---|
| Summer | Fall | Total | Summer | Fall | Total | |
| N | 124 | 148 | 272 | 124 | 148 | 272 |
| Mean | 162.61 | 108.61 | 133.23 | 0.61 | 0.52 | 0.56 |
| Min | 39.59 | 12.19 | 12.19 | 0.21 | 0.13 | 0.13 |
| Max | 366.10 | 302.97 | 366.10 | 0.94 | 0.94 | 0.94 |
| SD | 71.25 | 59.59 | 70.40 | 0.19 | 0.21 | 0.21 |
| Median | 158.36 | 94.50 | 120.67 | 0.63 | 0.52 | 0.57 |
| Metric | Time | Sensor | Veg Index | Training | Validation | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Int | Slope | r2 | rRMSE | RMSD | N | r2 | rRMSE | RMSD | ||||
| Biomass | Summer | LS7 | NDII7 | 60 | 104.06 | 343.18 | 0.69 | 22.84 | 39.27 | 20 | 0.81 | 21.25 | 34.87 |
| Summer | LS8 | NDII7 | 93 | 101.09 | 330.25 | 0.80 | 20.07 | 32.08 | 30 | 0.81 | 16.86 | 28.96 | |
| Summer | LS78 | NDII7 | 153 | 102.18 | 335.95 | 0.76 | 21.38 | 35.07 | 50 | 0.81 | 18.50 | 30.89 | |
| Fall | LS7 | NDTI | 78 | −56.45 | 1042.00 | 0.71 | 30.46 | 32.67 | 25 | 0.77 | 24.19 | 26.43 | |
| Fall | LS8 | NDTI | 99 | −58.04 | 1070.64 | 0.67 | 30.88 | 31.20 | 32 | 0.70 | 26.69 | 32.02 | |
| Fall | LS78 | NDTI | 177 | −55.30 | 1044.67 | 0.69 | 30.73 | 31.80 | 57 | 0.73 | 25.86 | 29.54 | |
| All-year | LS7 | NDTI | 120 | −36.53 | 944.63 | 0.67 | 29.32 | 40.52 | 40 | 0.76 | 25.94 | 37.26 | |
| All-year | LS8 | NDTI | 184 | −41.74 | 1028.00 | 0.74 | 26.34 | 35.38 | 62 | 0.77 | 27.22 | 35.32 | |
| All-year | LS78 | NDTI | 304 | −38.08 | 984.32 | 0.70 | 27.88 | 37.82 | 102 | 0.76 | 26.10 | 35.11 | |
| Cover | Summer | LS7 | NDII7 | 60 | 0.44 | 0.95 | 0.70 | 16.89 | 0.11 | 20 | 0.70 | 17.39 | 0.11 |
| Summer | LS8 | NDII7 | 93 | 0.43 | 0.94 | 0.78 | 16.07 | 0.10 | 30 | 0.75 | 13.00 | 0.08 | |
| Summer | LS78 | NDII7 | 153 | 0.44 | 0.94 | 0.75 | 16.44 | 0.10 | 50 | 0.72 | 14.84 | 0.09 | |
| Fall | LS7 | NDTI | 78 | −0.09 | 3.88 | 0.78 | 19.87 | 0.10 | 26 | 0.81 | 17.07 | 0.09 | |
| Fall | LS8 | NDTI | 99 | −0.10 | 3.97 | 0.72 | 21.73 | 0.11 | 32 | 0.72 | 22.71 | 0.13 | |
| Fall | LS78 | NDTI | 177 | −0.09 | 3.91 | 0.75 | 20.92 | 0.11 | 58 | 0.73 | 20.69 | 0.11 | |
| All-year | LS7 | NDTI | 120 | 0.07 | 2.70 | 0.65 | 22.85 | 0.13 | 40 | 0.72 | 21.00 | 0.12 | |
| All-year | LS8 | NDTI | 184 | 0.06 | 2.95 | 0.69 | 20.55 | 0.12 | 62 | 0.70 | 21.02 | 0.12 | |
| All-year | LS78 | NDTI | 304 | 0.07 | 2.82 | 0.67 | 21.71 | 0.12 | 102 | 0.70 | 20.74 | 0.12 | |
| Metric | Variable | Variable Source | Landsat 7 | Landsat 8 | ||
|---|---|---|---|---|---|---|
| r-Val | p-Val | r-Val | p-Val | |||
| Biomass | % Perennial Grass | LPI (canopy) | −0.21 | 0.001 | −0.15 | 0.006 |
| Biomass | % Litter | LPI (canopy) | 0.20 | 0.013 | 0.18 | 0.006 |
| Biomass | Rain Lag (days) | Sensor (Field) | 0.36 | 0.027 | 0.24 | 0.006 |
| Biomass | % Moss/Lichen | LPI (soil surface) | 0.13 | 0.014 | 0.004 | NS |
| Biomass | % Rock | LPI (soil surface) | 0.01 | 0.046 | −0.0578 | NS |
| Biomass | % Mean Utilization | Utilization | NS | NS | 0.10 | 0.003 |
| Cover | % Perennial Grass | LPI (canopy) | −0.407 | 0.000 | −0.3546 | 0.000 |
| Cover | % Annual Grass | LPI (canopy) | −0.174 | 0.018 | −0.2150 | 0.000 |
| Cover | % Annual Forb | LPI (canopy) | −0.264 | 0.000 | −0.1981 | 0.0322 |
| Cover | Field Data Lag (Days) | Sensor (Field) | −0.178 | 0.016 | −0.1800 | 0.0233 |
| Cover | % Brown and SD Color | LPI (color) | −0.178 | 0.016 | −0.1700 | NS |
| Cover | % Litter | LPI (canopy) | 0.315 | 0.000 | 0.2379 | 0.000 |
| Cover | Rain Lag (days) | Sensor (Weather Station) | 0.327 | 0.000 | 0.1815 | 0.000 |
| Cover | % Rock | LPI (soil surface) | 0.277 | 0.000 | 0.18317 | 0.000 |
| Cover | % Soil | LPI (soil surface) | 0.162 | 0.029 | 0.211 | 0.001 |
| Cover | % Moss/Lichen | LPI (soil surface) | 0.381 | 0.000 | 0.304 | 0.000 |
| Cover | % Green Color | LPI (color) | 0.178 | 0.016 | 0.1700 | NS |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jansen, V.S.; Kolden, C.A.; Schmalz, H.J. The Development of Near Real-Time Biomass and Cover Estimates for Adaptive Rangeland Management Using Landsat 7 and Landsat 8 Surface Reflectance Products. Remote Sens. 2018, 10, 1057. https://doi.org/10.3390/rs10071057
Jansen VS, Kolden CA, Schmalz HJ. The Development of Near Real-Time Biomass and Cover Estimates for Adaptive Rangeland Management Using Landsat 7 and Landsat 8 Surface Reflectance Products. Remote Sensing. 2018; 10(7):1057. https://doi.org/10.3390/rs10071057
Chicago/Turabian StyleJansen, Vincent S., Crystal A. Kolden, and Heidi J. Schmalz. 2018. "The Development of Near Real-Time Biomass and Cover Estimates for Adaptive Rangeland Management Using Landsat 7 and Landsat 8 Surface Reflectance Products" Remote Sensing 10, no. 7: 1057. https://doi.org/10.3390/rs10071057
APA StyleJansen, V. S., Kolden, C. A., & Schmalz, H. J. (2018). The Development of Near Real-Time Biomass and Cover Estimates for Adaptive Rangeland Management Using Landsat 7 and Landsat 8 Surface Reflectance Products. Remote Sensing, 10(7), 1057. https://doi.org/10.3390/rs10071057